Zinc Supplementation with or without Additional Micronutrients Does Not Affect Peripheral Blood Gene Expression or Serum Cytokine Level in Bangladeshi Children
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parent Trial
2.2. Sample Collection and Shipping
2.3. RNA Extraction
2.4. Library Preparation and RNA Sequencing
2.5. Bioinformatic Analysis of RNA Sequencing
2.6. RNA Sequencing Analyses
2.7. Simoa Analysis of Serum Cytokine Levels
3. Results
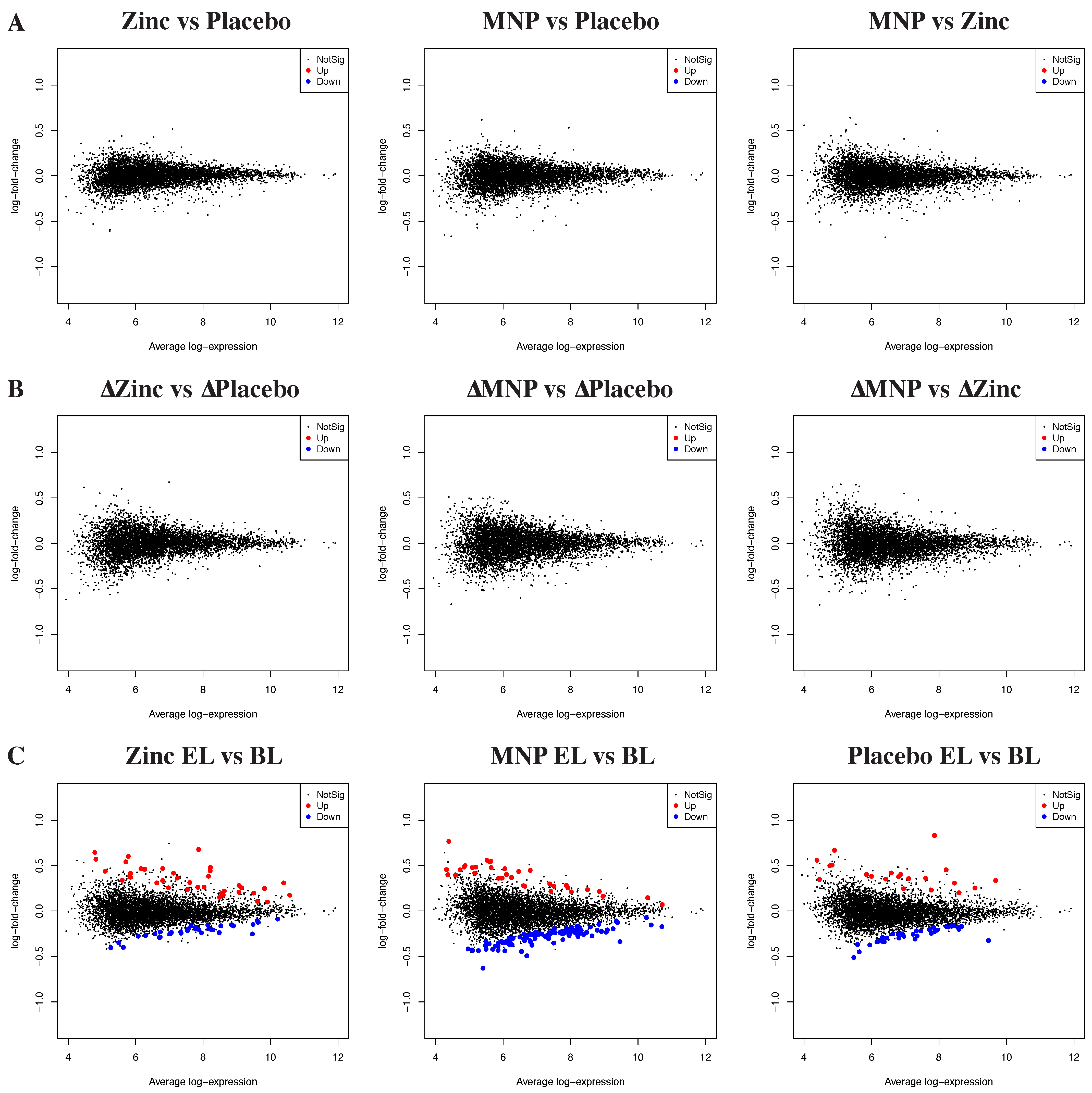
3.1. Neither Zinc MNPs nor Zinc Tablets Altered the Peripheral Blood Transcriptome
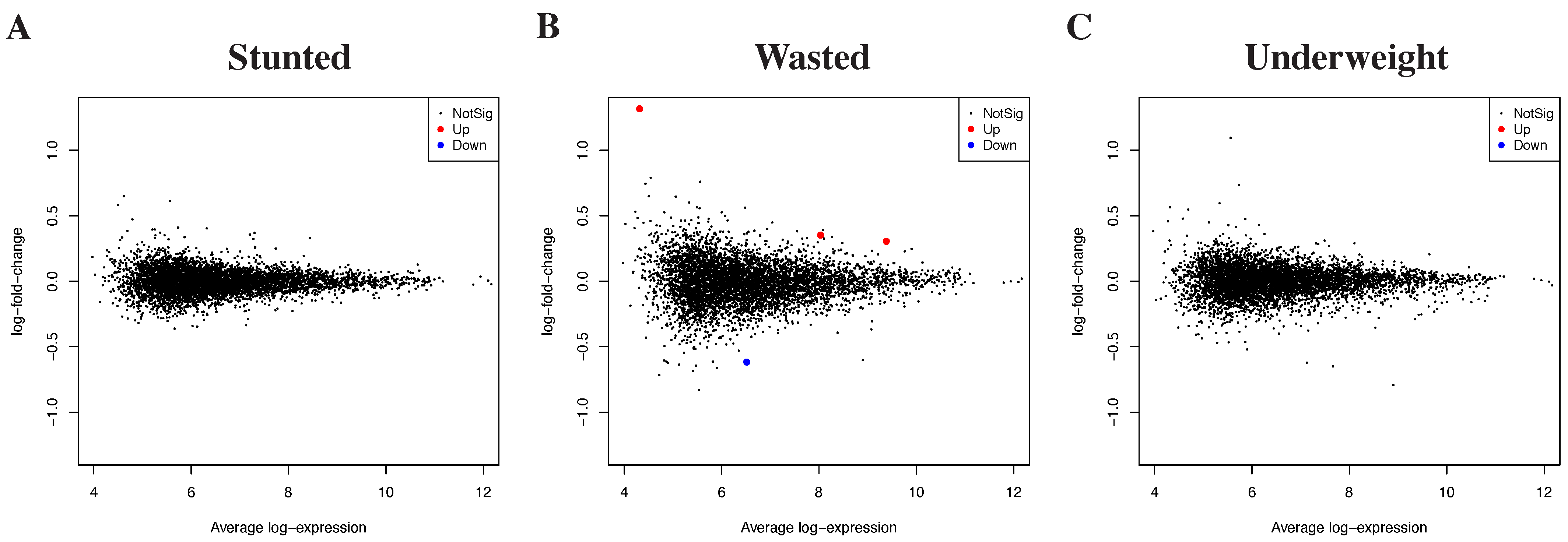
3.2. Differential Gene Expression by Growth Outcomes
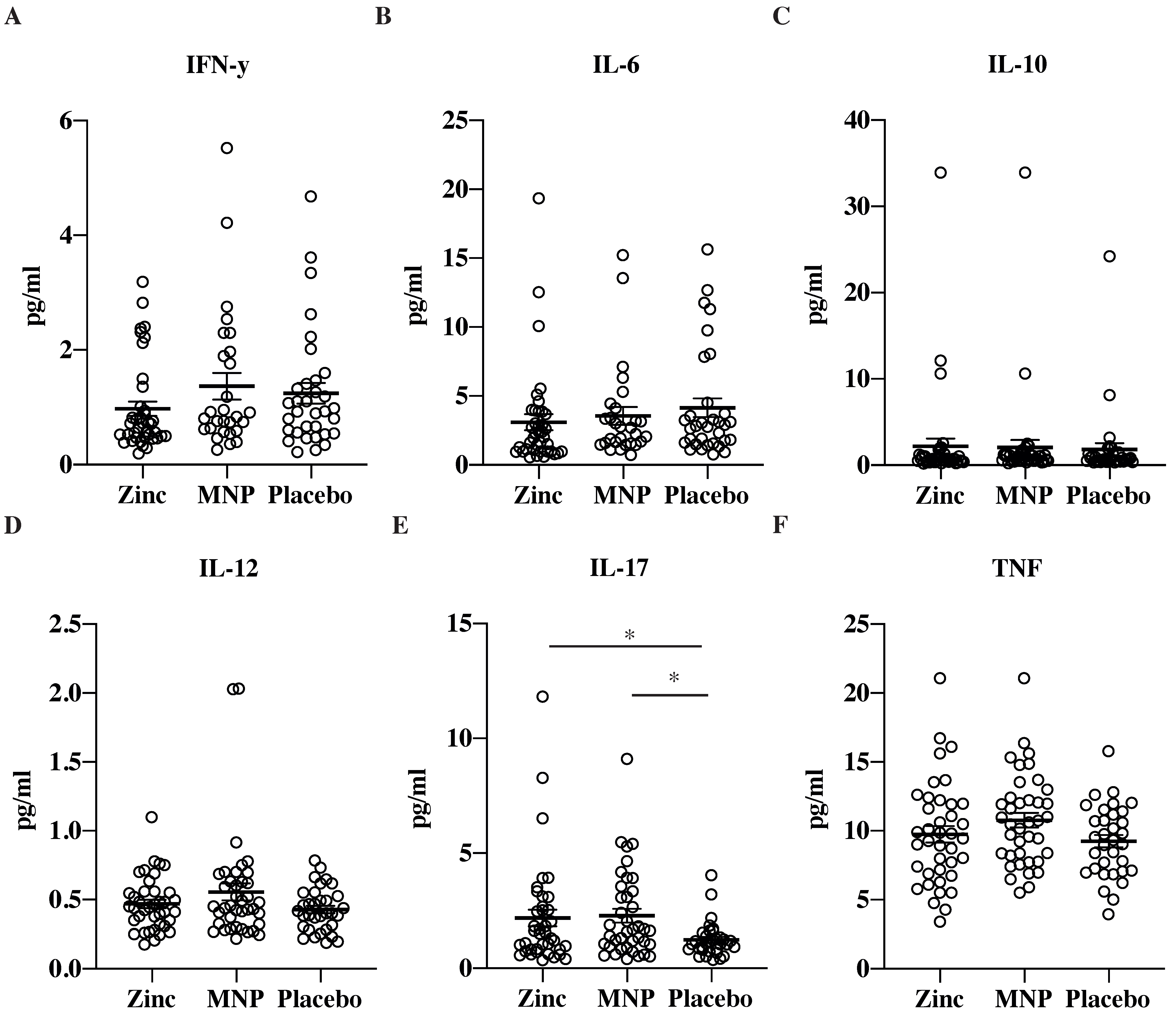
3.3. Simoa Analysis of Serum Cytokine Levels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wessells, K.R.; Brown, K.H. Estimating the global prevalence of zinc deficiency: Results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE 2012, 7, e50568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Brazier, A.K.M.; Lowe, N.M. Zinc deficiency in low- and middle-income countries: Prevalence and approaches for mitigation. J. Hum. Nutr. Diet 2020, 33, 624–643. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, U.; Kisenge, R.; Sudfeld, C.R.; Dhingra, P.; Somji, S.; Dutta, A.; Bakari, M.; Deb, S.; Devi, P.; Liu, E.; et al. Lower-Dose Zinc for Childhood Diarrhea—A Randomized, Multicenter Trial. N. Engl. J. Med. 2020, 383, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Integrated Management of Childhood Illness: Chart Booklet; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Brown, K.H.; Peerson, J.M.; Baker, S.K.; Hess, S.Y. Preventive zinc supplementation among infants, preschoolers, and older prepubertal children. Food Nutr. Bull. 2009, 30, S12–S40. [Google Scholar] [CrossRef] [PubMed]
