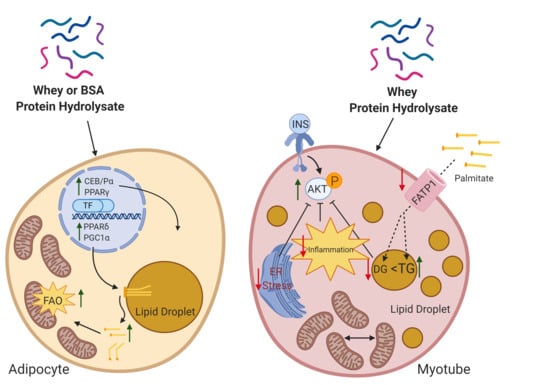
Whey Peptides Stimulate Differentiation and Lipid Metabolism in Adipocytes and Ameliorate Lipotoxicity-Induced Insulin Resistance in Muscle Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation of Protein and Peptide Mixtures
2.3. Cell Culture
2.4. Gene Expression Analysis
2.5. Immunoblotting Analysis
2.6. Lipid Analysis
2.7. Lipolysis Assay
2.8. Mitochondrial Analysis
2.9. Citrate Synthase Activity Assay
2.10. Statistical Analysis
3. Results
3.1. Profile of Peptides in Whey Protein Hydrolysate
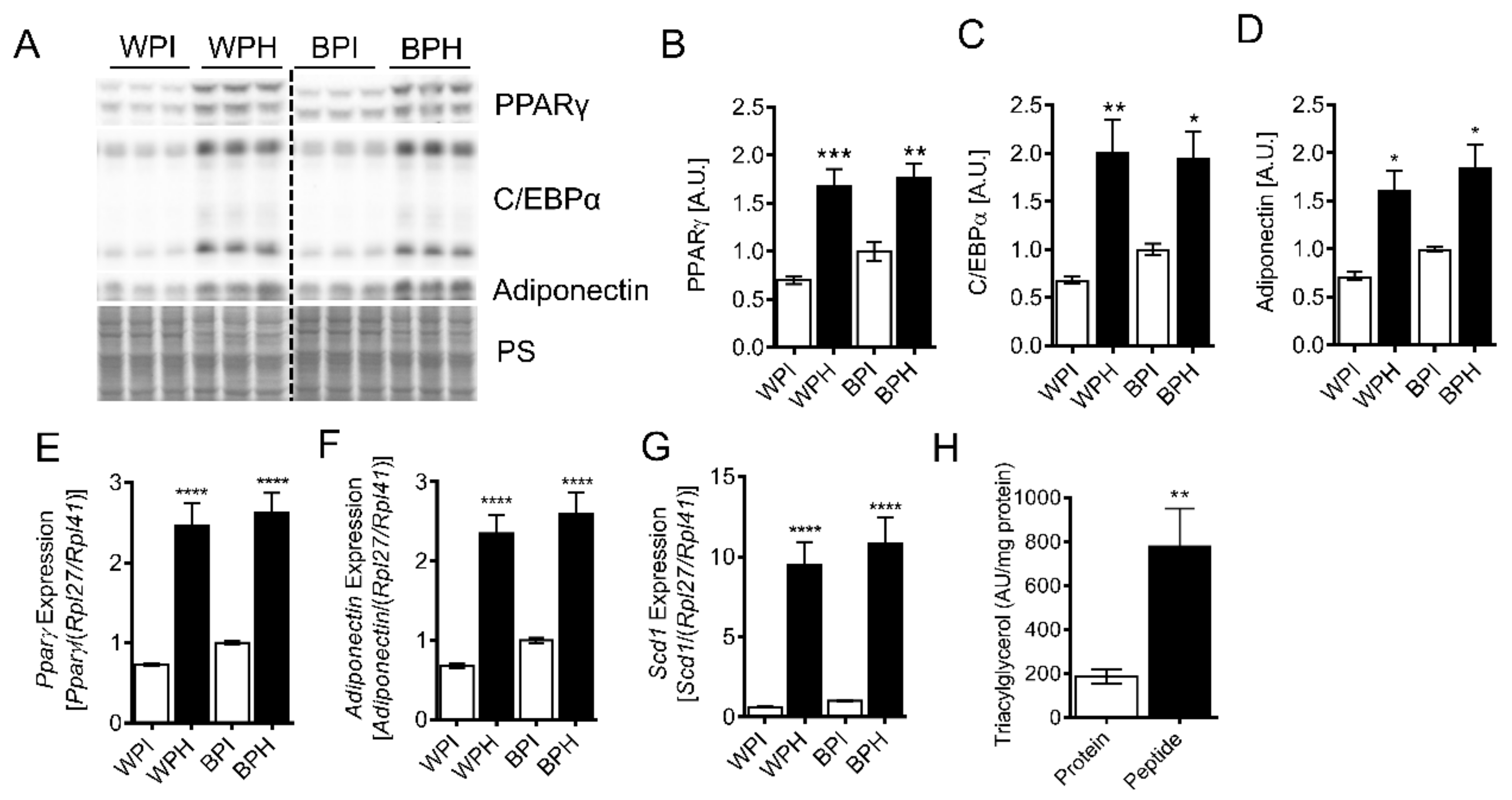
3.2. Whey Peptides Promote Differentiation of 3T3-L1 Adipocytes
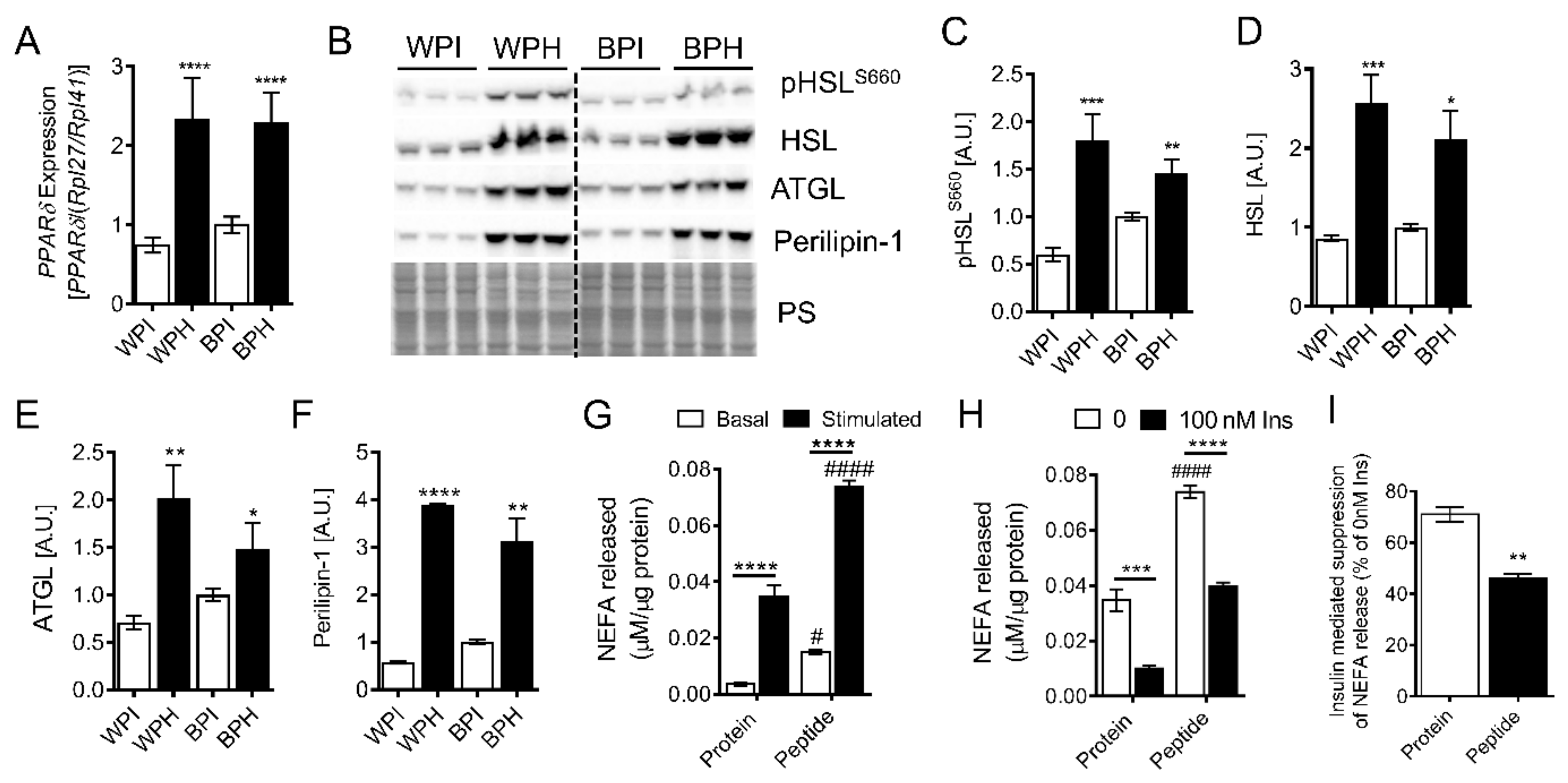
3.3. Whey Peptides Increase Lipolysis in 3T3-L1 Adipocytes
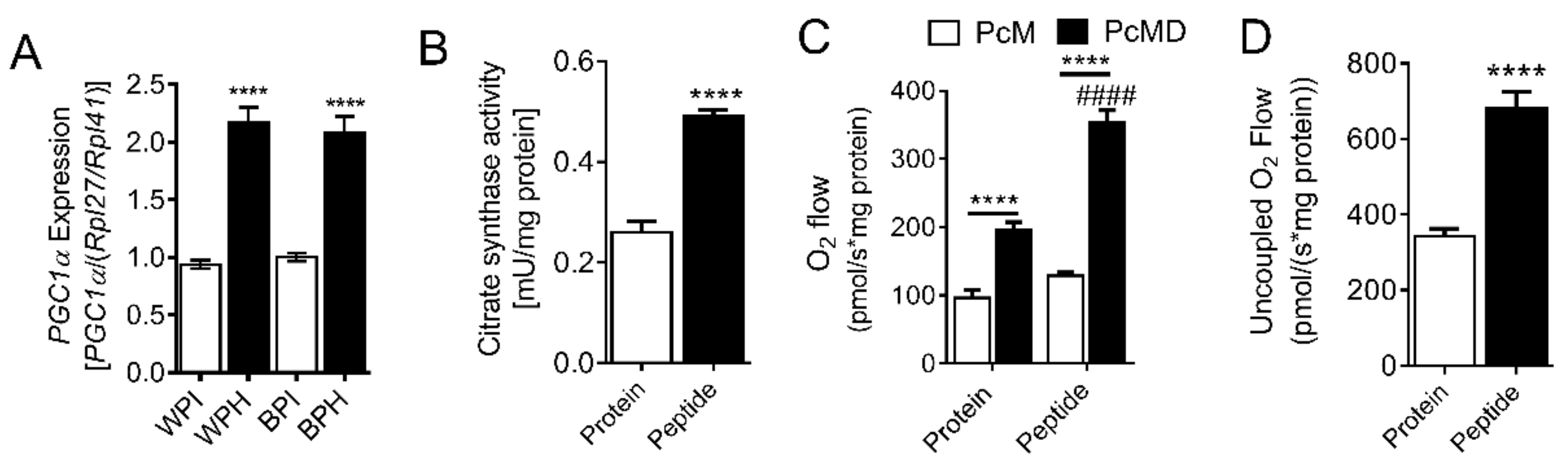
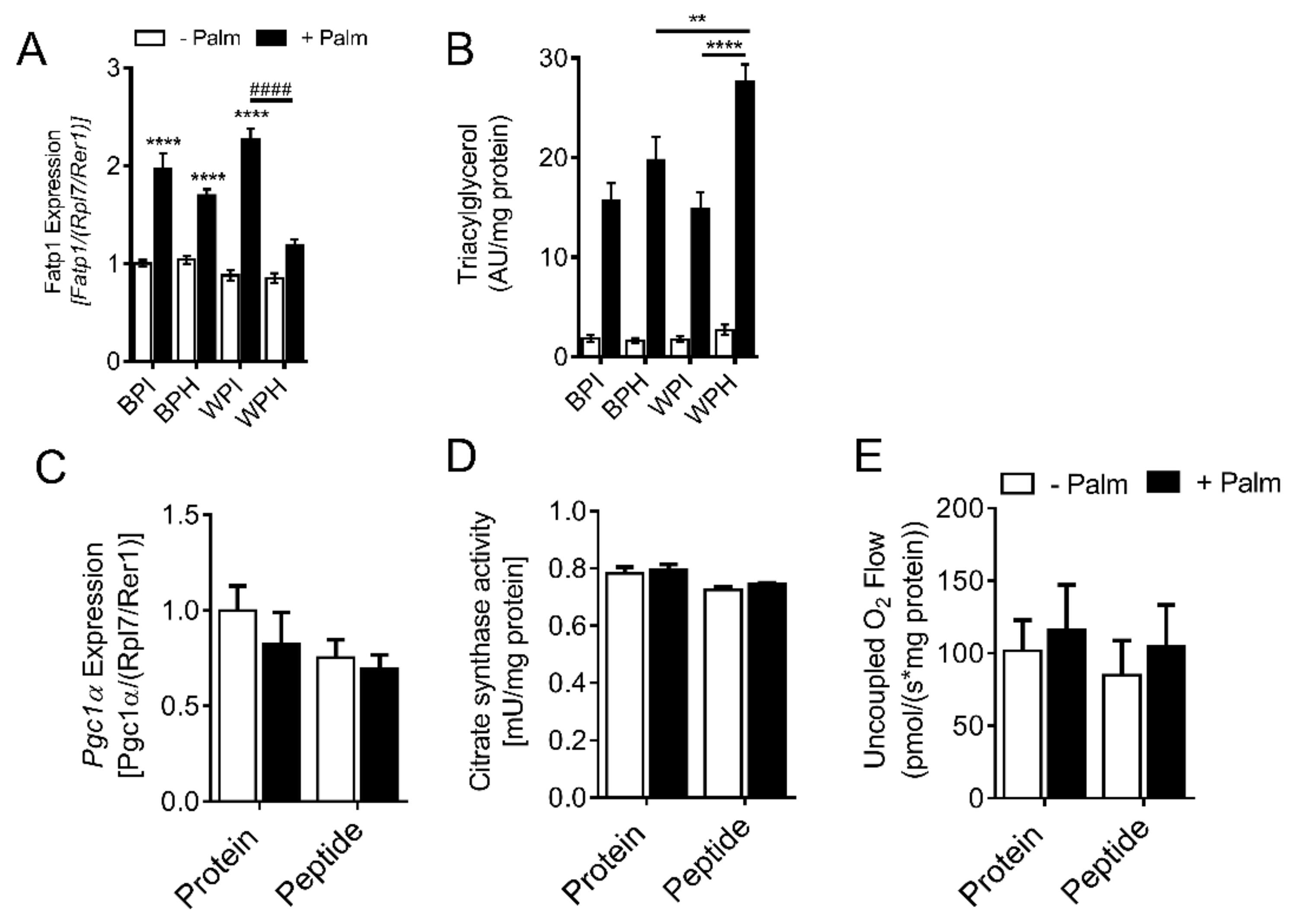
3.4. Whey Peptides Enhance Mitochondrial Fatty Acid Oxidation in 3T3-L1 Adipocytes
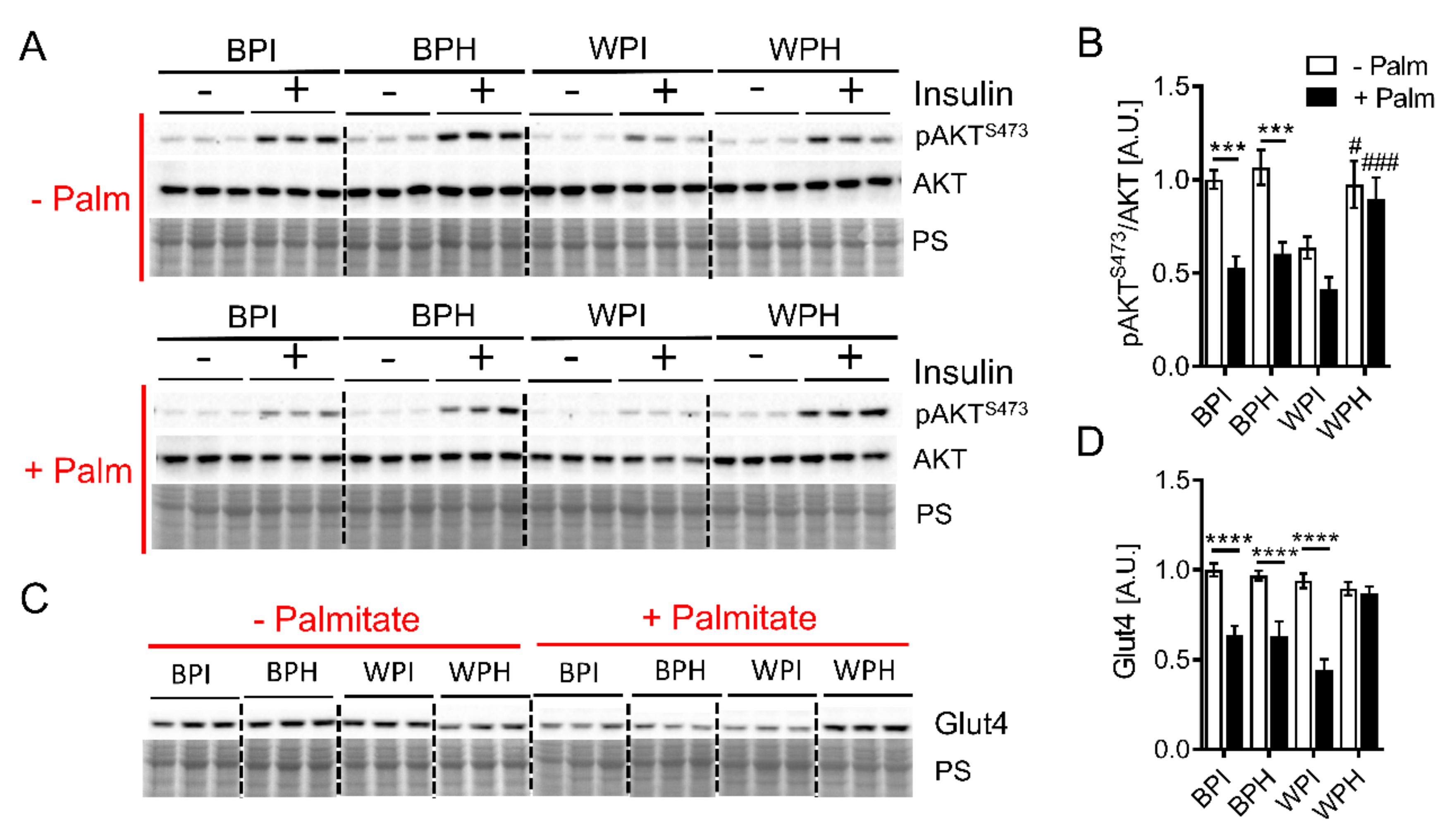
3.5. Whey Peptides Ameliorate Palmitate-Induced Insulin Resistance in C2C12 Myotubes
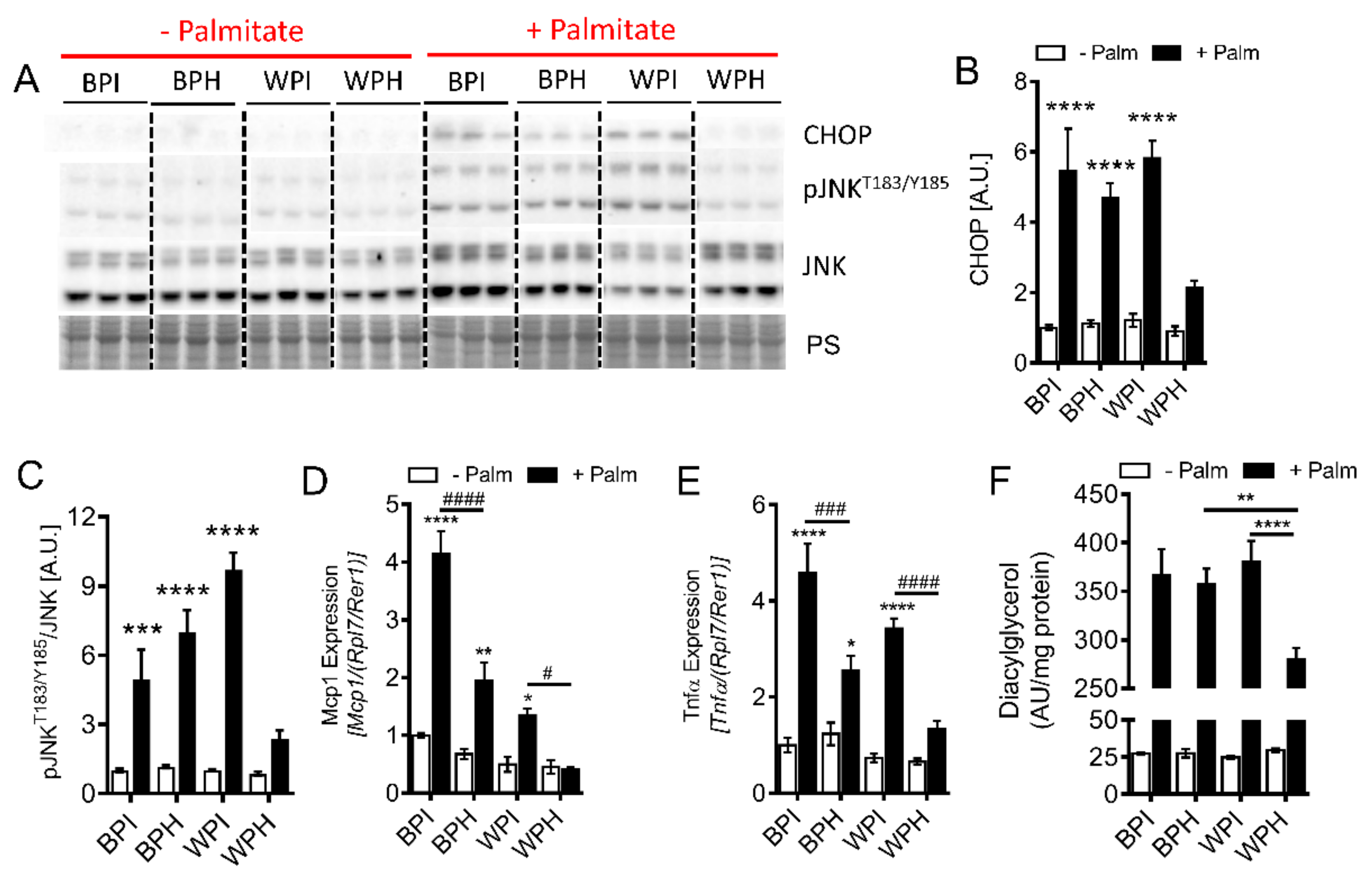
3.6. Whey Peptides Protect from Palmitate-Induced Inflammation and Endoplasmic Reticulum (ER) Stress, Which is Associated with Decreased DG Accumulation in C2C12 Myotubes
3.7. Whey Peptides Increase TG Accumulation in C2C12 Myotubes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeFronzo, R.A.; Tripathy, D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009, 32 (Suppl. 2), S157–S163. [Google Scholar] [CrossRef] [PubMed]
- Baron, A.D.; Brechtel, G.; Wallace, P.; Edelman, S.V. Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am. J. Physiol. 1988, 255 (6 Pt 1), E769–E774. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, J.M.; Stern, J.H.; Scherer, P.E. The cell biology of fat expansion. J. Cell. Biol. 2015, 208, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.E.; Marcucci, M.J.; Cline, G.W.; Bell, K.; Barucci, N.; Lee, D.; Goodyear, L.J.; Kraegen, E.W.; White, M.F.; Shulman, G.I. Free fatty acid-induced insulin resistance is associated with activation of protein kinase C theta and alterations in the insulin signaling cascade. Diabetes 1999, 48, 1270–1274. [Google Scholar] [CrossRef]
