Postprandial NMR-Based Metabolic Exchanges Reflect Impaired Phenotypic Flexibility across Splanchnic Organs in the Obese Yucatan Mini-Pig
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Procedure
2.2. Plasma Metabolomics
2.3. Statistical Analyses
2.4. Identification of Metabolites Exchanges
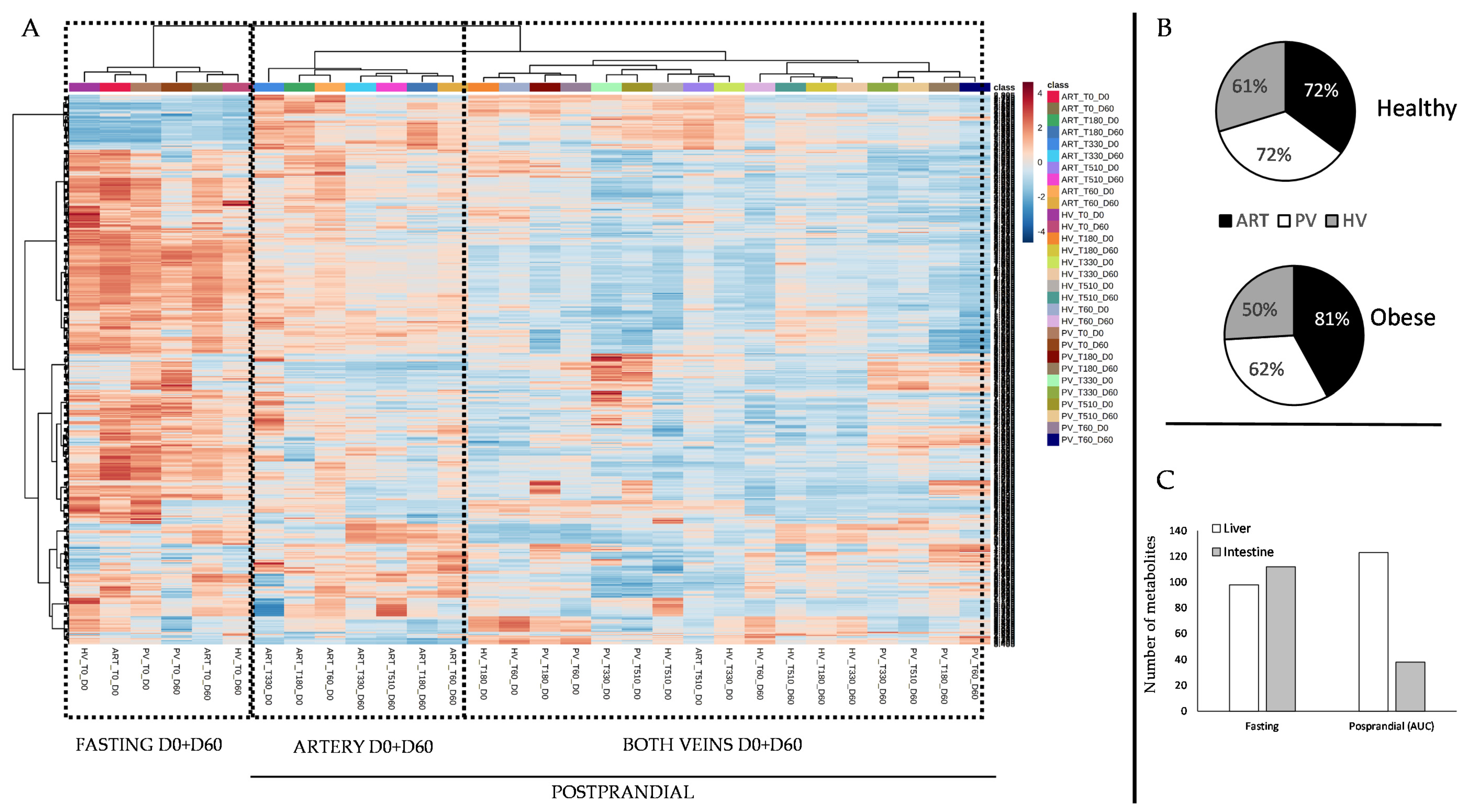
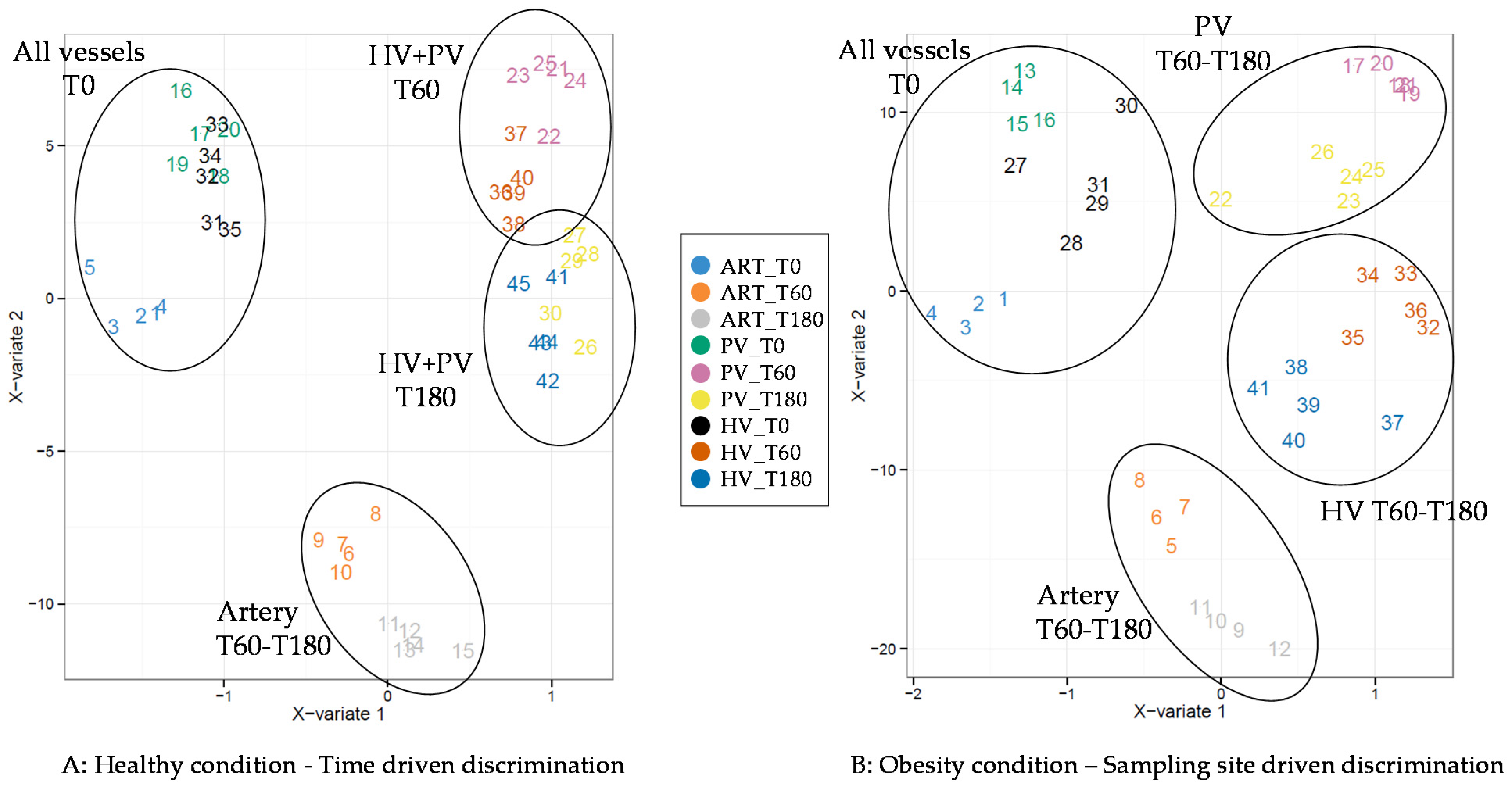
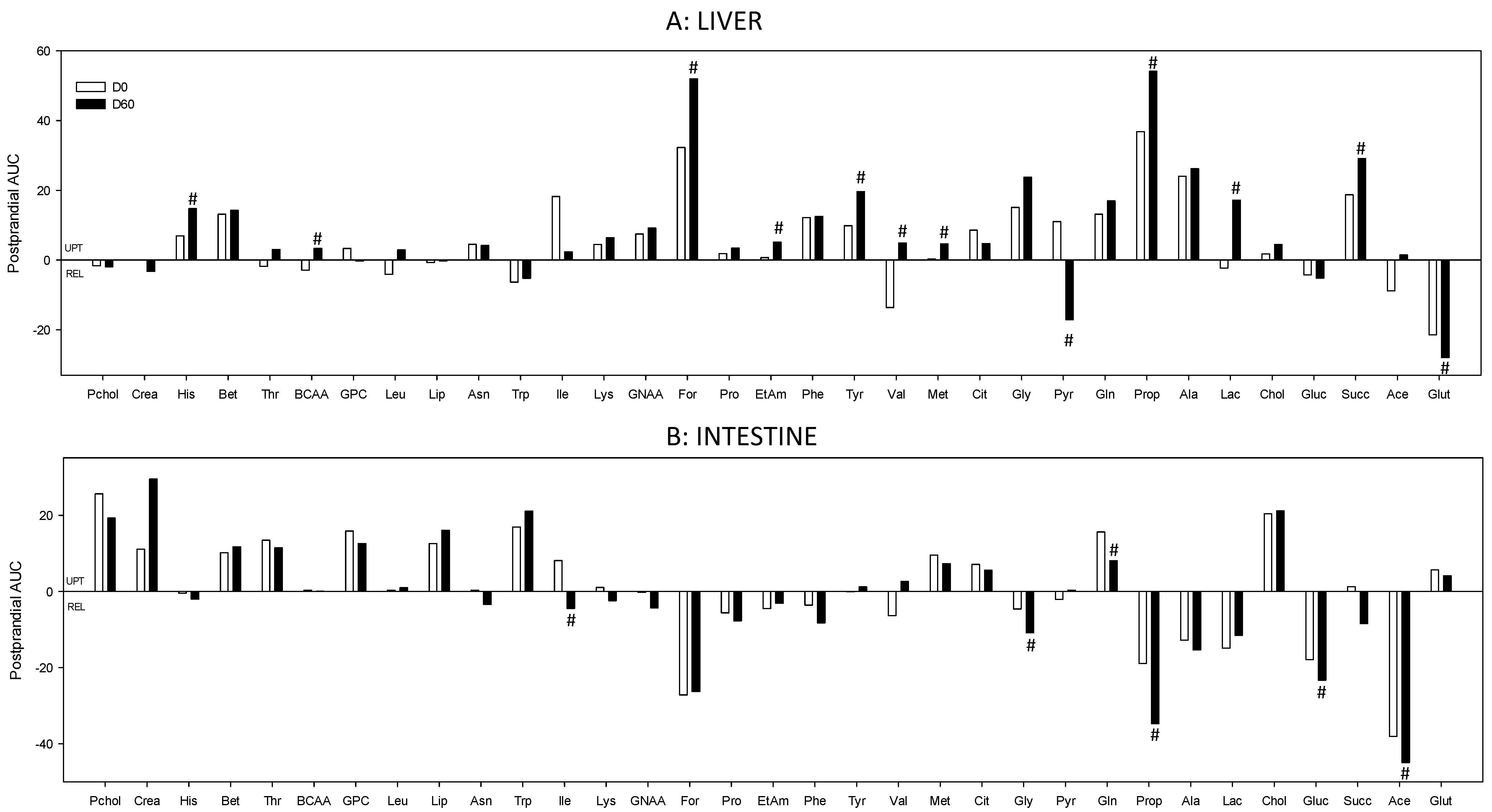
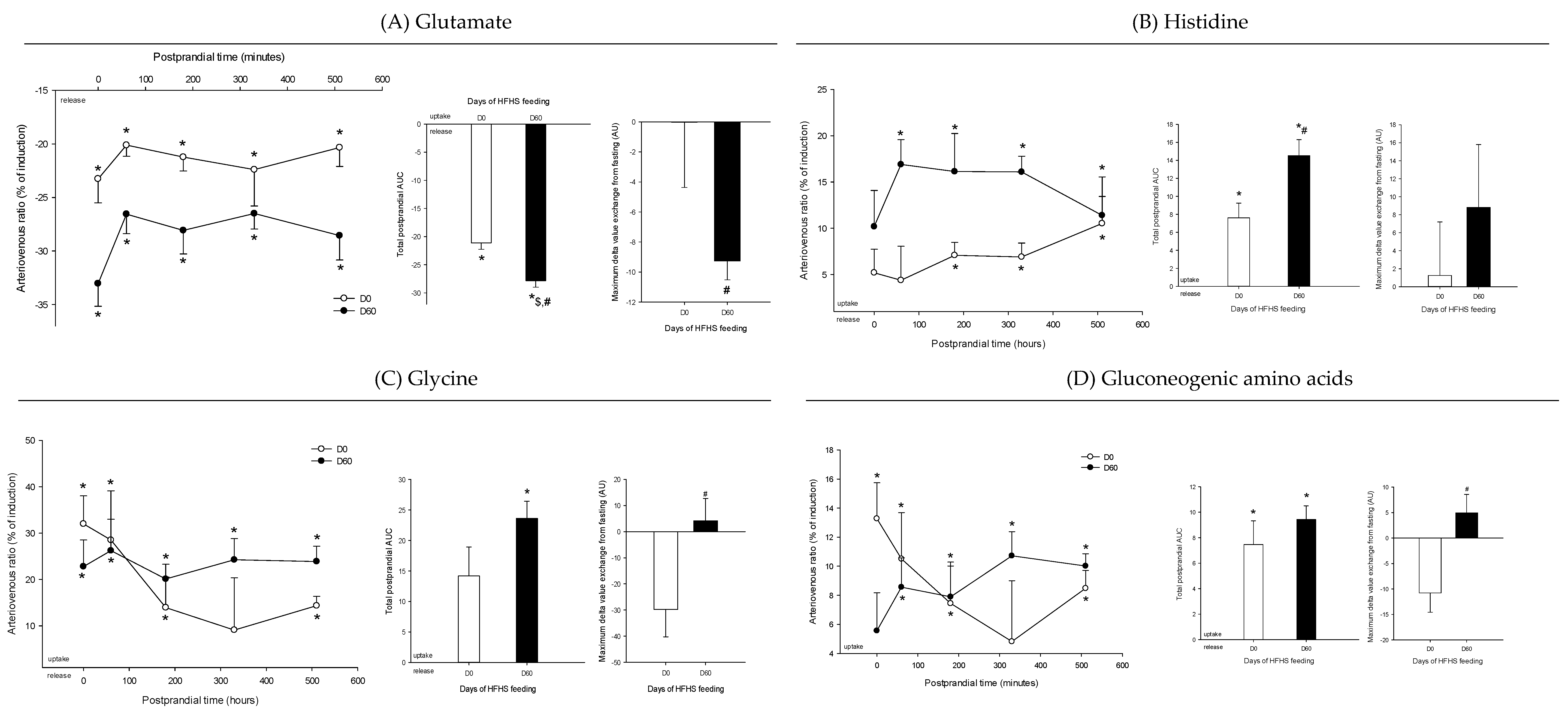
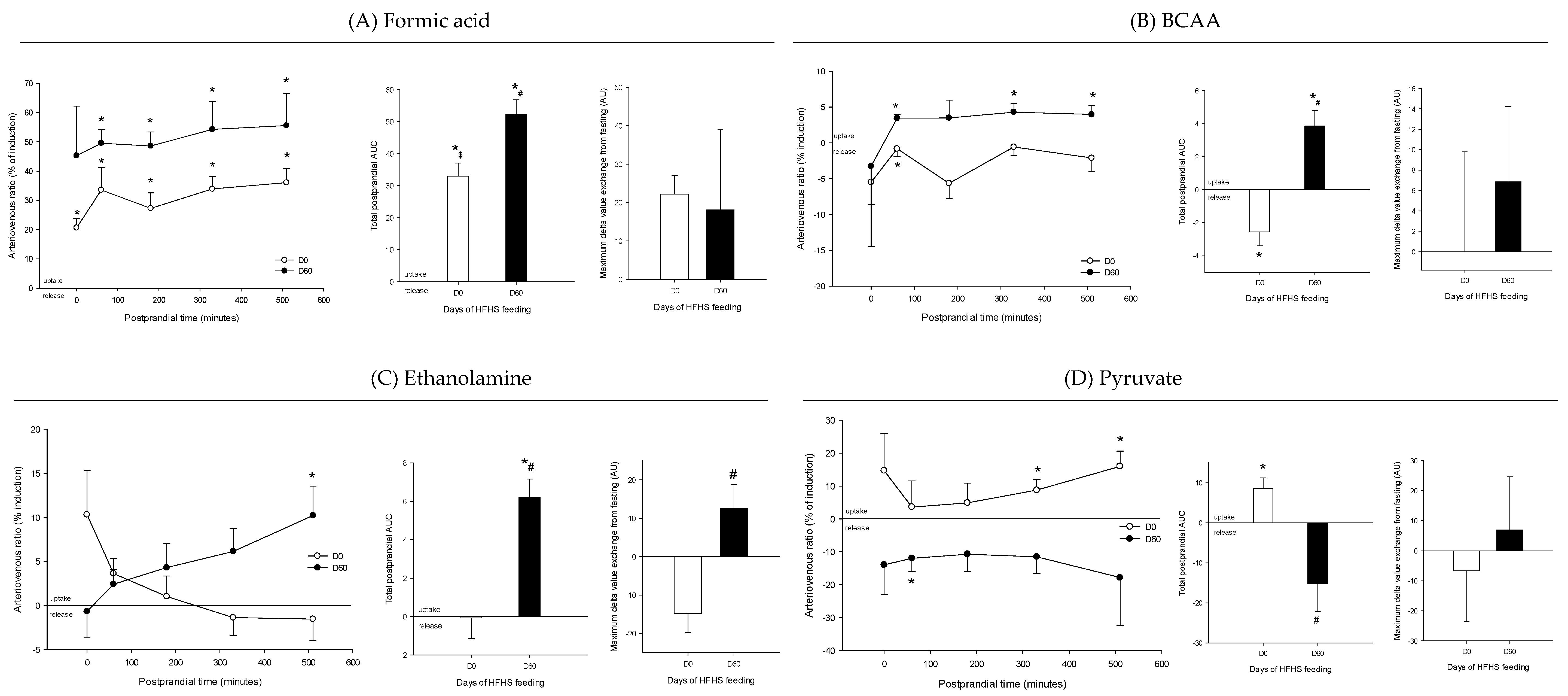
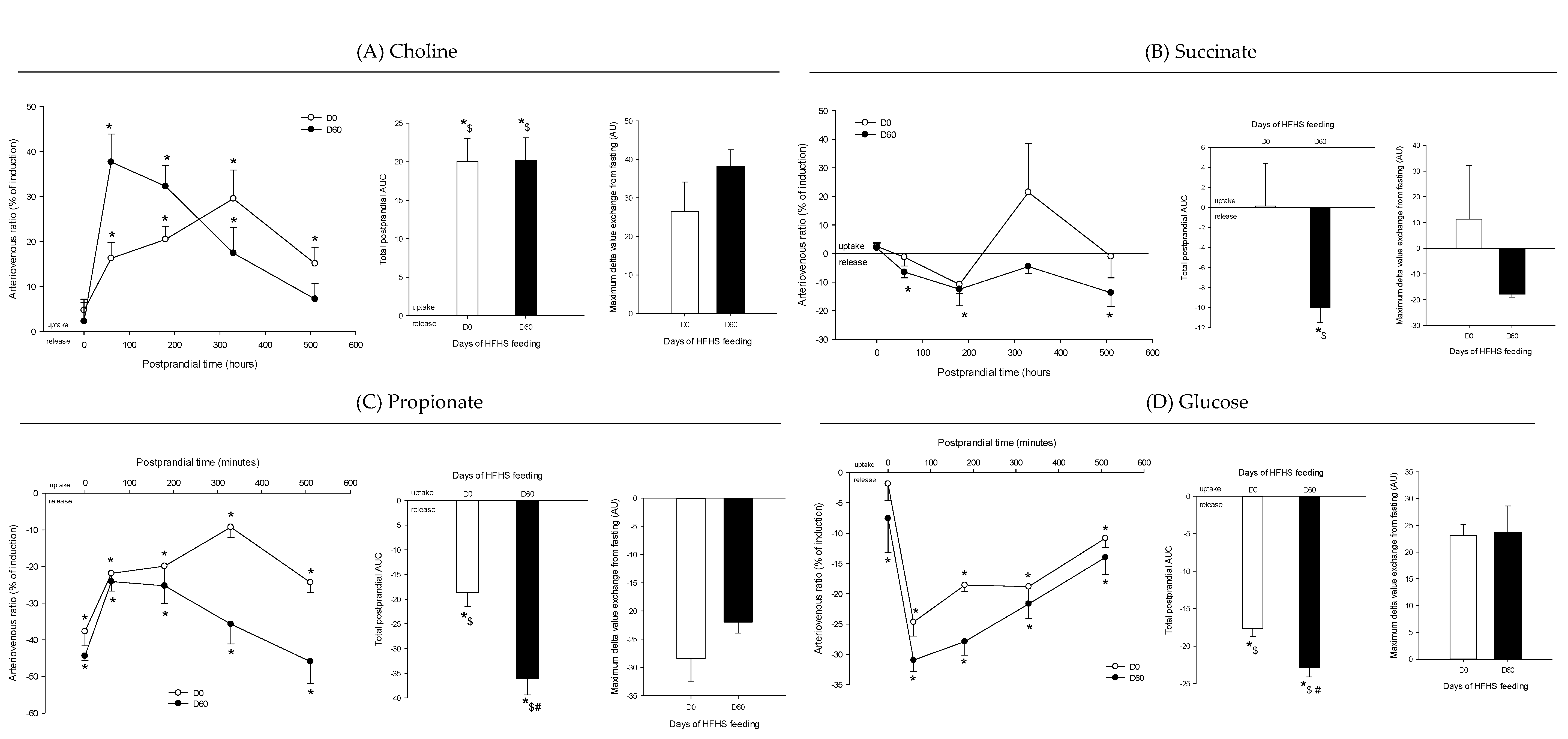
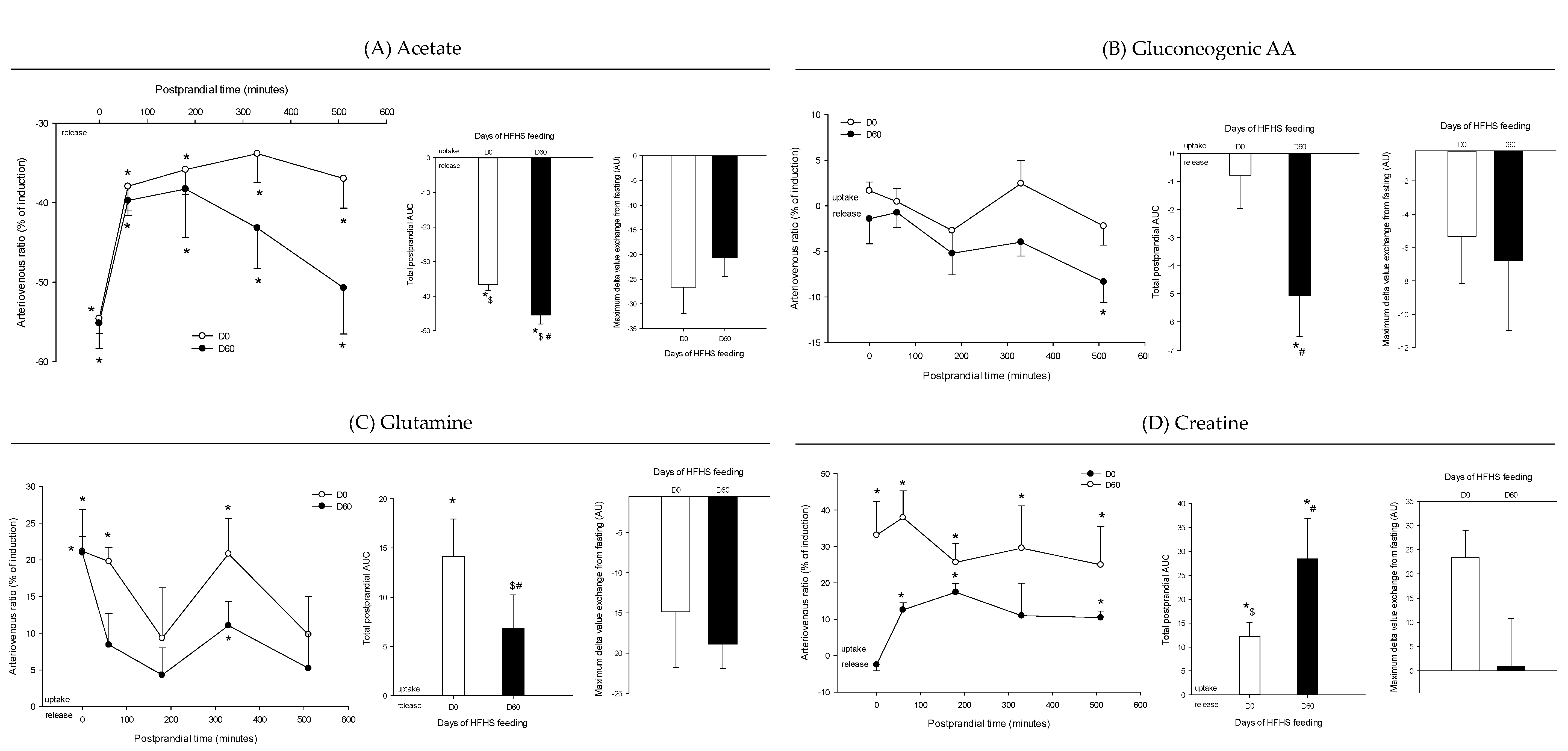
3. Results
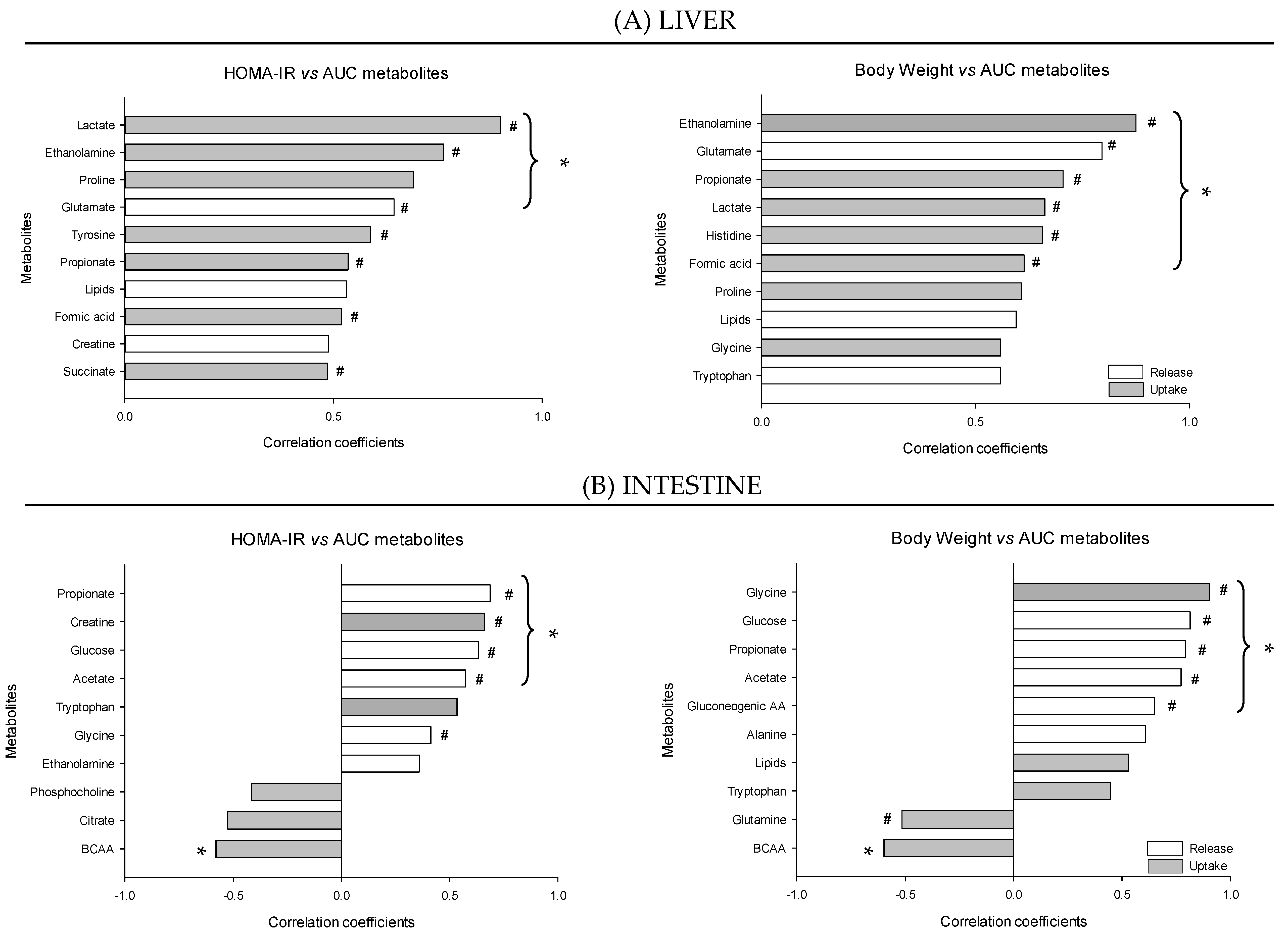
4. Discussion
4.1. The Metabolomes of the Splanchnic Area Are Greatly Altered Following Meals
4.2. Adaptive Changes Take Place in the Splanchnic Metabolism to Maintain Postprandial Glucose Homeostasis at the Onset of Obesity
4.3. The Loss of Postprandial Flexibility in the Splanchnic Organs as a Symptom of Early Metabolic Alterations
4.4. The Postprandial Metabolism of the Splanchnic Organs Adapts to Circulating Nutrient Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Ommen, B.; van der Greef, J.; Ordovas, J.M.; Daniel, H. Phenotypic flexibility as key factor in the human nutrition and health relationship. Genes Nutr. 2014, 9, 423. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, A.; Nadeau, A.; Despres, J.P.; Bouchard, C. Hyperinsulinemia and regulation of energy balance. Am. J. Clin. Nutr. 1995, 61, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Remond, D.; Bernalier-Donadille, A.; Rambeau, M.; Pujos-Guillot, E.; Comte, B.; Dardevet, D.; Savary-Auzeloux, I. Metabolic adaptations to HFHS overfeeding: How whole body and tissues postprandial metabolic flexibility adapt in Yucatan mini-pigs. Eur. J. Nutr. 2018, 57, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Savary-Auzeloux, I.; Mohamed, A.-B.; Cohade, B.; Dardevet, D.; David, J.; Hafnaoui, N.; Migné, C.; Pujos-Guillot, E.; Rémond, D.; Polakof, S. Profound changes in net energy and nitrogen metabolites fluxes within the splanchnic area during overfeeding of Yucatan mini pigs that remain euglycemic. Nutrients 2019, 11, 434. [Google Scholar]
- Polakof, S.; Remond, D.; David, J.; Dardevet, D.; Savary-Auzeloux, I. Time-course changes in circulating branched-chain amino acid levels and metabolism in obese Yucatan minipig. Nutrition 2018, 50, 66–73. [Google Scholar] [CrossRef]
- Zeng, Y.C.; David, J.; Remond, D.; Dardevet, D.; Savary-Auzeloux, I.; Polakof, S. Peripheral blood mononuclear cell metabolism acutely adapted to postprandial transition and mainly reflected metabolic adipose tissue adaptations to a high-fat diet in minipigs. Nutrients 2018, 10, 1816. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef]
