The Impact of Low-Level Iron Supplements on the Faecal Microbiota of Irritable Bowel Syndrome and Healthy Donors Using In Vitro Batch Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Ethics
2.3. Materials
2.4. Faecal Sample Preparation
2.5. Faecal Batch Culture Fermentation
2.6. Preparation of the Samples for Bacterial Metabolite Analysis, Ammonia Analysis and Flow Cytometry-Fluorescent In Situ Hybridisation (FISH)
2.7. Flow-FISH Analysis
2.8. Bacterial Quantification Using Flow-Fluorescent In Situ Hybridisation (FISH)
2.9. Gas Chromatography (GC) for Short Chain and Branched Chain Fatty Acid Analysis
2.9.1. Standard Solution Preparation
2.9.2. Ammonia Assay
2.9.3. Statistical Methods
3. Results
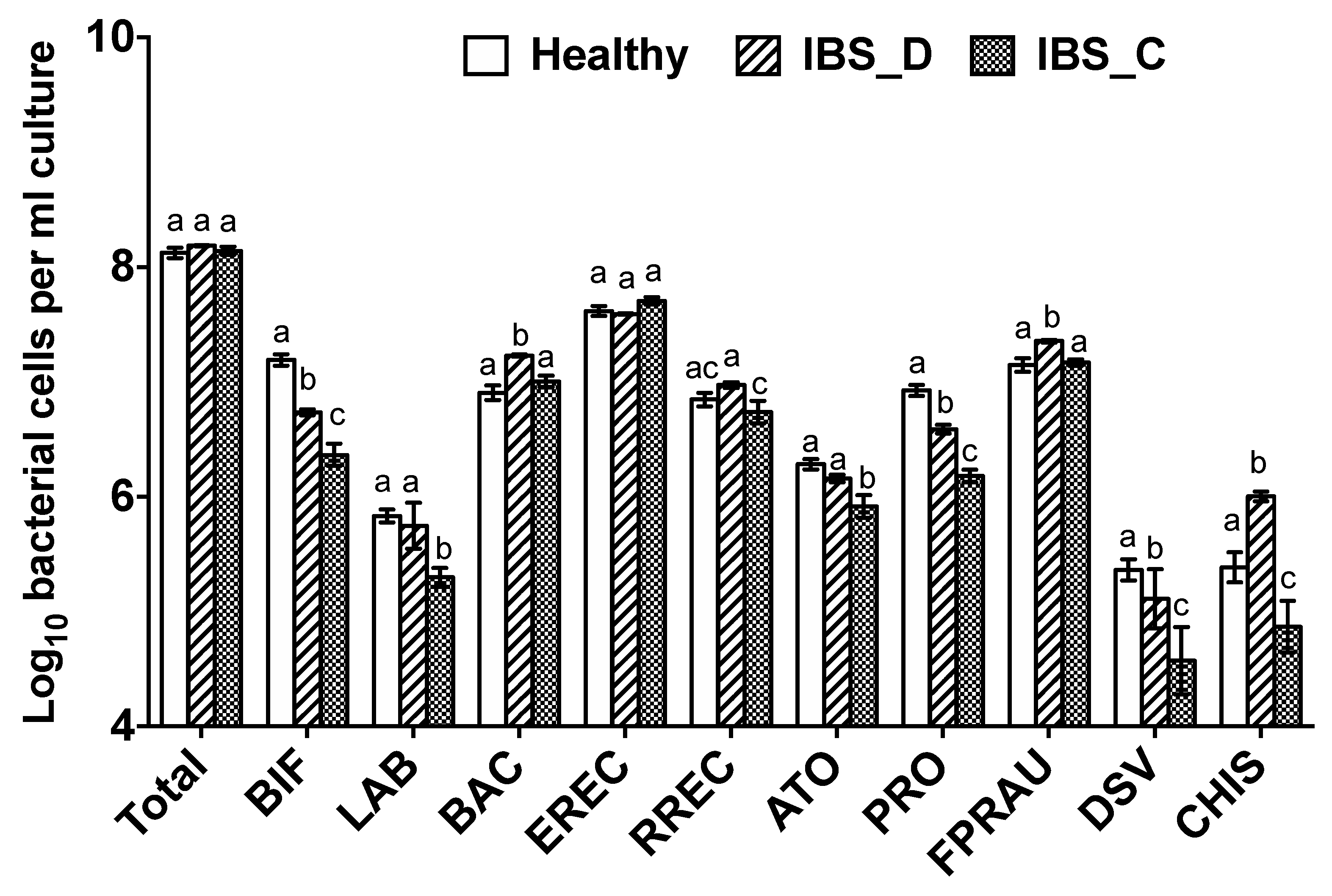
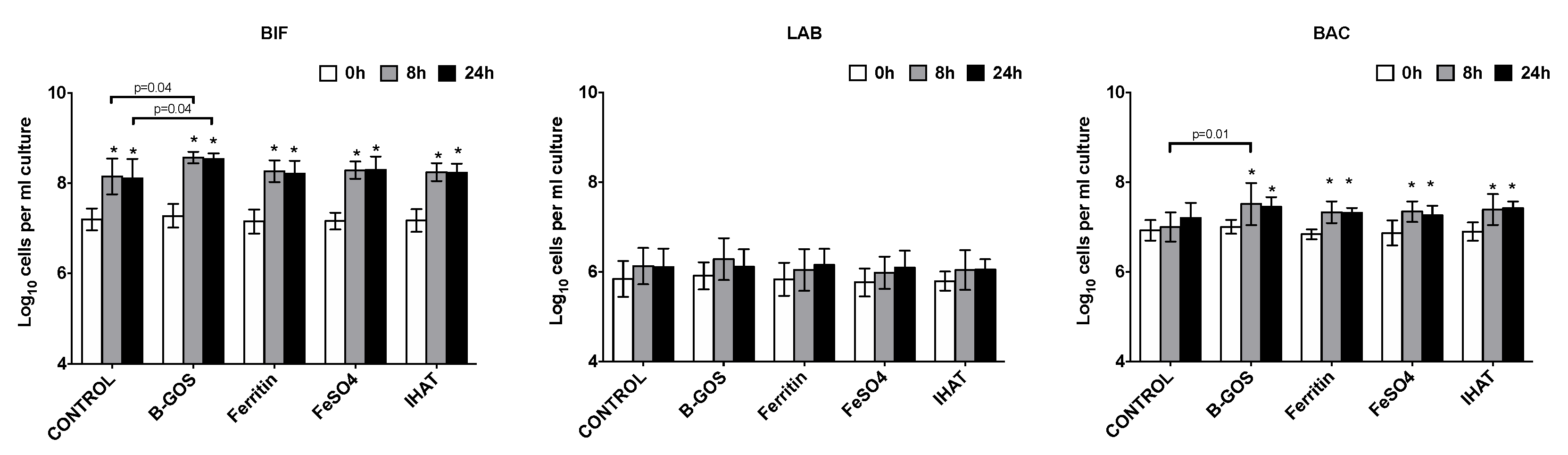
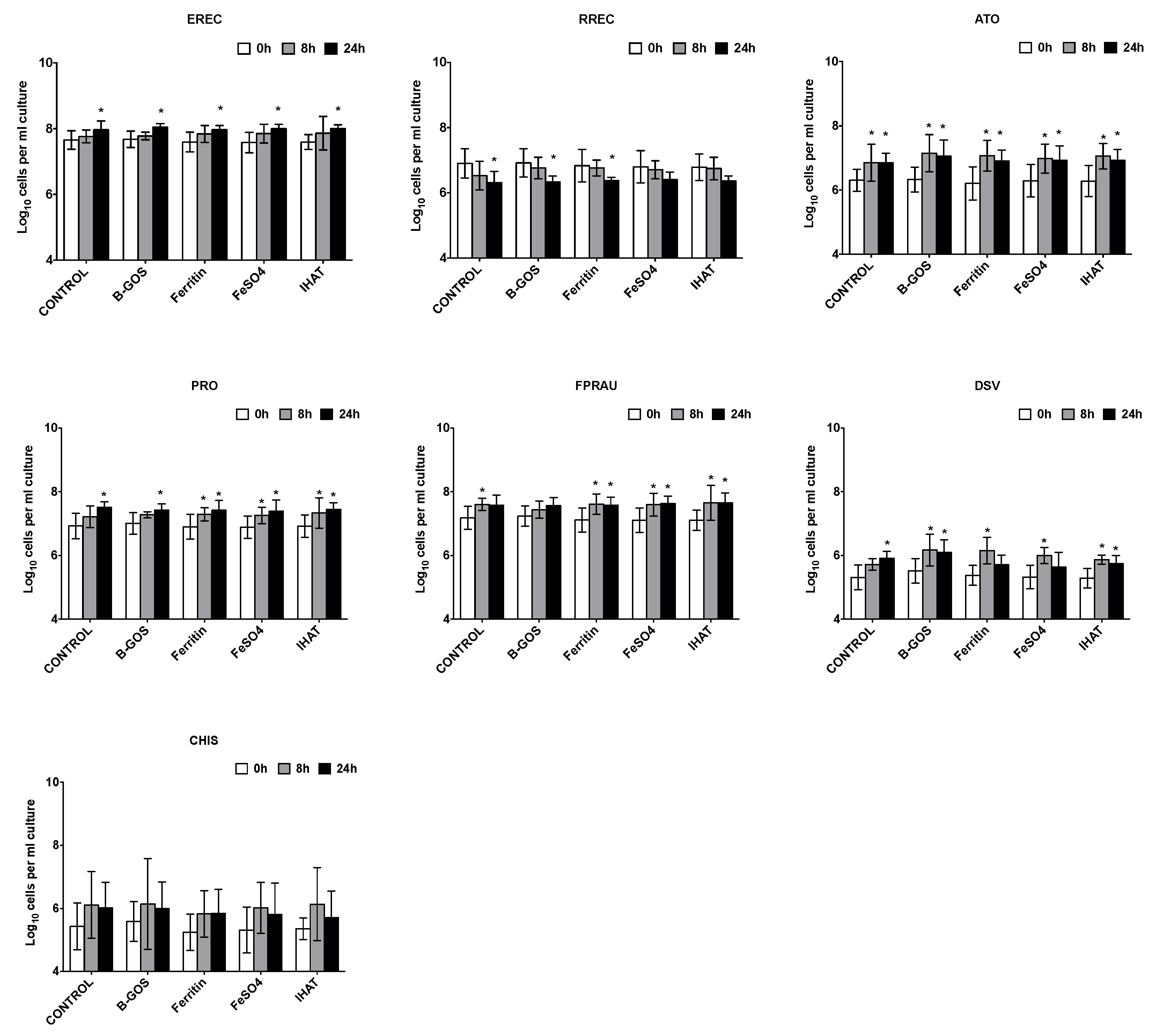
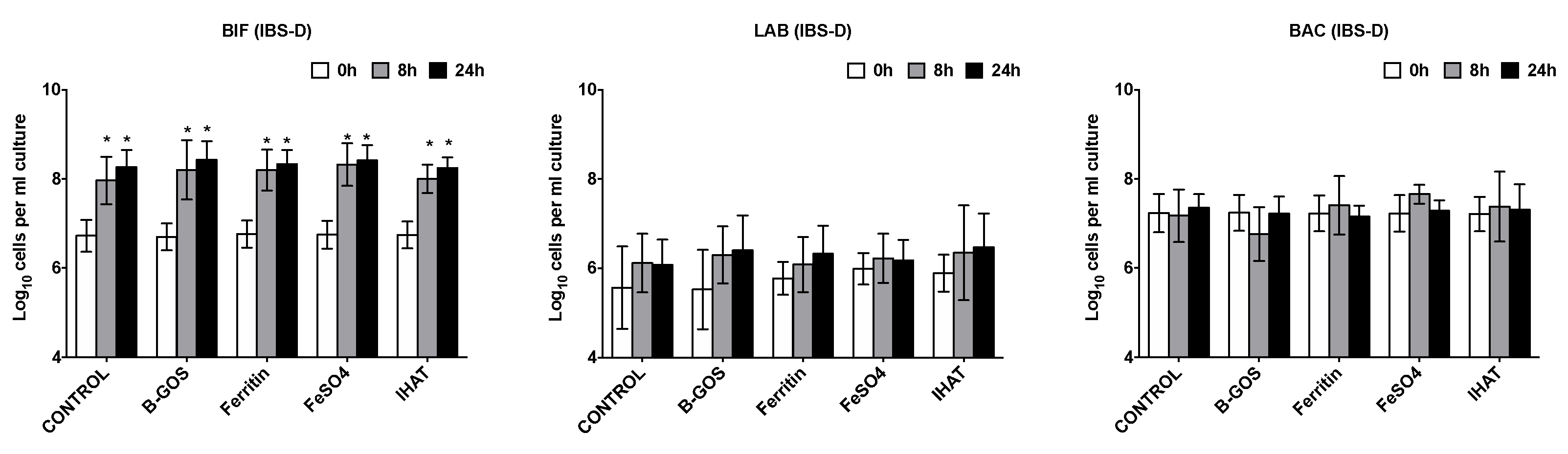
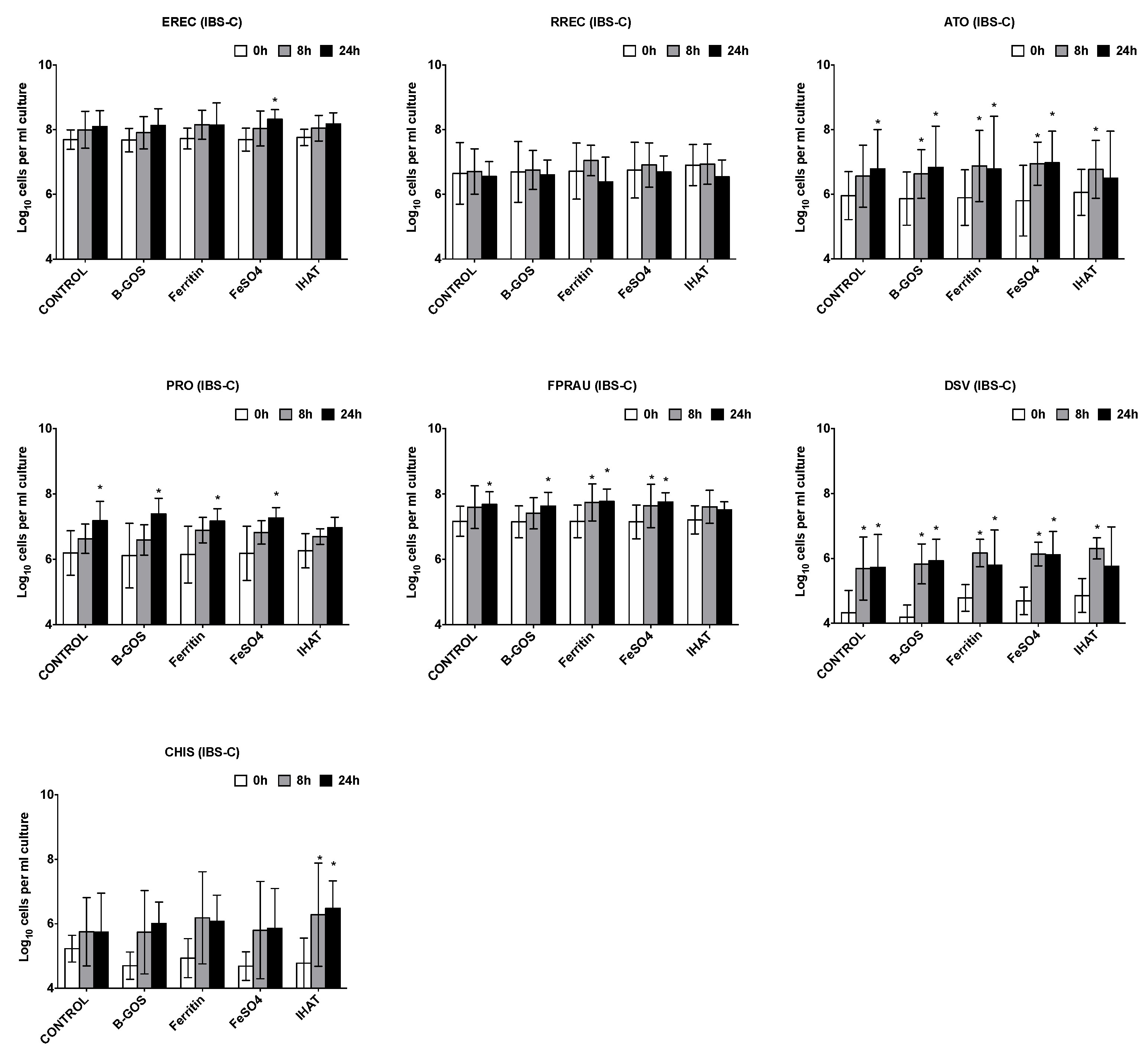
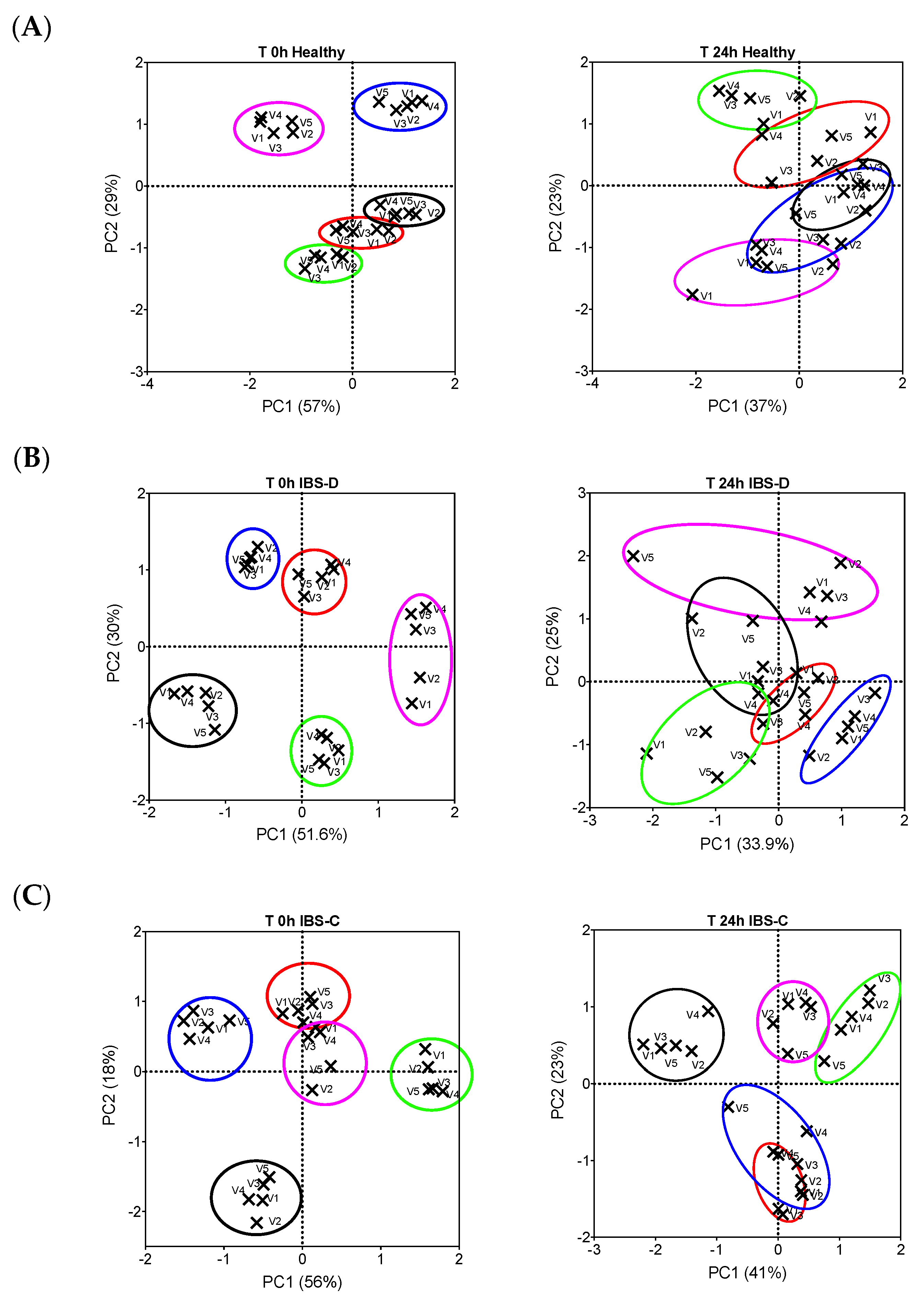
3.1. Bacterial Population Enumeration by Flow-FISH
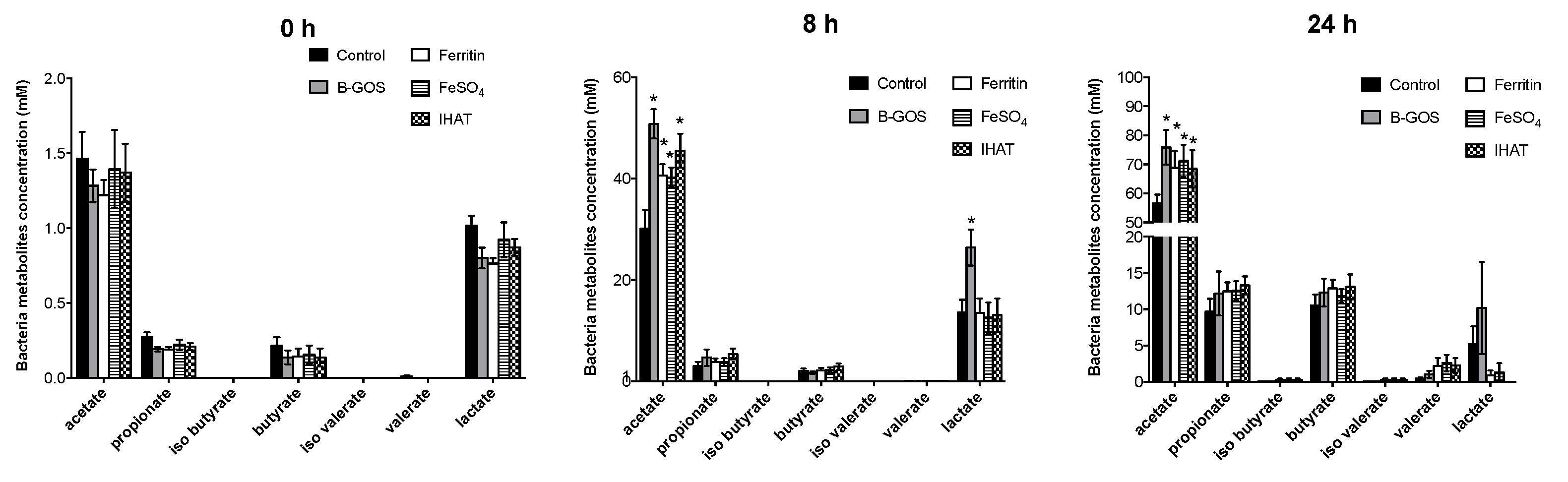
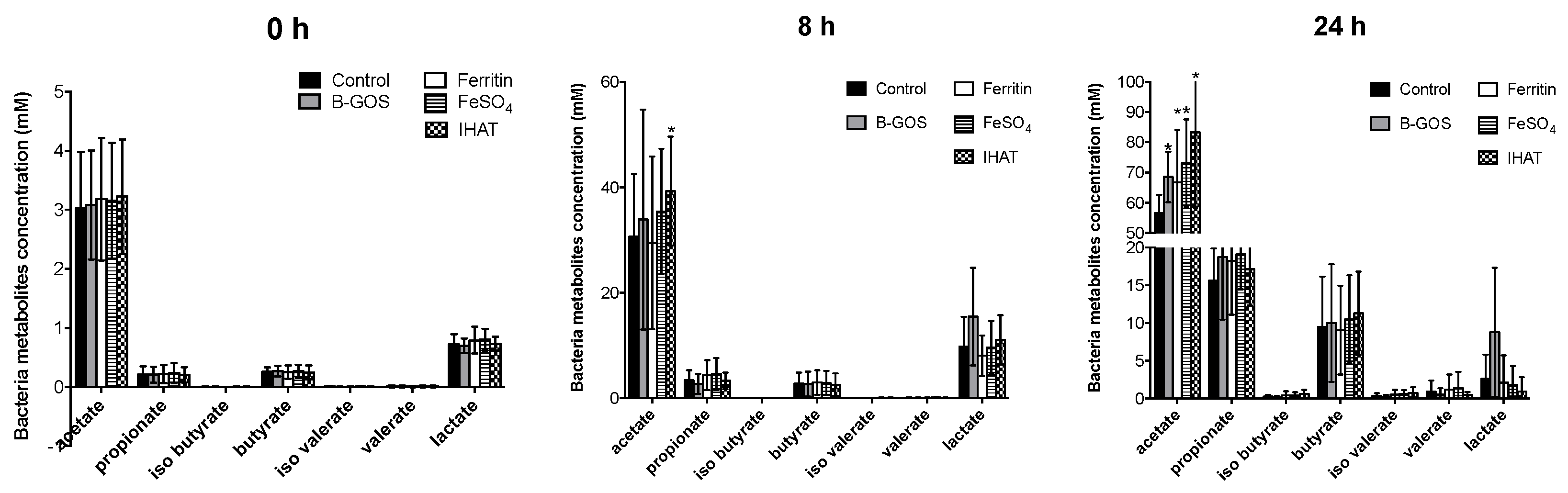
3.2. Bacterial Metabolites
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lopez, A.; Cacoub, P.; MacDougall, I.C.; Peyrin-Biroulet, L. Iron deficiency anaemia. Lancet 2016, 387, 907–916. [Google Scholar] [CrossRef]
- Prentice, A.M.; Mendoza, Y.A.; Pereira, D.; Cerami, C.; Wegmuller, R.; Constable, A.; Spieldenner, J. Dietary strategies for improving iron status: Balancing safety and efficacy. Nutr. Rev. 2017, 75, 49–60. [Google Scholar] [CrossRef]
- Strain, J.C.; Kevin, D. Introduction to Human Nutrition, 2nd ed.; Wiley-Blackwell: Chichester, UK, 2009; Chapter 9. [Google Scholar]
- Beard, J.L. Iron Biology in Immune Function, Muscle Metabolism and Neuronal Functioning. J. Nutr. 2001, 131, 568S–580S. [Google Scholar] [CrossRef]
- Latunde-Dada, G.O.; Pereira, D.I.A.; Tempest, B.; Ilyas, H.; Flynn, A.C.; Aslam, M.F.; Simpson, R.J.; Powell, J.J. A Nanoparticulate Ferritin-Core Mimetic Is Well Taken Up by HuTu 80 Duodenal Cells and Its Absorption in Mice Is Regulated by Body Iron. J. Nutr. 2014, 144, 1896–1902. [Google Scholar] [CrossRef] [Green Version]
- Girelli, D.; Ugolini, S.; Busti, F.; Marchi, G.; Castagna, A. Modern iron replacement therapy: Clinical and pathophysiological insights. Int. J. Hematol. 2018, 107, 16–30. [Google Scholar] [CrossRef] [Green Version]
- Goddard, A.F.; James, M.W.; McIntyre, A.S.; Scott, B.B.; Gastroenterology, O.B.O.T.B.S.O. Guidelines for the management of iron deficiency anaemia. Gut 2011, 60, 1309–1316. [Google Scholar] [CrossRef] [Green Version]
