Desaturase Activity and the Risk of Type 2 Diabetes and Coronary Artery Disease: A Mendelian Randomization Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.1.1. Individual-Level Data from EPIC-Potsdam
2.1.2. Summary-Level Data from DIAGRAM and CARDIoGRAM
2.2. DNA-Extraction, Genotyping and Quality Control
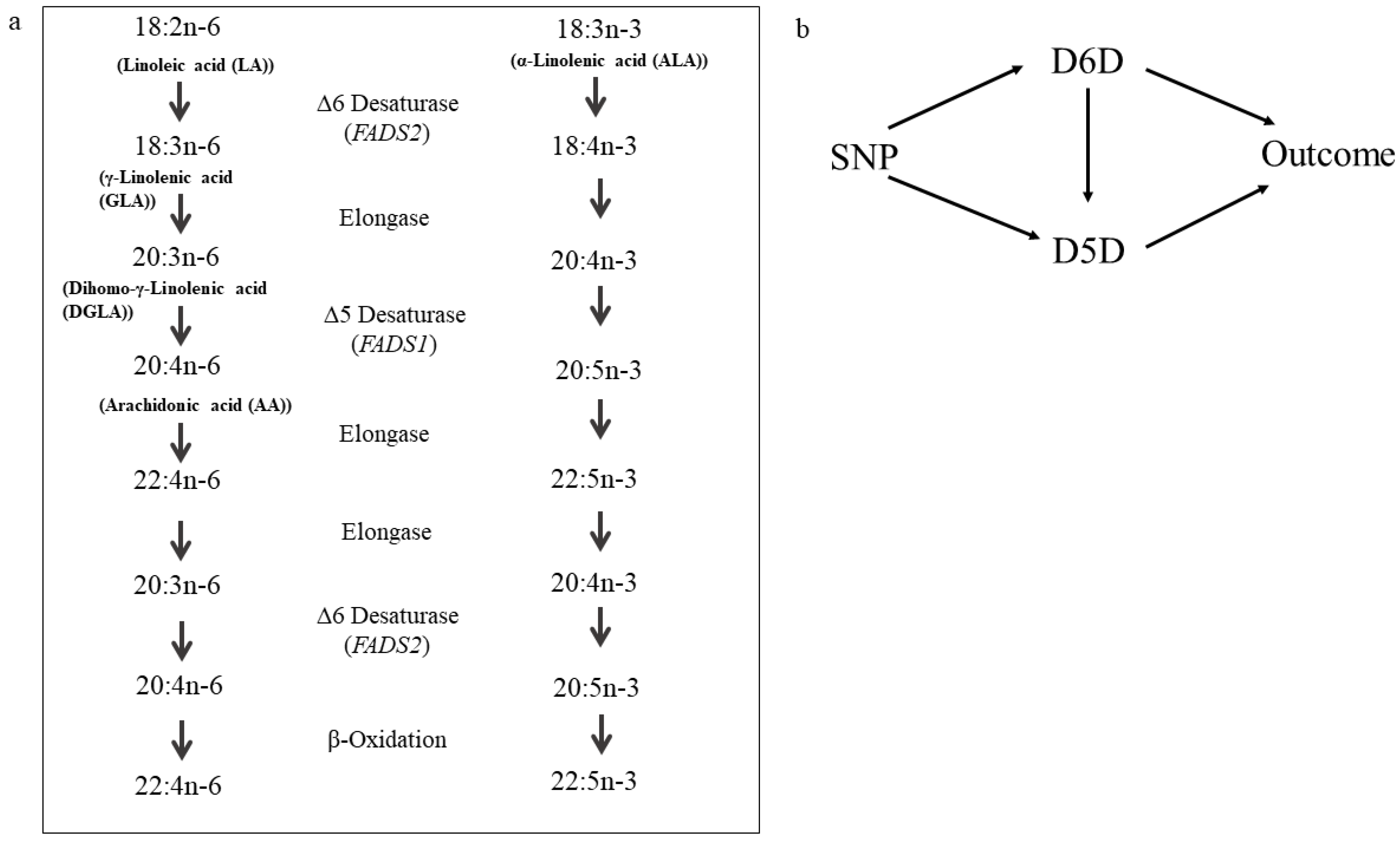
2.3. Determination of Desaturase Activities
2.4. Statistical Analysis
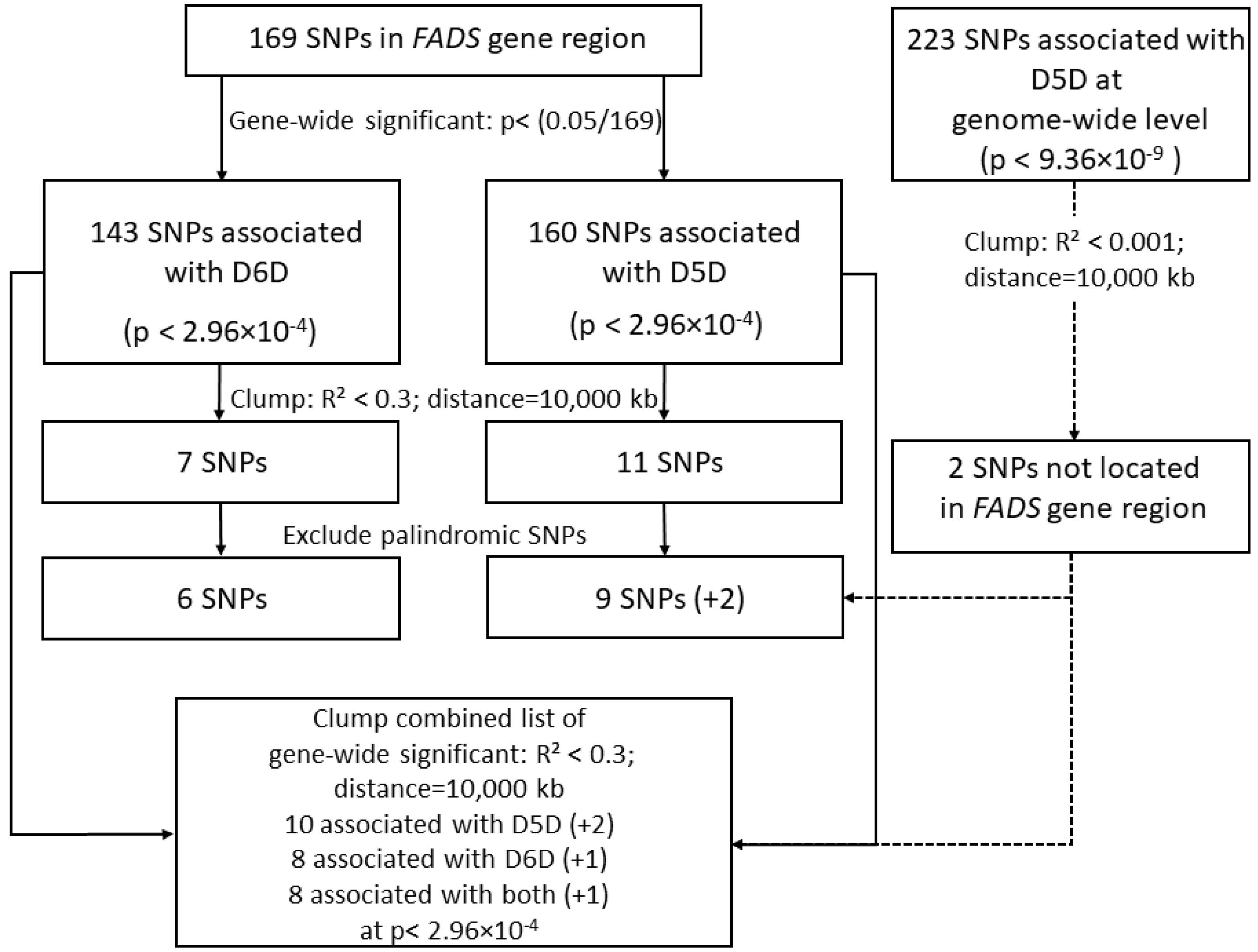
2.4.1. Selecting Genetic Instruments in Genome-Wide Association Study
2.4.2. Mendelian Randomization
2.4.3. Investigation of LD within the FADS-Gene Cluster
3. Results
3.1. Selection of Genetic Instruments
3.2. Mendelian Randomization
3.2.1. Univariable Mendelian Randomization
Causal Estimates for Desaturase Activities on Risk of Type 2 Diabetes
Causal Estimates for Desaturase Activities on Risk of Coronary Artery Disease
Sensitivity Analyses
3.2.2. Multivariable Mendelian Randomization
Estimates for Causal Direct Effects of Desaturase Activities on Risk of Type 2 Diabetes and Coronary Artery Disease
Sensitivity Analyses
3.2.3. Investigation of Confounding by Linkage Disequilibrium
4. Discussion
4.1. D6D and Risk of Type 2 Diabetes and Coronary Artery Disease
4.2. D5D and Risk of Type 2 Diabetes and Coronary Artery Disease
4.3. Biological Mechanisms
4.4. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Approval and Consent to Participate
Availability of Data and Materials
Code Availability
References
- Kröger, J.; Schulze, M.B. Recent insights into the relation of Delta5 desaturase and Delta6 desaturase activity to the development of type 2 diabetes. Curr. Opin. Lipidol. 2012, 23, 4–10. [Google Scholar] [CrossRef]