- Sébédio, J.-L.; Polakof, S. Using metabolomics to identify biomarkers for metabolic diseases: Analytical methods and applications. In Metabolomics as a Tool in Nutrition Research; Sebedio, J.L., Brennan, L., Eds.; Woodhead Publishing- Elsevier: Cambridge, UK, 2015; pp. 145–166. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Xu, F.; Tavintharan, S.; Sum, C.F.; Woon, K.; Lim, S.C.; Ong, C.N. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. J. Clin. Endocrinol. Metab. 2013, 98, E1060–E1065. [Google Scholar] [CrossRef]
- Polakof, S.; Dardevet, D.; Lyan, B.; Mosoni, L.; Gatineau, E.; Martin, J.F.; Pujos-Guillot, E.; Mazur, A.; Comte, B. Time course of molecular and metabolic events in the development of insulin resistance in fructose-fed rats. J. Proteome Res. 2016, 15, 1862–1874. [Google Scholar] [CrossRef]
- Polakof, S.; Rémond, D.; Rambeau, M.; Pujos-Guillot, E.; Sébédio, J.-L.; Dardevet, D.; Comte, B.; Savary-Auzeloux, I. Postprandial metabolic events in mini-pigs: New insights from a combined approach using plasma metabolomics, tissue gene expression, and enzyme activity. Metabolomics 2015, 11, 964–979. [Google Scholar] [CrossRef]
- Pimentel, G.; Burton, K.J.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergères, G. The postprandial metabolome—A source of Nutritional Biomarkers of Health. Curr. Opin. Food Sci. 2017, 16, 67–73. [Google Scholar] [CrossRef]
- Fiamoncini, J.; Rundle, M.; Gibbons, H.; Thomas, E.L.; Geillinger-Kästle, K.; Bunzel, D.; Trezzi, J.-P.; Kiselova-Kaneva, Y.; Wopereis, S.; Wahrheit, J.; et al. Plasma metabolome analysis identifies distinct human metabotypes in the postprandial state with different susceptibility to weight loss–mediated metabolic improvements. FASEB J. 2018, 32, 5447–5458. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.; Hendriks, H.F.; Cnubben, N.H.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M.; et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef]
- Baig, F.; Pechlaner, R.; Mayr, M. Caveats of untargeted metabolomics for biomarker discovery. J. Am. Coll. Cardiol. 2016, 68, 1294–1296. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451. [Google Scholar] [CrossRef]
- Jang, C.; Hui, S.; Zeng, X.; Cowan, A.J.; Wang, L.; Chen, L.; Morscher, R.J.; Reyes, J.; Frezza, C.; Hwang, H.Y.; et al. Metabolite exchange between mammalian organs quantified in pigs. Cell Metab. 2019, 30, 594–606. [Google Scholar] [CrossRef]
- Poupin, N.; Tremblay-Franco, M.; Amiel, A.; Canlet, C.; Rémond, D.; Debrauwer, L.; Dardevet, D.; Thiele, I.; Aurich, M.K.; Jourdan, F.; et al. Arterio-venous metabolomics exploration reveals major changes across liver and intestine in the obese Yucatan minipig. Sci. Rep. 2019, 9, 12527. [Google Scholar] [CrossRef]