- Cancelo-Hidalgo, M.J.; Castelo-Branco, C.; Palacios, S.; Haya-Palazuelos, J.; Ciria-Recasens, M.; Manasanch, J.; Pérez-Edo, L. Tolerability of different oral iron supplements: A systematic review. Curr. Med. Res. Opin. 2013, 29, 291–303. [Google Scholar] [CrossRef]
- Tolkien, Z.; Stecher, L.; Mander, A.P.; Pereira, D.I.; Powell, J.J. Ferrous Sulfate Supplementation Causes Significant Gastrointestinal Side-Effects in Adults: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0117383. [Google Scholar] [CrossRef] [Green Version]
- Jaeggi, T.; Kortman, G.A.M.; Moretti, D.; Chassard, C.; Holding, P.; Dostal, A.; Boekhorst, J.; Timmerman, H.M.; Swinkels, D.W.; Tjalsma, H.; et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 2015, 64, 731–742. [Google Scholar] [CrossRef]
- Power, S.E.; O’Toole, P.W.; Stanton, C.; Ross, R.P.; Fitzgerald, G.F. Intestinal microbiota, diet and health. Br. J. Nutr. 2014, 111, 387–402. [Google Scholar] [CrossRef]
- Lund, E.K.; Wharf, S.G.; Fairweather-Tait, S.J.; Johnson, I.T. Oral ferrous sulfate supplements increase the free radical–generating capacity of feces from healthy volunteers. Am. J. Clin. Nutr. 1999, 69, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Weinberg, E.D. The Lactobacillus Anomaly: Total Iron Abstinence. Perspect. Biol. Med. 1997, 40, 578–583. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Zimmermann, M.B.; Chassard, C.; Rohner, F.; N’Goran, E.K.; Nindjin, C.; Dostal, A.; Utzinger, J.; Ghattas, H.; Lacroix, C.; Hurrell, R.F. The effects of iron fortification on the gut microbiota in African children: A randomized controlled trial in Côte d’Ivoire. Am. J. Clin. Nutr. 2010, 92, 1406–1415. [Google Scholar] [CrossRef]
- Yilmaz, B.; Li, H. Gut Microbiota and Iron: The Crucial Actors in Health and Disease. Pharmaceuticals 2018, 11, 98. [Google Scholar] [CrossRef] [Green Version]
- Paganini, D.; Uyoga, M.A.; Zimmermann, M.B. Iron Fortification of Foods for Infants and Children in Low-Income Countries: Effects on the Gut Microbiome, Gut Inflammation, and Diarrhea. Nutrients 2016, 8, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, M.; Frank, D.N.; Sherlock, L.; Ir, D.; Robertson, C.E.; Krebs, N.F. Effect of Vitamin E With Therapeutic Iron Supplementation on Iron Repletion and Gut Microbiome in US Iron Deficient Infants and Toddlers. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 379–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soofi, S.; Cousens, S.; Iqbal, S.P.; Akhund, T.; Khan, J.; Ahmed, I.; Zaidi, A.K.M.; Bhutta, Z. Effect of provision of daily zinc and iron with several micronutrients on growth and morbidity among young children in Pakistan: A cluster-randomised trial. Lancet 2013, 382, 29–40. [Google Scholar] [CrossRef]