- Kim, J.Y.; van de Wall, E.; Laplante, M.; Azzara, A.; Trujillo, M.E.; Hofmann, S.M.; Schraw, T.; Durand, J.L.; Li, H.; Li, G.; et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 2007, 117, 2621–2637. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Scheja, L.; Heeren, J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019, 15, 507–524. [Google Scholar] [CrossRef]
- Freitas Lima, L.C.; Braga, V.A.; do Socorro de Franca Silva, M.; Cruz, J.C.; Sousa Santos, S.H.; de Oliveira Monteiro, M.M.; Balarini, C.M. Adipokines, diabetes and atherosclerosis: An inflammatory association. Front. Physiol. 2015, 6, 304. [Google Scholar] [CrossRef]
- Makki, K.; Froguel, P.; Wolowczuk, I. Adipose tissue in obesity-related inflammation and insulin resistance: Cells, cytokines, and chemokines. ISRN Inflamm. 2013, 2013, 139239. [Google Scholar] [CrossRef]
- Wu, H.; Ballantyne, C.M. Skeletal muscle inflammation and insulin resistance in obesity. J. Clin. Invest. 2017, 127, 43–54. [Google Scholar] [CrossRef]
- Anderson, E.J.; Lustig, M.E.; Boyle, K.E.; Woodlief, T.L.; Kane, D.A.; Lin, C.T.; Price, J.W., 3rd; Kang, L.; Rabinovitch, P.S.; Szeto, H.H.; et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Invest. 2009, 119, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Ayala, J.E.; Lee-Young, R.S.; Zhang, Z.; James, F.D.; Neufer, P.D.; Pozzi, A.; Zutter, M.M.; Wasserman, D.H. Diet-induced muscle insulin resistance is associated with extracellular matrix remodeling and interaction with integrin alpha2beta1 in mice. Diabetes 2011, 60, 416–426. [Google Scholar] [CrossRef]
- Koh, H.J.; Toyoda, T.; Didesch, M.M.; Lee, M.Y.; Sleeman, M.W.; Kulkarni, R.N.; Musi, N.; Hirshman, M.F.; Goodyear, L.J. Tribbles 3 mediates endoplasmic reticulum stress-induced insulin resistance in skeletal muscle. Nat. Commun. 2013, 4, 1871. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M., 2nd; Pratipanawatr, T.; Berria, R.; Wang, E.; DeFronzo, R.A.; Sullards, M.C.; Mandarino, L.J. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes 2004, 53, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Szendroedi, J.; Yoshimura, T.; Phielix, E.; Koliaki, C.; Marcucci, M.; Zhang, D.; Jelenik, T.; Muller, J.; Herder, C.; Nowotny, P.; et al. Role of diacylglycerol activation of PKCtheta in lipid-induced muscle insulin resistance in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 9597–9602. [Google Scholar] [CrossRef] [PubMed]