- Yakoob, M.Y.; Theodoratou, E.; Jabeen, A.; Imdad, A.; Eisele, T.P.; Ferguson, J.; Jhass, A.; Rudan, I.; Campbell, H.; Black, R.E.; et al. Preventive zinc supplementation in developing countries: Impact on mortality and morbidity due to diarrhea, pneumonia and malaria. BMC Public Health 2011, 11 (Suppl. S3), S23. [Google Scholar] [CrossRef] [Green Version]
- Kambe, T.; Tsuji, T.; Hashimoto, A.; Itsumura, N. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol. Rev. 2015, 95, 749–784. [Google Scholar] [CrossRef]
- Wessels, I.; Maywald, M.; Rink, L. Zinc as a Gatekeeper of Immune Function. Nutrients 2017, 9, 1286. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; He, L.; Cai, L. Transcriptome Sequencing: RNA-Seq. Methods Mol. Biol. 2018, 1754, 15–27. [Google Scholar] [CrossRef]
- Islam, M.M.; McDonald, C.M.; Krebs, N.F.; Westcott, J.; Rahman, A.E.; El Arifeen, S.; Ahmed, T.; King, J.C.; Black, R.E. Study Protocol for a Randomized, Double-Blind, Community-Based Efficacy Trial of Various Doses of Zinc in Micronutrient Powders or Tablets in Young Bangladeshi Children. Nutrients 2018, 10, 132. [Google Scholar] [CrossRef] [Green Version]
- World Health Organisation. WHO Child Growth Standards: Length/Height-for-Age, Weight-for-Age, Weight-for-Length, Weight-for-Height and Body Mass Index-for-Age. Methods and Development; World Health Organization: Geneva, Switzerland, 2006. [Google Scholar]
- Hashimshony, T.; Senderovich, N.; Avital, G.; Klochendler, A.; de Leeuw, Y.; Anavy, L.; Gennert, D.; Li, S.; Livak, K.J.; Rozenblatt-Rosen, O.; et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016, 17, 77. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Su, S.; Dong, X.; Amann-Zalcenstein, D.; Biben, C.; Seidi, A.; Hilton, D.J.; Naik, S.H.; Ritchie, M.E. scPipe: A flexible R/Bioconductor preprocessing pipeline for single-cell RNA-sequencing data. PLoS Comput. Biol. 2018, 14, e1006361. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Lun, A.T.L.; Smyth, G.K. No counts, no variance: Allowing for loss of degrees of freedom when assessing biological variability from RNA-seq data. Stat. Appl. Genet. Mol. Biol. 2017, 16, 83–93. [Google Scholar] [CrossRef]