- Rusu, I.G.; Suharoschi, R.; Vodnar, D.C.; Pop, C.R.; Socaci, S.A.; Vulturar, R.; Istrati, M.; Moroșan, I.; Farcas, A.C.; Kerezsi, A.D.; et al. Iron Supplementation Influence on the Gut Microbiota and Probiotic Intake Effect in Iron Deficiency—A Literature-Based Review. Nutrients 2020, 12, 1993. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Vanholder, R.; De Smet, R.; Lesaffer, G. p-Cresol: A toxin revealing many neglected but relevant aspects of uraemic toxicity. Nephrol. Dial. Transplant. 1999, 14, 2813–2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, W.R.; Hoyles, L.; Flint, H.J.; Dumas, M.-E. Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 2013, 16, 246–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dostal, A.; Fehlbaum, S.; Chassard, C.; Zimmermann, M.B.; Lacroix, C. Low iron availability in continuousin vitrocolonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol. Ecol. 2013, 83, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Glover, J.; Jacobs, A. Observations on iron in the jejunal lumen after a standard meal. Gut 1971, 12, 369–371. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, A.; Miller, F.; Worwood, M.; Beamish, M.R.; Wardrop, C.A. Ferritin in the Serum of Normal Subjects and Patients with Iron Deficiency and Iron Overload. BMJ 1972, 4, 206–208. [Google Scholar] [CrossRef] [Green Version]
- Kortman, G.A.M.; Dutilh, B.E.; Maathuis, A.J.H.; Engelke, U.F.; Boekhorst, J.; Keegan, K.P.; Nielsen, F.G.G.; Betley, J.R.; Weir, J.C.; Kingsbury, Z.; et al. Microbial Metabolism Shifts Towards an Adverse Profile with Supplementary Iron in the TIM-2 In vitro Model of the Human Colon. Front. Microbiol. 2016, 6, 1481. [Google Scholar] [CrossRef] [Green Version]
- Lönnerdal, B.; Bryant, A.; Liu, X.; Theil, E.C. Iron absorption from soybean ferritin in nonanemic women. Am. J. Clin. Nutr. 2006, 83, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.L.; Rodríguez-Ramiro, I.; Jones, E.R.; Jones, E.J.; Rodríguez-Celma, J.; Halsey, K.; Domoney, C.; Shewry, P.R.; Fairweather-Tait, S.; Balk, J. The stage of seed development influences iron bioavailability in pea (Pisum sativum L.). Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Perfecto, A.; Rodriguez-Ramiro, I.; Rodriguez-Celma, J.; Sharp, P.; Balk, J.; Fairweather-Tait, S. Pea Ferritin Stability under Gastric pH Conditions Determines the Mechanism of Iron Uptake in Caco-2 Cells. J. Nutr. 2018, 148, 1229–1235. [Google Scholar] [CrossRef]