- Forouhi, N.G.; Imamura, F.; Sharp, S.J.; Koulman, A.; Schulze, M.B.; Zheng, J.; Ye, Z.; Sluijs, I.; Guevara, M.; Huerta, J.M.; et al. Association of plasma phospholipid n-3 and n-6 polyunsaturated fatty acids with type 2 diabetes: The EPIC-interAct case-cohort study. PLoS Med. 2016, 13, e1002094. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.Y.; Marklund, M.; Imamura, F.; Tintle, N.; Ardisson Korat, A.V.; de Goede, J.; Zhou, X.; Yang, W.S.; de Oliveira Otto, M.C.; Kroger, J.; et al. Omega-6 fatty acid biomarkers and incident type 2 diabetes: Pooled analysis of individual-level data for 39 740 adults from 20 prospective cohort studies. Lancet Diabetes Endocrinol. 2017, 5, 965–974. [Google Scholar] [CrossRef]
- Warensjo, E.; Sundstrom, J.; Vessby, B.; Cederholm, T.; Riserus, U. Markers of dietary fat quality and fatty acid desaturation as predictors of total and cardiovascular mortality: A population-based prospective study. Am. J. Clin. Nutr. 2008, 88, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Vaarhorst, A.; Merry, A.H.; Dolle, M.E.; Hovenier, R.; Imholz, S.; Schouten, L.J.; Heijmans, B.T.; Muller, M.; Slagboom, P.E.; et al. Markers of endogenous desaturase activity and risk of coronary heart disease in the CAREMA cohort study. PLoS ONE 2012, 7, e41681. [Google Scholar] [CrossRef]
- Davey Smith, G.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, A.; Stohr, H.; White, K.; Weber, B.H. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics 2000, 66, 175–183. [Google Scholar] [CrossRef]
- De Toro-Martin, J.; Guenard, F.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.C. A common variant in ARHGEF10 alters delta-6 desaturase activity and influence susceptibility to hypertriglyceridemia. J. Clin. Lipidol. 2018, 12, 311–320. [Google Scholar] [CrossRef]