- Kardinaal, A.F.M.; Erk, M.J.v.; Dutman, A.E.; Stroeve, J.H.M.; Steeg, E.V.D.; Bijlsma, S.; Kooistra, T.; Ommen, B.V.; Wopereis, S. Quantifying phenotypic flexibility as the response to a high-fat challenge test in different states of metabolic health. FASEB J. 2015, 29, 4600–4613. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. (Stat. Method.) 1995, 289–300. [Google Scholar] [CrossRef]
- Liquet, B.; Cao, K.-A.L.; Hocini, H.; Thiébaut, R. A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinform. 2012, 13, 325. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.-A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comp. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.L.; Bergman, E.N. Simultaneous measurements of hepatic and portal venous blood flow in the sheep and dog. Am. J. Physiol. 1969, 216, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Secor, S.M. Specific dynamic action: A review of the postprandial metabolic response. J. Comp. Physiol. B 2009, 179, 1–56. [Google Scholar] [CrossRef]
- Fechner, E.; Bilet, L.; Peters, H.P.F.; Hiemstra, H.; Jacobs, D.M.; Op ’t Eyndt, C.; Kornips, E.; Mensink, R.P.; Schrauwen, P. Effects of a whole diet approach on metabolic flexibility, insulin sensitivity and postprandial glucose responses in overweight and obese adults—A randomized controlled trial. Clin. Nutr. 2019. [Google Scholar] [CrossRef]
- Serino, M.; Luche, E.; Gres, S.; Baylac, A.; Bergé, M.; Cenac, C.; Waget, A.; Klopp, P.; Iacovoni, J.; Klopp, C.; et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 2012, 61, 543–553. [Google Scholar]
- Wang, F.; Liu, F.; Cai, H.; Yang, L.; Sun, G. The efficacy of high fat load liquid meal on postprandial insulin level in postprandial insulin resistance population. FASEB J. 2016, 30, 904.12. [Google Scholar] [CrossRef]
- De Vegt, F.; Dekker, J.M.; Ruhé, H.G.; Stehouwer, C.D.A.; Nijpels, G.; Bouter, L.M.; Heine, R.J. Hyperglycaemia is associated with all-cause and cardiovascular mortality in the Hoorn population: The Hoorn Study. Diabetologia 1999, 42, 926–931. [Google Scholar] [CrossRef]
- Hu, L.; Kristensen, N.B.; Che, L.; Wu; Theil, P.K. Net absorption and liver metabolism of amino acids and heat production of portal-drained viscera and liver in multiparous sows during transition and lactation. J. Anim. Sci. Biotechnol. 2020, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.P.; Ellmerer, M.; Van Citters, G.W.; Bergman, R.N. Primacy of hepatic insulin resistance in the development of the metabolic syndrome induced by an isocaloric moderate-fat diet in the dog. Diabetes 2003, 52, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Gastaldelli, A.; Cusi, K.; Pettiti, M.; Hardies, J.; Miyazaki, Y.; Berria, R.; Buzzigoli, E.; Sironi, A.M.; Cersosimo, E.; Ferrannini, E.; et al. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology 2007, 133, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Hyotylainen, T.; Jerby, L.; Petaja, E.M.; Mattila, I.; Jantti, S.; Auvinen, P.; Gastaldelli, A.; Yki-Jarvinen, H.; Ruppin, E.; Oresic, M. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef]