- Pereira, D.I.; Bruggraber, S.F.; Faria, N.; Poots, L.K.; Tagmount, M.A.; Aslam, M.F.; Frazer, D.M.; Vulpe, C.D.; Anderson, G.J.; Powell, J.J. Nanoparticulate iron(III) oxo-hydroxide delivers safe iron that is well absorbed and utilised in humans. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1877–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, J.J.; Bruggraber, S.F.; Faria, N.; Poots, L.K.; Hondow, N.; Pennycook, T.J.; Latunde-Dada, G.O.; Simpson, R.J.; Brown, A.P.; Pereira, D.I. A nano-disperse ferritin-core mimetic that efficiently corrects anemia without luminal iron redox activity. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1529–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, D.I.; Aslam, M.F.; Frazer, D.M.; Schmidt, A.; Walton, G.E.; McCartney, A.L.; Gibson, G.R.; Anderson, G.J.; Powell, J.J. Dietary iron depletion at weaning imprints low microbiome diversity and this is not recovered with oral nano Fe(III). MicrobiologyOpen 2015, 4, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Matto, J.; Maunuksela, L.; Kajander, K.; Palva, A.; Korpela, R.; Kassinen, A.; Saarela, M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome—A longitudinal study in IBS and control subjects. FEMS Immunol. Med. Microbiol. 2005, 43, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.; Macfarlane, S.; Gibson, G. Validation of a Three-Stage Compound Continuous Culture System for Investigating the Effect of Retention Time on the Ecology and Metabolism of Bacteria in the Human Colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef]
- Eid, N.; Enani, S.; Walton, G.E.; Corona, G.; Costabile, A.; Gibson, G.R.; Rowland, I.; Spencer, J.P.E. The impact of date palm fruits and their component polyphenols, on gut microbial ecology, bacterial metabolites and colon cancer cell proliferation. J. Nutr. Sci. 2014, 3, e46. [Google Scholar] [CrossRef] [Green Version]
- Eid, N.; Osmanova, H.; Natchez, C.; Walton, G.; Costabile, A.; Gibson, G.; Rowland, I.; Spencer, J.P.E. Impact of palm date consumption on microbiota growth and large intestinal health: A randomised, controlled, cross-over, human intervention study. Br. J. Nutr. 2015, 114, 1226–1236. [Google Scholar] [CrossRef] [Green Version]
- Rigottier-Gois, L.; Bourhis, A.-G.; Gramet, G.; Rochet, V.; Doré, J. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 2003, 43, 237–245. [Google Scholar] [CrossRef]