- Marklund, M.; Morris, A.P.; Mahajan, A.; Ingelsson, E.; Lindgren, C.M.; Lind, L.; Riserus, U. Genome-wide association studies of estimated fatty acid desaturase activity in serum and adipose tissue in elderly individuals: Associations with insulin sensitivity. Nutrients 2018, 10, 1791. [Google Scholar] [CrossRef]
- O’Neill, C.M.; Minihane, A.M. The impact of fatty acid desaturase genotype on fatty acid status and cardiovascular health in adults. Proc. Nutr. Soc. 2017, 76, 64–75. [Google Scholar] [CrossRef]
- Brayner, B.; Kaur, G.; Keske, M.A.; Livingstone, K.M. FADS polymorphism, omega-3 fatty acids and diabetes risk: A systematic review. Nutrients 2018, 10, 758. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Larsson, S.C. Association of genetic variants related to plasma fatty acids with type 2 diabetes mellitus and glycaemic traits: A Mendelian randomisation study. Diabetologia 2020, 63, 116–123. [Google Scholar] [CrossRef]
- Yuan, S.; Back, M.; Bruzelius, M.; Mason, A.M.; Burgess, S.; Larsson, S. FADS polymorphism, omega-3 fatty acids and diabetes risk: A systematic review. Nutrients 2019, 11, 3001. [Google Scholar] [CrossRef] [PubMed]
- Kröger, J.; Zietemann, V.; Enzenbach, C.; Weikert, C.; Jansen, E.H.; Doring, F.; Joost, H.G.; Boeing, H.; Schulze, M.B. Erythrocyte membrane phospholipid fatty acids, desaturase activity, and dietary fatty acids in relation to risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Am. J. Clin. Nutr. 2011, 93, 127–142. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015, 181, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Boeing, H.; Wahrendorf, J.; Becker, N. EPIC-Germany-A source for studies into diet and risk of chronic diseases. European Investigation into Cancer and Nutrition. Ann. Nutr. Metab. 1999, 43, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Fritsche, A.; Weikert, C.; Boeing, H.; Joost, H.G.; Haring, H.U.; Schulze, M.B. Plasma fetuin-A levels and the risk of type 2 diabetes. Diabetes 2008, 57, 2762–2767. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef]