- Udenigwe, C.C.; Rouvinen-Watt, K. The Role of Food Peptides in Lipid Metabolism during Dyslipidemia and Associated Health Conditions. Int. J. Mol. Sci. 2015, 16, 9303–9313. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food. Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Wu, J. Milk-derived tripeptides IPP (Ile-Pro-Pro) and VPP (Val-Pro-Pro) promote adipocyte differentiation and inhibit inflammation in 3T3-F442A cells. PLoS ONE 2015, 10, e0117492. [Google Scholar] [CrossRef]
- Sawada, Y.; Sakamoto, Y.; Toh, M.; Ohara, N.; Hatanaka, Y.; Naka, A.; Kishimoto, Y.; Kondo, K.; Iida, K. Milk-derived peptide Val-Pro-Pro (VPP) inhibits obesity-induced adipose inflammation via an angiotensin-converting enzyme (ACE) dependent cascade. Mol. Nutr. Food. Res. 2015, 59, 2502–2510. [Google Scholar] [CrossRef]
- Jahandideh, F.; Chakrabarti, S.; Davidge, S.T.; Wu, J. Egg white hydrolysate shows insulin mimetic and sensitizing effects in 3T3-F442A preadipocytes. PLoS ONE 2017, 12, e0185653. [Google Scholar] [CrossRef]
- Li, S.; Liu, L.; He, G.; Wu, J. Molecular targets and mechanisms of bioactive peptides against metabolic syndromes. Food. Funct. 2018, 9, 42–52. [Google Scholar] [CrossRef] [PubMed]
- De Campos Zani, S.C.; Wu, J.; Chan, C.B. Egg and Soy-Derived Peptides and Hydrolysates: A Review of Their Physiological Actions against Diabetes and Obesity. Nutrients 2018, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Ewert, J.; Luz, A.; Volk, V.; Stressler, T.; Fischer, L. Enzymatic production of emulsifying whey protein hydrolysates without the need of heat inactivation. J. Sci. Food. Agric. 2019, 99, 3443–3450. [Google Scholar] [CrossRef]
- Ichinoseki-Sekine, N.; Kakigi, R.; Miura, S.; Naito, H. Whey peptide ingestion suppresses body fat accumulation in senescence-accelerated mouse prone 6 (SAMP6). Eur. J. Nutr. 2015, 54, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Song, J.; Du, M.; Mao, X. Bovine alpha-Lactalbumin Hydrolysates (alpha-LAH) Ameliorate Adipose Insulin Resistance and Inflammation in High-Fat Diet-Fed C57BL/6J Mice. Nutrients 2018, 10, 242. [Google Scholar] [CrossRef]