- Rochet, V.; Rigottier-Gois, L.; Rabot, S.; Doré, J. Validation of fluorescent in situ hybridization combined with flow cytometry for assessing interindividual variation in the composition of human fecal microflora during long-term storage of samples. J. Microbiol. Methods 2004, 59, 263–270. [Google Scholar] [CrossRef]
- Grimaldi, R.; Cela, D.; Swann, J.R.; Vulevic, J.; Gibson, G.R.; Tzortzis, G.; Costabile, A. In vitrofermentation of B-GOS: Impact on faecal bacterial populations and metabolic activity in autistic and non-autistic children. FEMS Microbiol. Ecol. 2017, 93, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langendijk, P.S.; Schut, F.; Jansen, G.J.; Raangs, G.C.; Kamphuis, G.R.; Wilkinson, M.H.; Welling, G.W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 1995, 61, 3069–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manz, W.; Amann, R.; Vancanneyt, M.; Schleifer, K.-H.; Ludwig, W. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 1996, 142, 1097–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmsen, H.J.; Elfferich, P.M.; Schut, F.; Welling, G.W. A 16S rRNA-targeted Probe for Detection of Lactobacilli and Enterococci in Faecal Samples by FluorescentIn SituHybridization. Microb. Ecol. Health Dis. 1999, 11, 3–12. [Google Scholar] [CrossRef]
- Franks, A.H.; Harmsen, H.J.; Raangs, G.C.; Jansen, G.J.; Schut, F.; Welling, G.W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 1998, 64, 3336–3345. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Duncan, S.H.; Leitch, E.C.M.; Child, M.W.; Flint, H.J. pH and Peptide Supply Can Radically Alter Bacterial Populations and Short-Chain Fatty Acid Ratios within Microbial Communities from the Human Colon. Appl. Environ. Microbiol. 2005, 71, 3692–3700. [Google Scholar] [CrossRef] [Green Version]
- Harmsen, H.J.M.; Wildeboer-Veloo, A.C.M.; Grijpstra, J.; Knol, J.; Degener, J.E.; Welling, G.W. Development of 16S rRNA-Based Probes for theCoriobacterium Group and the Atopobium Cluster and Their Application for Enumeration of Coriobacteriaceaein Human Feces from Volunteers of Different Age Groups. Appl. Environ. Microbiol. 2000, 66, 4523–4527. [Google Scholar] [CrossRef] [Green Version]
- Hold, G.L.; Schwiertz, A.; Aminov, R.I.; Blaut, M.; Flint, H.J. Oligonucleotide Probes That Detect Quantitatively Significant Groups of Butyrate-Producing Bacteria in Human Feces. Appl. Environ. Microbiol. 2003, 69, 4320–4324. [Google Scholar] [CrossRef] [Green Version]
- Devereux, R.; Kane, M.D.; Winfrey, J.; Stahl, D.A. Genus- and Group-Specific Hybridization Probes for Determinative and Environmental Studies of Sulfate-Reducing Bacteria. Syst. Appl. Microbiol. 1992, 15, 601–609. [Google Scholar] [CrossRef]