- Giusi-Perier, A.; Fiszlewicz, M.; Rerat, A. Influence of diet composition on intestinal volatile fatty acid and nutrient absorption in unanesthetized pigs. J. Anim. Sci. 1989, 67, 386–402. [Google Scholar] [CrossRef]
- Bach Knudsen, K.E.; Jorgensen, H.; Canibe, N. Quantification of the absorption of nutrients derived from carbohydrate assimilation: Model experiment with catheterised pigs fed on wheat- or oat-based rolls. Br. J. Nutr. 2000, 84, 449–458. [Google Scholar] [CrossRef]
- Hiyoshi, T.; Fujiwara, M.; Yao, Z. Postprandial hyperglycemia and postprandial hypertriglyceridemia in type 2 diabetes. J. Biomed. Res. 2017. [Google Scholar] [CrossRef]
- Foster, D.W. Banting lecture 1984. From glycogen to ketones: And back. Diabetes 1984, 33, 1188–1199. [Google Scholar] [CrossRef]
- Azzout-Marniche, D.; Gaudichon, C.; Blouet, C.; Bos, C.; Mathe, V.; Huneau, J.F.; Tome, D. Liver glyconeogenesis: A pathway to cope with postprandial amino acid excess in high-protein fed rats? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R1400–R1407. [Google Scholar] [CrossRef]
- Toye, A.A.; Dumas, M.E.; Blancher, C.; Rothwell, A.R.; Fearnside, J.F.; Wilder, S.P.; Bihoreau, M.T.; Cloarec, O.; Azzouzi, I.; Young, S.; et al. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia 2007, 50, 1867–1879. [Google Scholar] [CrossRef]
- Bazotte, R.B.; Silva, L.G.; Schiavon, F.P. Insulin resistance in the liver: Deficiency or excess of insulin? Cell Cycle 2014, 13, 2494–2500. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.B.; Remond, D.; Chambon, C.; Sayd, T.; Hebraud, M.; Capel, F.; Cohade, B.; Hafnaoui, N.; Bechet, D.; Coudy-Gandilhon, C.; et al. A mix of dietary fermentable fibers improves lipids handling by the liver of overfed minipigs. J. Nutr. Biochem. 2018, 65, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Arroyo, S.F.; Sumpton, D.; Mackay, G.M.; Martin-Castillo, B.; Camps, J.; Joven, J.; Menendez, J.A.; Vazquez, A.; METTEN Study Group. Stratification of cancer and diabetes based on circulating levels of formate and glucose. Cancer Metab. 2019, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.E.; Larson, M.G.; Ghorbani, A.; Cheng, S.; Chen, M.H.; Keyes, M.; Rhee, E.P.; Clish, C.B.; Vasan, R.S.; Gerszten, R.E.; et al. Metabolomic profiles of body mass index in the Framingham Heart Study reveal distinct cardiometabolic phenotypes. PLoS ONE 2016, 11, e0148361. [Google Scholar] [CrossRef]
- Costallat, B.L.; Miglioli, L.; Silva, P.A.C.; Novo, N.F.; Duarte, J.L.G. Resistência à insulina com a suplementação de creatina em animais de experimentação. Revista Brasileira Medicina Esporte 2007, 13, 22–26. [Google Scholar] [CrossRef]