- Mohammed-Geba, K.; Arrutia, F.; Do-Huu, H.; Borrell, Y.J.; Galal-Khallaf, A.; Ardura, A.; Riera, F.A.; Garcia-Vazquez, E. VY6, a beta-lactoglobulin-derived peptide, altered metabolic lipid pathways in the zebra fish liver. Food. Funct. 2016, 7, 1968–1974. [Google Scholar] [CrossRef]
- Morifuji, M.; Sakai, K.; Sanbongi, C.; Sugiura, K. Dietary whey protein increases liver and skeletal muscle glycogen levels in exercise-trained rats. Br. J. Nutr. 2005, 93, 439–445. [Google Scholar] [CrossRef][Green Version]
- Kanda, A.; Morifuji, M.; Fukasawa, T.; Koga, J.; Kanegae, M.; Kawanaka, K.; Higuchi, M. Dietary whey protein hydrolysates increase skeletal muscle glycogen levels via activation of glycogen synthase in mice. J. Agric. Food. Chem. 2012, 60, 11403–11408. [Google Scholar] [CrossRef]
- Morifuji, M.; Koga, J.; Kawanaka, K.; Higuchi, M. Branched-chain amino acid-containing dipeptides, identified from whey protein hydrolysates, stimulate glucose uptake rate in L6 myotubes and isolated skeletal muscles. J. Nutr. Sci. Vitaminol. 2009, 55, 81–86. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- D’Souza, K.; Kane, D.A.; Touaibia, M.; Kershaw, E.E.; Pulinilkunnil, T.; Kienesberger, P.C. Autotaxin is Regulated by Glucose and Insulin in Adipocytes. Endocrinology 2017, 158, 791–803. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, K.; Nzirorera, C.; Cowie, A.M.; Varghese, G.P.; Trivedi, P.; Eichmann, T.O.; Biswas, D.; Touaibia, M.; Morris, A.J.; Aidinis, V.; et al. Autotaxin-Lysophosphatidic Acid Signaling Contributes to Obesity-Induced Insulin Resistance in Muscle and Impairs Mitochondrial Metabolism. J. Lipid. Res. 2018, 59, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.J.; Rios, L.; Trivedi, P.; D’Souza, K.; Cowie, A.; Nzirorera, C.; Webster, D.; Brunt, K.; Legare, J.F.; Hassan, A.; et al. Validation of optimal reference genes for quantitative real time PCR in muscle and adipose tissue for obesity and diabetes research. Sci. Rep. 2017, 7, 3612. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid. Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Breitkopf, S.B.; Ricoult, S.J.H.; Yuan, M.; Xu, Y.; Peake, D.A.; Manning, B.D.; Asara, J.M. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 2017, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Boudina, S.; Sena, S.; O’Neill, B.T.; Tathireddy, P.; Young, M.E.; Abel, E.D. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation 2005, 112, 2686–2695. [Google Scholar] [CrossRef]
- Agyei, D.; Tsopmo, A.; Udenigwe, C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018, 410, 3463–3472. [Google Scholar] [CrossRef]
- Larsen, S.; Nielsen, J.; Hansen, C.N.; Nielsen, L.B.; Wibrand, F.; Stride, N.; Schroder, H.D.; Boushel, R.; Helge, J.W.; Dela, F.; et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012, 590, 3349–3360. [Google Scholar] [CrossRef]
- Sadeghi, A.; Seyyed Ebrahimi, S.S.; Golestani, A.; Meshkani, R. Resveratrol Ameliorates Palmitate-Induced Inflammation in Skeletal Muscle Cells by Attenuating Oxidative Stress and JNK/NF-kappaB Pathway in a SIRT1-Independent Mechanism. J. Cell. Biochem. 2017, 118, 2654–2663. [Google Scholar] [CrossRef]
- Perry, B.D.; Rahnert, J.A.; Xie, Y.; Zheng, B.; Woodworth-Hobbs, M.E.; Price, S.R. Palmitate-induced ER stress and inhibition of protein synthesis in cultured myotubes does not require Toll-like receptor 4. PLoS ONE 2018, 13, e0191313. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Shulman, G.I. Mechanisms for insulin resistance: Common threads and missing links. Cell 2012, 148, 852–871. [Google Scholar] [CrossRef]
- Xu, S.P.; Mao, X.Y.; Ren, F.Z.; Che, H.L. Attenuating effect of casein glycomacropeptide on proliferation, differentiation, and lipid accumulation of in vitro Sprague-Dawley rat preadipocytes. J. Dairy. Sci. 2011, 94, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Kim, I.H.; Choi, J.W.; Lee, M.K.; Nam, T.J. The anti-obesity effects of a tuna peptide on 3T3-L1 adipocytes are mediated by the inhibition of the expression of lipogenic and adipogenic genes and by the activation of the Wnt/beta-catenin signaling pathway. Int. J. Mol. Med. 2015, 36, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X. PPARs: Diverse regulators in energy metabolism and metabolic diseases. Cell. Res. 2010, 20, 124–137. [Google Scholar] [CrossRef]