- Daims, H.; Brühl, A.; Amann, R.; Schleifer, K.-H.; Wagner, M. The Domain-specific Probe EUB338 is Insufficient for the Detection of all Bacteria: Development and Evaluation of a more Comprehensive Probe Set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993, 14, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.; Calder, A.; Stewart, C.; Smith, A. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 1989, 9, 5–8. [Google Scholar] [CrossRef]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human Microbiome Analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerós, A.S.; Simmons, A.; Drakesmith, H.; Aulicino, A.; Frost, J.N. The battle for iron in enteric infections. Immunology 2020, 161, 186–199. [Google Scholar] [CrossRef]
- Rocha, E.R.; De Uzeda, M.; Brock, J.H. Effect of ferric and ferrous iron chelators on growth of Bacteroides fragilis under anaerobic conditions. FEMS Microbiol. Lett. 1991, 84, 45–50. [Google Scholar] [CrossRef]
- Mevissen-Verhage, E.A.E.; Marcelis, J.H.; Harmsen-Van Amerongen, W.C.; De Vos, N.M.; Verhoef, J. Effect of iron on neonatal gut flora during the first three months of life. Eur. J. Clin. Microbiol. Infect. Dis. 1985, 4, 273–278. [Google Scholar] [CrossRef]
- Pereira, D.I.; Mohammed, N.I.; Ofordile, O.; Camara, F.; Baldeh, B.; Mendy, T.; Sanyang, C.; Jallow, A.T.; Hossain, I.; Wason, J.; et al. A novel nano-iron supplement to safely combat iron deficiency and anaemia in young children: The IHAT-GUT double-blind, randomised, placebo-controlled trial protocol. Gates Open Res. 2018, 2, 48. [Google Scholar] [CrossRef]
- Pereira, D.I.A.; Mergler, B.I.; Faria, N.; Bruggraber, S.F.A.; Aslam, M.F.; Poots, L.K.; Prassmayer, L.; Lönnerdal, B.; Brown, A.P.; Powell, J.J. Caco-2 Cell Acquisition of Dietary Iron(III) Invokes a Nanoparticulate Endocytic Pathway. PLoS ONE 2013, 8, e81250. [Google Scholar] [CrossRef] [Green Version]
- Alleyne, M.; Horne, M.K.; Miller, J.L. Individualized Treatment for Iron-deficiency Anemia in Adults. Am. J. Med. 2008, 121, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Clavel, T.; Smirnov, K.; Schmidt, A.; Lagkouvardos, I.; Walker, A.; Lucio, M.; Michalke, B.; Schmitt-Kopplin, P.; Fedorak, R.; et al. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut 2017, 66, 863–871. [Google Scholar] [CrossRef]