- Alves, A.; Bassot, A.; Bulteau, A.L.; Pirola, L.; Morio, B. Glycine Metabolism and Its Alterations in Obesity and Metabolic Diseases. Nutrients 2019, 11, 1356. [Google Scholar] [CrossRef]
- Hsieh, J.; Hayashi, A.A.; Webb, J.; Adeli, K. Postprandial dyslipidemia in insulin resistance: Mechanisms and role of intestinal insulin sensitivity. Atheroscler. Suppl. 2008, 9, 7–13. [Google Scholar] [CrossRef]
- Veilleux, A.; Mayeur, S.; Berube, J.C.; Beaulieu, J.F.; Tremblay, E.; Hould, F.S.; Bosse, Y.; Richard, D.; Levy, E. Altered intestinal functions and increased local inflammation in insulin-resistant obese subjects: A gene-expression profile analysis. BMC Gastroenterol. 2015, 15, 1–12. [Google Scholar] [CrossRef]
- Barber, T.; Vina, J.R.; Vina, J.; Cabo, J. Decreased urea synthesis in cafeteria-diet-induced obesity in the rat. Biochem. J. 1985, 230, 675–681. [Google Scholar] [CrossRef]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.C.; Fasshauer, M.; Filatova, N.; Grundell, L.A.; Zielinski, E.; Zhou, J.Y.; Scherer, T.; Lindtner, C.; White, P.J.; Lapworth, A.L.; et al. Brain insulin lowers circulating BCAA levels by inducing hepatic BCAA catabolism. Cell Metab. 2014, 20, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, F.; Sun, D.; Wang, X.; Zhang, X.; Zhang, J.; Yan, F.; Huang, C.; Xie, H.; Lin, C.; et al. Branched-chain amino acids exacerbate obesity-related hepatic glucose and lipid metabolic disorders via attenuating Akt2 signaling. Diabetes 2020, 69, 1164–1177. [Google Scholar] [CrossRef] [PubMed]


| Postprandial Time (Minutes) | ||||||
|---|---|---|---|---|---|---|
| 0 | 60 | 180 | 330 | 510 | ||
| Lysine | Day 0 | 8.97 ± 4.10 | 5.81 ± 1.67 * | 2.08 ± 1.48 | 4.13 ± 0.86 * | 6.26 ± 1.51 * |
| Day 60 | 3.88 ± 3.35 | 6.57 ± 0.48 * | 4.02 ± 2.28 | 7.68 ± 1.24 * | 8.91 ± 1.45 * | |
| Threonine | Day 0 | −0.63 ± 3.73 | 2.61 ± 1.78 | −2.44 ± 0.96 | −4.69 ± 3.16 | −0.36 ± 1.30 |
| Day 60 | 5.07 ± 3.66 | 5.21 ± 1.98 | 0.62 ± 1.76 | 3.39 ± 1.60 | 3.76 ± 4.54 | |
| Citrate | Day 0 | 30.87 ± 21.17 | 12.45 ± 1.52 * | 8.66 ± 6.59 | 0.86 ± 10.14 | 17.66 ± 5.89 * |
| Day 60 | 7.48 ± 7.95 | 1.74 ± 0.77 | 4.63 ± 11.26 | 7.53 ± 1.39 * | 10.62 ± 7.47 | |
| Isoleucine | Day 0 | 4.97 ± 3.38 | 10.94 ± 2.26 * | 26.12 ± 10.22 * | 19.64 ± 9.22 | 21.38 ± 7.98 * |
| Day 60 | 26.38 ± 22.41 | 3.10 ± 1.99 | −3.29 ± 10.12 | 5.19 ± 2.52 | 6.30 ± 4.31 | |
| Proline | Day 0 | 15.17 ± 10.74 | 2.59 ± 3.13 | 2.10 ± 3.54 | 1.18 ± 1.72 | 0.06 ± 1.54 |
| Day 60 | 0.76 ± 1.02 | 0.59 ± 0.87 | 3.05 ± 1.46 | 3.65 ± 2.25 | 8.80 ± 1.98 * | |
| Acetate | Day 0 | 1.06 ± 10.49 | −1.68 ± 2.30 | 1.17 ± 0.80 * | −16.05 ± 4.62 | −13.71 ± 5.11 |
| Day 60 | 4.92 ± 10.97 | 5.18 ± 8.58 | −1.40 ± 11.18 | 3.55 ± 11.85 | 21.64 ± 22.29 | |