- Langenberg, C.; Sharp, S.J.; Franks, P.W.; Scott, R.A.; Deloukas, P.; Forouhi, N.G.; Froguel, P.; Groop, L.C.; Hansen, T.; Palla, L.; et al. Gene-lifestyle interaction and type 2 diabetes: The EPIC interact case-cohort study. PLoS Med. 2014, 11, e1001647. [Google Scholar] [CrossRef]
- Goldstein, J.I.; Crenshaw, A.; Carey, J.; Grant, G.B.; Maguire, J.; Fromer, M.; O’Dushlaine, C.; Moran, J.L.; Chambert, K.; Stevens, C.; et al. zCall: A rare variant caller for array-based genotyping: Genetics and population analysis. Bioinformatics 2012, 28, 2543–2545. [Google Scholar] [CrossRef] [PubMed]
- Grove, M.L.; Yu, B.; Cochran, B.J.; Haritunians, T.; Bis, J.C.; Taylor, K.D.; Hansen, M.; Borecki, I.B.; Cupples, L.A.; Fornage, M.; et al. Best practices and joint calling of the HumanExome BeadChip: The CHARGE Consortium. PLoS ONE 2013, 8, e68095. [Google Scholar] [CrossRef]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; He, J.; Zhao, S.; Wu, H.; Zhong, X.; Sheng, Q.; Samuels, D.C.; Shyr, Y.; Long, J. Illumina human exome genotyping array clustering and quality control. Nat. Protoc. 2014, 9, 2643–2662. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar]
- Loh, P.R.; Palamara, P.F.; Price, A.L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016, 48, 811–816. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Rayner, N.W.; Robertson, N.; Mahajan, A.; McCarthy, M.I. A Suite of Programs for Pre- and Postimputation Data Checking. Available online: www.well.ox.ac.uk/~wrayner/tools (accessed on 15 April 2019).