- Chassard, C.; Dapoigny, M.; Scott, K.P.; Crouzet, L.; Del’Homme, C.; Marquet, P.; Martin, J.C.; Pickering, G.; Ardid, D.; Eschalier, A.; et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012, 35, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Parkes, G.C.; Rayment, N.B.; Hudspith, B.N.; Petrovska, L.; Lomer, M.C.; Brostoff, J.; Whelan, K.; Sanderson, J.D. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Rajilić–Stojanović, M.; Biagi, E.; Heilig, H.G.; Kajander, K.; Kekkonen, R.A.; Tims, S.; De Vos, W.M. Global and Deep Molecular Analysis of Microbiota Signatures in Fecal Samples From Patients With Irritable Bowel Syndrome. Gastroenterology 2011, 141, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Liu, R.; Lee, A.; Long, Y.; Du, L.; Lai, S.; Chen, X.; Wang, L.; Si, J.; Owyang, C.; et al. Altered Intestinal Microbiota with Increased Abundance of Prevotella Is Associated with High Risk of Diarrhea-Predominant Irritable Bowel Syndrome. Gastroenterol. Res. Pract. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Jia, Q.; Song, L.; Duan, L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome. Medicine 2019, 98, e14513. [Google Scholar] [CrossRef]
- Ellermann, M.; Gharaibeh, R.Z.; Maharshak, N.; Peréz-Chanona, E.; Jobin, C.; Carroll, I.M.; Arthur, J.C.; Plevy, S.E.; Fodor, A.A.; Brouwer, C.; et al. Dietary iron variably modulates assembly of the intestinal microbiota in colitis-resistant and colitis-susceptible mice. Gut Microbes 2020, 11, 32–50. [Google Scholar] [CrossRef] [Green Version]
- Mahalhal, A.; Williams, J.M.; Johnson, S.; Ellaby, N.; Duckworth, C.A.; Burkitt, M.D.; Liu, X.; Hold, G.L.; Campbell, B.J.; Pritchard, D.M.; et al. Oral iron exacerbates colitis and influences the intestinal microbiome. PLoS ONE 2018, 13, e0202460. [Google Scholar] [CrossRef] [Green Version]




| Probe Name | Sequence |
|---|---|
| Non-Eub | ACTCCTAGGGAGGCAGA |
| Eub338 ‡ | GCTGCCTCCCGTAGGAGT |
| Eub338 ‡ | GCAGCCACCCGTAGGTGT |
| Eub338 ‡ | GCTGCCACCCGTAGGTGT |
| Bif164 | CATCCGGCATTACCACCC |
| Lab158 | GGTATTAGCAYCTGTTTGGA |
| Bac303 | CCAATGTGGGGGACCTT |
| Erec482 | GCTTCTTAGTCARGTACCG |
| Rrec584 | TCAGACTTGCCGYACCGC |
| Ato291 | GGTCGGTCTCTCAACCC |
| Prop853 | ATTGCGTTAACTCCGGCAC |
| Fprau655 | CGCCTACCTCTGCACTAC |
| DSV687 | TACGGATTTCACTCCT |
| Chis150 | TTATGCGGTATTAATCTYCCTTT |
| Time (hours) | Acetate | Propionate | Butyrate | Iso Butyrate | Iso Valerate | Valerate | Lactate | BCFA | Ammonia mM | |
|---|---|---|---|---|---|---|---|---|---|---|
| B-GOS | 0 | 2.20 ± 1.05 | 0.21 ± 0.09 | 0.2 ± 0.11 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.72 ± 0.13 | 0.02 ± 0.03 | 18.9 3± 4.38 |
| FeSO4 | 0 | 2.26 ± 1.08 | 0.23 ± 0.11 | 0.2 ± 0.12 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.02 | 0.8 ± 0.21 | 0.02 ± 0.03 | 20.4 ± 4.27 |