| Tryptophan | Day 0 | −12.34 ± 7.34 | −9.32 ± 7.77 | −3.51 ± 7.79 | −9.03 ± 5.57 | 6.81 ± 6.89 |
| Day 60 | 0.65 ± 9.26 | 5.50 ± 5.19 | −0.91 ± 9.36 | −5.34 ± 4.20 | −16.44 ± 9.22 | |
| Creatine | Day 0 | −0.94 ± 2.83 | 2.15 ± 3.22 | −5.08 ± 1.21 * | 2.37 ± 2.46 | 2.10 ± 1.91 |
| Day 60 | −4.79 ± 7.77 | 2.53 ± 2.36 | −5.16 ± 2.49 | −2.75 ± 2.52 | −5.53 ± 3.65 | |
| Betaine | Day 0 | 11.87 ± 2.11 * | 15.29 ± 4.93 * | 5.94 ± 2.83 | 17.69 ± 8.61 | 11.30 ± 2.32 * |
| Day 60 | 15.79 ± 4.41 * | 14.92 ± 2.32 * | 10.70 ± 1.94 * | 16.54 ± 3.10 * | 12.88 ± 2.43 * | |
| Phosphocholine | Day 0 | −2.29 ± 4.76 | 3.74 ± 4.53 | −7.12 ± 2.74 | −2.58 ± 2.90 | 4.75 ± 3.29 |
| Day 60 | 5.87 ± 4.86 | 3.51 ± 2.78 | −7.04 ± 4.61 | −0.06 ± 3.08 | −2.58 ± 4.65 | |
| Alanine | Day 0 | 51.82 ± 15.81 * | 26.21 ± 4.81 * | 25.36 ± 7.37 * | 20.55 ± 0.97 * | 24.69 ± 1.24 * |
| Day 60 | 23.52 ± 7.30 * | 20.34 ± 2.21 * | 26.50 ± 3.85 * | 28.78 ± 4.14 * | 30.99 ± 3.74 * | |
| Asparagine | Day 0 | 18.39 ± 9.83 | 5.54 ± 3.80 | 6.01 ± 3.38 | 3.54 ± 4.07 | 0.84 ± 3.13 |
| Day 60 | −0.15 ± 2.64 | 2.09 ± 1.82 | 3.17 ± 1.09 * | 4.72 ± 2.07 | 10.95 ± 7.37 | |
| Methionine | Day 0 | 12.27 ± 6.82 | 8.98 ± 1.52 * | 4.90 ± 2.30 | 33.92 ± 4.22 * | 36.07 ± 4.86 * |
| Day 60 | −2.62 ± 3.97 | 4.45 ± 0.60 * | 4.97 ± 2.48 | 3.28 ± 2.54 | 6.90 ± 3.38 | |
| Lipids | Day 0 | −0.46 ± 2.30 | 0.44 ± 0.61 | 0.44 ± 1.65 | −0.91 ± 0.81 | −2.75 ± 1.25 |
| Day 60 | −0.80 ± 2.53 | 0.23 ± 1.79 | 0.81 ± 3.64 | −1.00 ± 1.89 | −1.37 ± 1.20 | |
| Glycero-Phosphocholine | Day 0 | 1.59 ± 3.92 | 4.11 ± 3.07 | 0.61 ± 1.54 | 4.12 ± 2.06 | 5.68 ± 1.37 * |
| Day 60 | 4.14 ± 2.65 | 4.19 ± 2.52 | −3.27 ± 2.14 | 0.50 ± 2.59 | −2.16 ± 3.37 | |
| Glutamine | Day 0 | 19.13 ± 9.29 * | 10.06 ± 3.71 | 11.42 ± 3.90 * | 13.83 ± 3.56 * | 17.78 ± 4.40 * |
| Day 60 | 8.75 ± 6.11 | 12.56 ± 0.51 * | 16.02 ± 5.74 * | 20.83 ± 0.72 * | 23.92 ± 5.11 | |
| Valine | Day 0 | −9.98 ± 13.40 | −6.63 ± 2.56 | −19.59 ± 6.02 * | −10.05 ± 3.39 * | −11.89 ± 5.61 |
| Day 60 | −9.99 ± 8.82 | 4.06 ± 1.81 | 9.75 ± 8.69 | 5.08 ± 2.03 | 5.76 ± 2.78 | |
| Leucine | Day 0 | −4.25 ± 7.91 | −1.69 ± 1.16 | −6.50 ± 2.69 | −2.97 ± 1.41 | −3.25 ± 2.54 |
| Day 60 | −4.04 ± 5.91 | 3.25 ± 0.95 * | 3.88 ± 3.77 | 3.67 ± 1.3 * | 2.84 ± 2.02 | |
| Tyrosine | Day 0 | 15.49 ± 4.37 * | 8.92 ± 2.96 * | 9.83 ± 3.76 * | 7.55 ± 8.06 | 16.10 ± 3.99 * |
| Day 60 | 8.91 ± 7.53 | 17.21 ± 3.49 * | 20.15 ± 3.58 * | 22.39 ± 2.77 * | 20.90 ± 4.00 * | |
| Phenylalanine | Day 0 | 12.11 ± 8.13 | 12.45 ± 4.40 * | 8.16 ± 2.17 * | 14.17 ± 3.47 * | 15.69 ± 3.86 * |
| Day 60 | −24.07 ± 11.72 | 20.06 ± 3.20 * | 15.18 ± 2.65 * | 20.22 ± 1.80 * | 12.62 ± 6.05 | |
| Glucose | Day 0 | −6.91 ± 1.38 * | −6.28 ± 1.75 * | −4.48 ± 0.27 * | −3.69 ± 1.90 | −2.19 ± 1.40 |