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Yavorska, O.O.; Burgess, S. MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017, 46, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Spiller, W.; Del Greco, M.F.; Sheehan, N.; Thompson, J.; Minelli, C.; Davey Smith, G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int. J. Epidemiol. 2018, 47, 1264–1278. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, E.; Spiller, W.; Bowden, J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable mendelian randomisation. BioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.04.02.021980v1 (accessed on 3 April 2020). [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2015, 43, D662–D669. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Rees, J.M.B.; Wood, A.M.; Burgess, S. Extending the MR-Egger method for multivariable Mendelian randomization to correct for both measured and unmeasured pleiotropy. Stat. Med. 2017, 36, 4705–4718. [Google Scholar] [CrossRef]
- Morrison, J.; Knoblauch, N.; Marcus, J.H.; Stephens, M.; He, X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 2020, 52, 740–747. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guasch-Ferre, M.; Li, Y.; Hu, F.B. Dietary intake and biomarkers of linoleic acid and mortality: Systematic review and meta-analysis of prospective cohort studies. Am. J. Clin. Nutr. 2020, 112, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Marklund, M.; Wu, J.H.Y.; Imamura, F.; Del Gobbo, L.C.; Fretts, A.; de Goede, J.; Shi, P.; Tintle, N.; Wennberg, M.; Aslibekyan, S.; et al. Biomarkers of dietary omega-6 fatty acids and incident cardiovascular disease and mortality: An individual-level pooled analysis of 30 cohort studies. Circulation 2019, 139, 2422–2436. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Das, U.N. A defect in Delta6 and Delta5 desaturases may be a factor in the initiation and progression of insulin resistance, the metabolic syndrome and ischemic heart disease in South Asians. Lipids Health Dis. 2010, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Kröger, J.; Jacobs, S.; Jansen, E.H.; Fritsche, A.; Boeing, H.; Schulze, M.B. Erythrocyte membrane fatty acid fluidity and risk of type 2 diabetes in the EPIC-Potsdam study. Diabetologia 2015, 58, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ding, E.L.; Toledo, E.T.; Campos, H.; Baylin, A.; Hu, F.B.; Sun, Q. A novel fatty acid lipophilic index and risk of CHD in US men: The health professionals follow-up study. Br. J. Nutr. 2013, 110, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.L.; West, K.L. Mechanisms by which dietary fatty acids modulate plasma lipids. J. Nutr. 2005, 135, 2075–2078. [Google Scholar] [CrossRef]
- Gleim, S.; Stitham, J.; Tang, W.H.; Martin, K.A.; Hwa, J. An eicosanoid-centric view of atherothrombotic risk factors. Cell. Mol. Life Sci. 2012, 69, 3361–3380. [Google Scholar] [CrossRef]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhang, H.; Yu, K. Power calculation for the general two-sample Mendelian randomization analysis. Genet. Epidemiol. 2020, 44, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.P.; Nakamura, M.; Clarke, S.D. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 1999, 274, 37335–37339. [Google Scholar] [CrossRef]
- Cho, H.P.; Nakamura, M.T.; Clarke, S.D. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J. Biol. Chem. 1999, 274, 471–477. [Google Scholar] [CrossRef]