- Roberts, L.D.; Murray, A.J.; Menassa, D.; Ashmore, T.; Nicholls, A.W.; Griffin, J.L. The contrasting roles of PPARdelta and PPARgamma in regulating the metabolic switch between oxidation and storage of fats in white adipose tissue. Genome. Biol. 2011, 12, R75. [Google Scholar] [CrossRef]
- Matsusue, K.; Peters, J.M.; Gonzalez, F.J. PPARbeta/delta potentiates PPARgamma-stimulated adipocyte differentiation. FASEB J. 2004, 18, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.L. Rationally designed PPARdelta-specific agonists and their therapeutic potential for metabolic syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, 3284–3285. [Google Scholar] [CrossRef] [PubMed]
- Baer, D.J.; Stote, K.S.; Paul, D.R.; Harris, G.K.; Rumpler, W.V.; Clevidence, B.A. Whey protein but not soy protein supplementation alters body weight and composition in free-living overweight and obese adults. J. Nutr. 2011, 141, 1489–1494. [Google Scholar] [CrossRef]
- Lopes Gomes, D.; Moehlecke, M.; Lopes da Silva, F.B.; Dutra, E.S.; D’Agord Schaan, B.; Baiocchi de Carvalho, K.M. Whey Protein Supplementation Enhances Body Fat and Weight Loss in Women Long After Bariatric Surgery: A Randomized Controlled Trial. Obes. Surg. 2017, 27, 424–431. [Google Scholar] [CrossRef]
- Tranberg, B.; Hellgren, L.I.; Lykkesfeldt, J.; Sejrsen, K.; Jeamet, A.; Rune, I.; Ellekilde, M.; Nielsen, D.S.; Hansen, A.K. Whey protein reduces early life weight gain in mice fed a high-fat diet. PLoS ONE 2013, 8, e71439. [Google Scholar] [CrossRef]
- Lu, J.; Zeng, Y.; Hou, W.; Zhang, S.; Li, L.; Luo, X.; Xi, W.; Chen, Z.; Xiang, M. The soybean peptide aglycin regulates glucose homeostasis in type 2 diabetic mice via IR/IRS1 pathway. J. Nutr. Biochem. 2012, 23, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Soga, M.; Ohashi, A.; Taniguchi, M.; Matsui, T.; Tsuda, T. The di-peptide Trp-His activates AMP-activated protein kinase and enhances glucose uptake independently of insulin in L6 myotubes. FEBS Open. Bio. 2014, 4, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.J.; Kim, C.S.; Choi, M.S.; Park, T.; Sung, M.K.; Yun, J.W.; Yoo, H.; Mine, Y.; Yu, R. The Soy Peptide Phe-Leu-Val Reduces TNFalpha-Induced Inflammatory Response and Insulin Resistance in Adipocytes. J. Med. Food. 2016, 19, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef]