| Day 60 | −5.82 ± 2.57 | −5.70 ± 2.10 | −4.80 ± 3.54 | −5.36 ± 1.93 | −4.87 ± 2.19 | |
| Postprandial Time (Minutes) | ||||||
|---|---|---|---|---|---|---|
| 0 | 60 | 180 | 330 | 510 | ||
| Lysine | Day 0 | 1.08 ± 1.22 | 1.79 ± 1.36 | −2.42 ± 3.24 | 4.62 ± 1.74t | −1.32 ± 2.1 |
| Day 60 | −1.09 ± 3.82 | 0.96 ± 1.38 | −3.12 ± 1.73 | −2.34 ± 1.16 | −6.81 ± 2.09 * | |
| Threonine | Day 0 | 10.53 ± 1.88 * | 16.22 ± 1.85 * | 8.56 ± 3.52t | 20.03 ± 4.32 * | 8.93 ± 5.06 |
| Day 60 | 5.97 ± 3.11 | 16.57 ± 4.02 * | 11.55 ± 0.94 * | 12.5 ± 2.32 * | 4.40 ± 2.79 | |
| Citrate | Day 0 | 4.12 ± 2.64 | 0.83 ± 3.73 | −9.27 ± 10.05 | 42.08 ± 27.92 | 2.13 ± 10.86 |
| Day 60 | 5.51 ± 2.66 | 9.65 ± 4.54t | 2.18 ± 3.58 | 10.26 ± 6.4 | −3.52 ± 6.86 | |
| Isoleucine | Day 0 | 18.41 ± 6.24 * | 0.16 ± 7.15 | 2.65 ± 7.3 | 18.88 ± 6.04 * | 2.84 ± 8.25 |
| Day 60 | −4.48 ± 4.20 | 2.95 ± 2.58 | 1.49 ± 10.46 | −10.25 ± 3.84t | −9.55 ± 7.47 | |
| Valine | Day 0 | −8.30 ± 3.03t | 14.95 ± 8.59 | 1.58 ± 5.99 | −23.38 ± 11.61 | −1.88 ± 11.28 |
| Day 60 | 6.20 ± 6.84 | 0.03 ± 1.20 | −1.47 ± 5.02 | 5.81 ± 3.93 | 6.52 ± 8.09 | |
| Leucine | Day 0 | −2.73 ± 1.57 | 10.95 ± 3.66 * | 3.18 ± 1.67 | −5.73 ± 3.12 | −1.81 ± 4.28 |
| Day 60 | 2.61 ± 4.83 | 3.28 ± 1.14 * | −1.61 ± 1.99 | 1.79 ± 2.08 | 1.21 ± 3.27 | |
| BCAA | Day 0 | −1.44 ± 1.22 | 9.34 ± 2.61 * | 2.35 ± 0.90t | −4.60 ± 3.16 | −1.44 ± 3.41 |
| Day 60 | 2.03 ± 4.97 | 2.48 ± 1.11t | −2.16 ± 1.59 | 0.44 ± 1.61 | −0.13 ± 2.29 | |
| Tryptophan | Day 0 | 8.37 ± 5.91 | 12.87 ± 10.36 | 18.66 ± 7.73 | 26.83 ± 5.00t | 16.55 ± 15.69 * |
| Day 60 | 12.35 ± 1.85 | 26.8 ± 6.19 | 30.38 ± 12.89 | 12.02 ± 9.50 | 32.67 ± 11.17 | |
| Betaine | Day 0 | 1.31 ± 1.74 | 19.29 ± 2.68 * | 16.22 ± 2.58 * | 9.60 ± 4.30 | 4.66 ± 2.15 |
| Day 60 | 2.05 ± 2.62 | 23.74 ± 2.22 * | 14.25 ± 1.06 * | 10.15 ± 6.25 | 4.53 ± 4.29 | |
| Phosphocholine | Day 0 | 9.27 ± 1.87 * | 16.01 ± 3.32 * | 24.78 ± 7.04 * | 39.95 ± 9.35 * | 21.66 ± 7.24 * |
| Day 60 | 8.00 ± 3.16t | 34.4 ± 8.98 * | 22.05 ± 6.45 * | 17.73 ± 4.78 * | 10.11 ± 6.58 | |
| Alanine | Day 0 | −6.74 ± 1.81 * | −13.2 ± 0.93 * | −15.79 ± 2.1 * | −10.59 ± 2.01 * | −12.95 ± 1.59 * |
| Day 60 | −7.71 ± 4.07 | −12.89 ± 0.65 * | −16.04 ± 2.00 * | −15.73 ± 2.17 * | −17.95 ± 2.63 * | |
| Asparagine | Day 0 | 6.64 ± 4.36 | 6.73 ± 3.8 | −10.33 ± 6.11 | 7.79 ± 6.85 | −3.01 ± 5.40 |
| Day 60 | 5.73 ± 4.10 | −1.89 ± 1.79 | −8.88 ± 2.23 * | 0.47 ± 1.90 | −6.5 ± 3.73 | |
| Formic | Day 0 | −31.91 ± 3.80 * | −26.38 ± 1.81 * | −25.4 ± 5.39 * | −29.63 ± 0.98 * | −21.06 ± 2.61 * |
| Day 60 | −17.67 ± 10.34 | −33.63 ± 3.61 * | −22.35 ± 7.93 * | −23.58 ± 8.58 * | −26.17 ± 5.52 * | |
| Lipids | Day 0 | 1.38 ± 1.95 | 21.6 ± 2.97 * | 16.28 ± 3.19 * | 9.45 ± 2.80 * | 9.5 ± 2.38 * |
| Day 60 | 7.44 ± 3.32t | 21.71 ± 2.92 * | 17.36 ± 2.53 * | 16.06 ± 1.47 * | 12.12 ± 2.5 * | |