- Smyth, G.K.; Michaud, J.; Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust Hyperparameter Estimation Protects against Hypervariable Genes and Improves Power to Detect Differential Expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Rivnak, A.J.; Rissin, D.M.; Kan, C.W.; Song, L.; Fishburn, M.W.; Piech, T.; Campbell, T.G.; DuPont, D.R.; Gardel, M.; Sullivan, S.; et al. A fully-automated, six-plex single molecule immunoassay for measuring cytokines in blood. J. Immunol. Methods 2015, 424, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S. Zinc in human health: Effect of zinc on immune cells. Mol. Med. 2008, 14, 353–357. [Google Scholar] [CrossRef]
- Prasad, A.S.; Bao, B.; Beck, F.W.; Sarkar, F.H. Zinc activates NF-kappaB in HUT-78 cells. J. Lab. Clin. Med. 2001, 138, 250–256. [Google Scholar] [CrossRef]
- Liu, M.J.; Bao, S.; Galvez-Peralta, M.; Pyle, C.J.; Rudawsky, A.C.; Pavlovicz, R.E.; Killilea, D.W.; Li, C.; Nebert, D.W.; Wewers, M.D.; et al. ZIP8 regulates host defense through zinc-mediated inhibition of NF-kappaB. Cell Rep. 2013, 3, 386–400. [Google Scholar] [CrossRef] [Green Version]
- Aydemir, T.B.; Liuzzi, J.P.; McClellan, S.; Cousins, R.J. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J. Leukoc. Biol. 2009, 86, 337–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cousins, R.J.; Blanchard, R.K.; Popp, M.P.; Liu, L.; Cao, J.; Moore, J.B.; Green, C.L. A global view of the selectivity of zinc deprivation and excess on genes expressed in human THP-1 mononuclear cells. Proc. Natl. Acad. Sci. USA 2003, 100, 6952–6957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, M.S.; Langkamp-Henken, B.; Chang, S.M.; Shankar, M.N.; Cousins, R.J. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc. Natl. Acad. Sci. USA 2011, 108, 20970–20975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar, A.H.; Prasad, A.S. Zinc and immune function: The biological basis of altered resistance to infection. Am. J. Clin. Nutr. 1998, 68, 447S–463S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kewcharoenwong, C.; Schuster, G.U.; Wessells, K.R.; Hinnouho, G.M.; Barffour, M.A.; Kounnavong, S.; Brown, K.H.; Hess, S.Y.; Samer, W.; Tussakhon, I.; et al. Daily Preventive Zinc Supplementation Decreases Lymphocyte and Eosinophil Concentrations in Rural Laotian Children from Communities with a High Prevalence of Zinc Deficiency: Results of a Randomized Controlled Trial. J. Nutr. 2020, 150, 2204–2213. [Google Scholar] [CrossRef]
- Long, J.M.; Mondal, P.; Westcott, J.E.; Miller, L.V.; Islam, M.M.; Ahmed, M.; Mahfuz, M.; Ahmed, T.; Krebs, N.F. Zinc Absorption from Micronutrient Powders Is Low in Bangladeshi Toddlers at Risk of Environmental Enteric Dysfunction and May Increase Dietary Zinc Requirements. J. Nutr. 2019, 149, 98–105. [Google Scholar] [CrossRef]
- Gautam, A.; Donohue, D.; Hoke, A.; Miller, S.A.; Srinivasan, S.; Sowe, B.; Detwiler, L.; Lynch, J.; Levangie, M.; Hammamieh, R.; et al. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS ONE 2019, 14, e0225137. [Google Scholar] [CrossRef]

| Zinc MNPs | Zinc Tablets | Placebo | Difference Between Groups (p-Value, ANOVA) | |
|---|---|---|---|---|
| N | 28 | 39 | 33 | |
| Age (months) | 9.8 (0.94) | 9.7 (0.87) | 9.7 (0.85) | 0.93 |
| Sex N, (F%) | 12 (42.9%) | 20 (51.3%) | 17 (51.5%) | 0.75 |
| WLZ at baseline | −0.66 (0.85) | −0.40 (0.88) | −0.61 (0.95) | 0.85 |
| WLZ after 24 weeks | −1.0 (0.87) | −0.68 (0.86) | −0.90 (0.94) | 0.32 |
| LAZ at baseline | −1.45 (0.84) | −1.23 (1.10) | −1.36 (1.08) | 0.67 |
| LAZ after 24 weeks | −1.70 (0.78) | −1.41 (1.10) | −1.50 (1.09) | 0.52 |
| WAZ at baseline | −1.29 (0.94) | −0.99 (1.08) | −1.22 (1.09) | 0.42 |