- Listenberger, L.L.; Han, X.; Lewis, S.E.; Cases, S.; Farese, R.V., Jr.; Ory, D.S.; Schaffer, J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 2003, 100, 3077–3082. [Google Scholar] [CrossRef]
- Sitnick, M.T.; Basantani, M.K.; Cai, L.; Schoiswohl, G.; Yazbeck, C.F.; Distefano, G.; Ritov, V.; Delany, J.P.; Schreiber, R.; Stolz, D.B.; et al. Skeletal muscle triacylglycerol hydrolysis does not influence metabolic complications of obesity. Diabetes 2013, 62, 3350–3361. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; He, J.; Watkins, S.; Kelley, D.E. Skeletal muscle lipid content and insulin resistance: Evidence for a paradox in endurance-trained athletes. J. Clin. Endocrinol. Metab. 2001, 86, 5755–5761. [Google Scholar] [CrossRef]
| Target | Primer Sequence (5’ to 3’) |
|---|---|
| Pparγ F | ACATAAAGTCCTTCCCGCTGA |
| Pparγ R | TCGAAACTGGCACCCTTGAAAA |
| Adipoq F | AGCCGCTTATGTGTATCGC |
| Adipoq R | GTCCCGGAATGTTGCAGTAGAAC |
| Scd1 F | TTACGACCGGAAGAAAGTT |
| Scd1 R | ATTAACACCCCGATAGCAATA |
| Pparδ F | TCATTGAGCCCAAGTTCGAGT |
| Pparδ R | CCGGTCTCCACACAAAATGAT |
| Pgc1α F | TTTGCCCAGATCTTCCTGAAC |
| Pgc1α R | TCGCTACACCACTTCAATCCA |
| Mcp1 F | TCGGAACCAAATGAGATCAGA |
| Mcp1 R | CAGATTTACGGGTCAACTTC |
| Tnfα F | CATCCATTCTCTACCCAGCCC |
| Tnfα R | CATGAGAGGCCCACAGTCCA |
| Fatp1 F | CCTCTGGGCACCATTCTATATTC |
| Fatp1 R | ACACTAGCCACATCCAAGTGA |
| Rpl27 F | ACGGTGGAGCCTTATGTGAC |
| Rpl27 R | TCCGTCAGAGGGACTGTCTT |
| Rpl41 F | GCCATGAGAGCGAAGTGG |
| Rpl41 R | CTCCTGCAGGCGTCGTAG |
| Rpl7 F | ACGGTGGAGCCTTATGTGAC |
| Rpl7 R | TCCGTCAGAGGGACTGTCTT |
| Rer1 F | GCCTTGGGAATTTACCACCT |
| Rer1 R | CTTCGAATGAAGGGACGAAA |
| ID | Whey Protein Groups Identified | Major Protein UniProtKB Accession Number | Number of Peptides Identified | % of Total Number of Peptides |
|---|---|---|---|---|
| 1 | Alpha-lactalbumin | P00711 | 69 | 26.2 |
| 2 | Alpha-S2-casein | P02663 | 1 | 0.4 |