| Methionine | Day 0 | 7.15 ± 2.66 * | 8.06 ± 1.50 * | −3.58 ± 3.87 | 16.68 ± 8.97 * | 8.38 ± 6.08 |
| Day 60 | 15.06 ± 3.60 * | 5.84 ± 2.85 | −0.45 ± 1.28 | 10.42 ± 2.62 * | 3.99 ± 3.63 | |
| Glutamate | Day 0 | 8.49 ± 0.54 * | 9.56 ± 1.54 * | 1.15 ± 3.04 | 8.15 ± 2.93 * | 4.37 ± 3.74 |
| Day 60 | 9.99 ± 4.23t | 9.32 ± 2.97 * | 0.65 ± 2.56 | 4.25 ± 0.89 * | 1.09 ± 1.98 | |
| Lactate | Day 0 | 15.06 ± 8.44 | 7.48 ± 9.83 | −27.27 ± 10.61t | −11.56 ± 10.07 | −30.78 ± 14.73 |
| Day 60 | −1.53 ± 8.96 | −16.06 ± 12.96 | −4.54 ± 14.18 | 10.54 ± 29.75 | −24.19 ± 5.68 * | |
| Pyruvate | Day 0 | −2.38 ± 2.9 | 4.9 ± 6.06 | −1.57 ± 3.73 | 0.69 ± 2.47 | −5.27 ± 4.76 |
| Day 60 | −0.85 ± 6.94 | 7.34 ± 8.25 | 7.85 ± 6.43 | −1.06 ± 6.09 | −0.43 ± 7.74 | |
| Tyrosine | Day 0 | 2.51 ± 3.64 | 1.46 ± 3.66 | −4.91 ± 2.19 | 8.61 ± 8.00 | −6.76 ± 3.88 |
| Day 60 | 8.65 ± 6.07 | 1.48 ± 4.57 | −0.58 ± 6.02 | 0.49 ± 5.72 | 1.68 ± 6.97 | |
| Histidine | Day 0 | 0.89 ± 2.05 | 3.79 ± 2.88 | −3.51 ± 2.46 | 3.37 ± 2.88 | −6.62 ± 2.98t |
| Day 60 | 0.00 ± 1.64 | 4.55 ± 2.35 | −5.08 ± 2.25t | −2.41 ± 1.45 | −5.50 ± 3.45 | |
| Ethanolamine | Day 0 | −4.79 ± 2.86 | 5.05 ± 2.36t | −8.8 ± 3.18 * | −5.01 ± 1.07 * | −5.82 ± 2.72t |
| Day 60 | −1.14 ± 4.21 | −1.82 ± 3.69 | −8.8 ± 2.82 * | 1.47 ± 4.16 | −4.98 ± 3.69 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tremblay-Franco, M.; Poupin, N.; Amiel, A.; Canlet, C.; Rémond, D.; Debrauwer, L.; Dardevet, D.; Jourdan, F.; Savary-Auzeloux, I.; Polakof, S. Postprandial NMR-Based Metabolic Exchanges Reflect Impaired Phenotypic Flexibility across Splanchnic Organs in the Obese Yucatan Mini-Pig. Nutrients 2020, 12, 2442. https://doi.org/10.3390/nu12082442
Tremblay-Franco M, Poupin N, Amiel A, Canlet C, Rémond D, Debrauwer L, Dardevet D, Jourdan F, Savary-Auzeloux I, Polakof S. Postprandial NMR-Based Metabolic Exchanges Reflect Impaired Phenotypic Flexibility across Splanchnic Organs in the Obese Yucatan Mini-Pig. Nutrients. 2020; 12(8):2442. https://doi.org/10.3390/nu12082442
Chicago/Turabian StyleTremblay-Franco, Marie, Nathalie Poupin, Aurélien Amiel, Cécile Canlet, Didier Rémond, Laurent Debrauwer, Dominique Dardevet, Fabien Jourdan, Isabelle Savary-Auzeloux, and Sergio Polakof. 2020. "Postprandial NMR-Based Metabolic Exchanges Reflect Impaired Phenotypic Flexibility across Splanchnic Organs in the Obese Yucatan Mini-Pig" Nutrients 12, no. 8: 2442. https://doi.org/10.3390/nu12082442
APA StyleTremblay-Franco, M., Poupin, N., Amiel, A., Canlet, C., Rémond, D., Debrauwer, L., Dardevet, D., Jourdan, F., Savary-Auzeloux, I., & Polakof, S. (2020). Postprandial NMR-Based Metabolic Exchanges Reflect Impaired Phenotypic Flexibility across Splanchnic Organs in the Obese Yucatan Mini-Pig. Nutrients, 12(8), 2442. https://doi.org/10.3390/nu12082442