| WAZ after 24 weeks | −1.53 (0.91) | −1.16 (0.99) | −1.36 (1.01) | 0.33 |
| Any diarrhea within last 2 weeks at endline | 1 (3.6%) | 2 (5.1%) | 2 (6.1%) | 0.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayman, T.; Hickey, P.; Amann-Zalcenstein, D.; Bennett, C.; Ataide, R.; Sthity, R.A.; Khandaker, A.M.; Islam, K.M.; Stracke, K.; Yassi, N.; et al. Zinc Supplementation with or without Additional Micronutrients Does Not Affect Peripheral Blood Gene Expression or Serum Cytokine Level in Bangladeshi Children. Nutrients 2021, 13, 3516. https://doi.org/10.3390/nu13103516
Hayman T, Hickey P, Amann-Zalcenstein D, Bennett C, Ataide R, Sthity RA, Khandaker AM, Islam KM, Stracke K, Yassi N, et al. Zinc Supplementation with or without Additional Micronutrients Does Not Affect Peripheral Blood Gene Expression or Serum Cytokine Level in Bangladeshi Children. Nutrients. 2021; 13(10):3516. https://doi.org/10.3390/nu13103516
Chicago/Turabian StyleHayman, Thomas, Peter Hickey, Daniela Amann-Zalcenstein, Cavan Bennett, Ricardo Ataide, Rahvia Alam Sthity, Afsana Mim Khandaker, Kazi Munisul Islam, Katharina Stracke, Nawaf Yassi, and et al. 2021. "Zinc Supplementation with or without Additional Micronutrients Does Not Affect Peripheral Blood Gene Expression or Serum Cytokine Level in Bangladeshi Children" Nutrients 13, no. 10: 3516. https://doi.org/10.3390/nu13103516
APA StyleHayman, T., Hickey, P., Amann-Zalcenstein, D., Bennett, C., Ataide, R., Sthity, R. A., Khandaker, A. M., Islam, K. M., Stracke, K., Yassi, N., Watson, R., Long, J., Westcott, J., Krebs, N. F., King, J. C., Black, R. E., Islam, M. M., McDonald, C. M., & Pasricha, S.-R. (2021). Zinc Supplementation with or without Additional Micronutrients Does Not Affect Peripheral Blood Gene Expression or Serum Cytokine Level in Bangladeshi Children. Nutrients, 13(10), 3516. https://doi.org/10.3390/nu13103516