| 3 | Beta-casein | P02666 | 21 | 8.0 |
| 4 | Kappa-casein | P02668 | 17 | 6.5 |
| 5 | Serum albumin (BSA) | P02769 | 16 | 6.1 |
| 6 | Osteopontin | P31096 | 1 | 0.4 |
| 7 | Serotransferrin | Q29443 | 2 | 0.8 |
| 8 | Ig-like domain-containing protein | F1MLW7 | 2 | 0.8 |
| 9 | Alpha-amylase | Q3MHH8 | 2 | 0.8 |
| 10 | Folate receptor alpha | P02702 | 2 | 0.8 |
| 11 | Beta-lactoglobulin | P02754 | 115 | 43.7 |
| 12 | Lactotransferrin | P24627 | 2 | 0.8 |
| 13 | NPC intracellular cholesterol transporter 2 | P79345 | 4 | 1.5 |
| 14 | Glycosylation-dependent cell adhesion molecule 1 | P80195 | 2 | 0.8 |
| 15 | Lactadherin | G3MYW7 | 1 | 0.4 |
| 16 | Sortilin related VPS10 domain containing receptor 1 | A0A3Q1LSH9 | 1 | 0.4 |
| 17 | Multiple coagulation factor deficiency 2 | Q3MHJ4 | 1 | 0.4 |
| 18 | Uncharacterized protein | A0A3Q1M3L6 | 4 | 1.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Souza, K.; Mercer, A.; Mawhinney, H.; Pulinilkunnil, T.; Udenigwe, C.C.; Kienesberger, P.C. Whey Peptides Stimulate Differentiation and Lipid Metabolism in Adipocytes and Ameliorate Lipotoxicity-Induced Insulin Resistance in Muscle Cells. Nutrients 2020, 12, 425. https://doi.org/10.3390/nu12020425
D’Souza K, Mercer A, Mawhinney H, Pulinilkunnil T, Udenigwe CC, Kienesberger PC. Whey Peptides Stimulate Differentiation and Lipid Metabolism in Adipocytes and Ameliorate Lipotoxicity-Induced Insulin Resistance in Muscle Cells. Nutrients. 2020; 12(2):425. https://doi.org/10.3390/nu12020425
Chicago/Turabian StyleD’Souza, Kenneth, Angella Mercer, Hannah Mawhinney, Thomas Pulinilkunnil, Chibuike C. Udenigwe, and Petra C. Kienesberger. 2020. "Whey Peptides Stimulate Differentiation and Lipid Metabolism in Adipocytes and Ameliorate Lipotoxicity-Induced Insulin Resistance in Muscle Cells" Nutrients 12, no. 2: 425. https://doi.org/10.3390/nu12020425
APA StyleD’Souza, K., Mercer, A., Mawhinney, H., Pulinilkunnil, T., Udenigwe, C. C., & Kienesberger, P. C. (2020). Whey Peptides Stimulate Differentiation and Lipid Metabolism in Adipocytes and Ameliorate Lipotoxicity-Induced Insulin Resistance in Muscle Cells. Nutrients, 12(2), 425. https://doi.org/10.3390/nu12020425