- Vaittinen, M.; Mannisto, V.; Kakela, P.; Agren, J.; Tiainen, M.; Schwab, U.; Pihlajamaki, J. Interorgan cross talk between fatty acid metabolism, tissue inflammation, and FADS2 genotype in humans with obesity. Obesity 2017, 25, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.S.; Thompson, J.R. Beyond Mendelian randomization: How to interpret evidence of shared genetic predictors. J. Clin. Epidemiol. 2016, 69, 208–216. [Google Scholar] [CrossRef]
| EPIC-Potsdam | |
|---|---|
| N | 1853 |
| Sex (% men) | 37.1 |
| Age in years; median (interquartile range) | 49.0 (15.5) |
| Waist circumference in cm; mean (SD) | 85.3 (12.6) |
| Δ6-desaturase activity (18:3n-6/18:2n-6); median (interquartile range) | 0.005 (0.003) |
| Δ5-desaturase activity (20:4n-6/20:3n-6); mean (SD) | 8.80 (1.91) |
| Lipid medication (%) | 3.72 |
| T2DM | CAD | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | N (SNPs) * | OR (95% CI) | p-Value | N (SNPs) † | OR (95% CI) | p-Value | ||
| D6D | instruments from FADS | IVW | 6 | 1.08 (1.06–1.09) | <0.001 | 6 | 1.06 (1.02–1.11) | 0.008 |
| MVIVW | 10 | 1.03 (0.94–1.12) | 0.528 | 10 | 1.00 (0.93–1.07) | 0.971 | ||
| MVIVW ‡ | 10 | 1.03 (0.99–1.16) | 10 | 1.01 (0.91–1.12) | ||||
| instruments from FADS and genome-wide hits | MVIVW | 12 | 1.03 (0.95–1.10) | 0.514 | 12 | 1.00 (0.95–1.06) | 0.907 | |
| MVIVW ‡ | 12 | 1.01 (0.94–1.10) | 12 | 1.00 (0.96–1.15) | ||||
| D5D | instruments from FADS | IVW | 9 | 1.03 (1.01–1.04) | <0.001 | 9 | 1.03 (1.01–1.05) | 0.017 |
| MVIVW | 10 | 1.00 (0.96–1.04) | 0.824 | 10 | 1.04 (1.01–1.08) | 0.021 | ||
| MVIVW ‡ | 10 | 1.02 (0.98–1.04) | 10 | 1.04 (1.01–1.15) | ||||
| instruments from FADS and genome-wide hits | IVW | 11 | 1.04 (1.02–1.06) | <0.001 | 11 | 1.03 (1.01–1.06) | 0.017 | |
| MVIVW | 12 | 1.00 (0.96–1.03) | 0.845 | 12 | 1.03 (0.99–1.06) | 0.108 | ||
| MVIVW ‡ | 12 | 1.02 (0.99–1.05) | 12 | 1.04 (0.99–1.07) | ||||
| T2DM | CAD | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Method | N (SNPs) | OR (95% CI) | p-Value | Intercept (SE), p-Value | N (SNPs) | OR (95% CI) | p-Value | Intercept (SE), p-Value | |
| D6D | MR-Egger | 3 † | 0.87 (0.55–1.37) | 0.538 | 0.050 (0.045), 0.267 | 3 † | 1.26 (0.21–7.70) | 0.804 | −0.022 (0.176), 0.902 |
| IVW * | 3 † | 1.12 (1.06–1.18) | <0.001 | 3 † | 1.12 (1.04–1.21) | 0.002 | |||
| MVIVW * | 3 † | 0.74 (0.34–1.62) | 0.453 | 3 † | 1.12 (0.34–3.69) | 0.850 | |||
| D5D | IVW * | 2 ‡ | 1.04 (0.99–1.08) | 0.087 | 2 ‡ | 1.04 (0.98–1.11) | 0.236 | ||
| MVIVW * | 2 ‡ | 1.01 (0.95–1.07) | 0.790 | 2 ‡ | 1.00 (0.93–1.08) | 0.996 | |||
| MVIVW * | 3 † | 1.29 (0.75–2.21) | 0.362 | 3 † | 1.00 (0.48–2.09) | 1.000 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäger, S.; Cuadrat, R.; Hoffmann, P.; Wittenbecher, C.; Schulze, M.B. Desaturase Activity and the Risk of Type 2 Diabetes and Coronary Artery Disease: A Mendelian Randomization Study. Nutrients 2020, 12, 2261. https://doi.org/10.3390/nu12082261
Jäger S, Cuadrat R, Hoffmann P, Wittenbecher C, Schulze MB. Desaturase Activity and the Risk of Type 2 Diabetes and Coronary Artery Disease: A Mendelian Randomization Study. Nutrients. 2020; 12(8):2261. https://doi.org/10.3390/nu12082261
Chicago/Turabian StyleJäger, Susanne, Rafael Cuadrat, Per Hoffmann, Clemens Wittenbecher, and Matthias B. Schulze. 2020. "Desaturase Activity and the Risk of Type 2 Diabetes and Coronary Artery Disease: A Mendelian Randomization Study" Nutrients 12, no. 8: 2261. https://doi.org/10.3390/nu12082261
APA StyleJäger, S., Cuadrat, R., Hoffmann, P., Wittenbecher, C., & Schulze, M. B. (2020). Desaturase Activity and the Risk of Type 2 Diabetes and Coronary Artery Disease: A Mendelian Randomization Study. Nutrients, 12(8), 2261. https://doi.org/10.3390/nu12082261