| IHAT | 0 | 2.24 ± 1.06 | 0.22 ± 0.09 | 0.18 ± 0.11 | 0.01 ± 0.01 | 0 ± 0.01 | 0.01 ± 0.01 | 0.76 ± 0.13 | 0.02 ± 0.03 | 20.14 ± 5.29 |
| negative | 0 | 2.26 ± 1.01 | 0.24 ± 0.11 | 0.2 3 ± 0.10 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.02 | 0.8 ± 0.20 | 0.03 ± 0.03 | 19.01 ± 4.59 |
| pea ferritin | 0 | 2.18 ± 1.11 | 0.21 ± 0.09 | 0.2 ± 0.12 | 0 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.73 ± 0.15 | 0.02 ± 0.03 | 19.62 ± 5.01 |
| B-GOS | 8 | 39.37 ± 15.44 | 3.18 ± 2.57 | 1.95 ± 1.60 | 0.01 ± 0.01 | 0.01 ± 0.02 | 0.0 6± 0.06 | 20.46 ± 10.59 * | 0.08 ± 0.08 | 23.99 ± 6.71 |
| FeSO4 | 8 | 36.92 ± 8.05 * | 3.96 ± 2.04 | 2.28 ± 1.58 | 0.02 ± 0.02 | 0.02 ± 0.03 | 0.1 ± 0.08 | 11.14 ± 5.51 | 0.13 ± 0.11 | 29.29 ± 7.36 |
| IHAT | 8 | 38.85 ± 9.31 * | 4.55 ± 2.46 | 2.47 ± 1.75 | 0.02 ± 0.04 | 0.03 ± 0.06 | 0.09 ± 0.10 | 10.59 ± 5.28 | 0.14 ± 0.16 | 30.44 ± 8.69 |
| negative | 8 | 30.28 ± 8.71 | 3.39 ± 2.81 | 2.19 ± 1.58 | 0.01 ± 0.02 | 0.01 ± 0.02 | 0.09 ± 0.08 | 11.52 ± 5.51 | 0.11 ± 0.11 | 29.38 ± 9.47 |
| pea ferritin | 8 | 34.79 ± 11.42 | 4.24 ± 2.58 | 2.48 ± 1.73 | 0.01 ± 0.02 | 0.02 ± 0.02 | 0.09 ± 0.08 | 10.86 ± 5.29 | 0.12 ± 0.11 | 28.57 ± 8.67 |
| B-GOS | 24 | 71.96 ± 9.58 * | 14.71 ± 6.73 | 12.03 ± 7.18 | 0.09 ± 0.10 | 0.13 ± 0.15 | 0.83 ± 1.02 | 10.2 ± 11.55 | 1.04 ± 1.06 | 44.68 ± 14.93 * |
| FeSO4 | 24 | 70.56 ± 11.81 * | 16.34 ± 4.43 * | 11.53 ± 3.64 | 0.29 ± 0.36 | 0.36 ± 0.41 | 1.95 ± 2.23 | 1.55 ± 2.66 * | 2.59 ± 2.57 | 67.72 ± 24.62 |
| IHAT | 24 | 70.8 ± 19.00 * | 14.93 ± 3.66 | 11.61 ± 4.15 | 0.34 ± 0.43 | 0.4 ± 0.53 | 1.57 ± 1.86 | 0.91 ± 2.38 * | 2.31 ± 1.96 | 64.24 ± 21.95 |
| negative | 24 | 56.95 ± 5.56 | 12.39 ± 4.84 | 9.39 ± 4.23 | 0.12 ± 0.16 | 0.18 ± 0.22 | 0.89 ± 1.39 | 5.27 ± 6.24 | 1.19 ± 1.49 | 60.52 ± 17.46 |
| pea ferritin | 24 | 65.89 ± 12.21 * | 15.3 ± 5.07 | 11.45 ± 4.24 | 0.27 ± 0.39 | 0.32 ± 0.43 | 1.63 ± 2.09 | 1.35 ± 2.49 * | 2.21 ± 2.41 | 65.9 9± 23.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poveda, C.; Pereira, D.I.A.; Lewis, M.C.; Walton, G.E. The Impact of Low-Level Iron Supplements on the Faecal Microbiota of Irritable Bowel Syndrome and Healthy Donors Using In Vitro Batch Cultures. Nutrients 2020, 12, 3819. https://doi.org/10.3390/nu12123819
Poveda C, Pereira DIA, Lewis MC, Walton GE. The Impact of Low-Level Iron Supplements on the Faecal Microbiota of Irritable Bowel Syndrome and Healthy Donors Using In Vitro Batch Cultures. Nutrients. 2020; 12(12):3819. https://doi.org/10.3390/nu12123819
Chicago/Turabian StylePoveda, Carlos, Dora I. A. Pereira, Marie C. Lewis, and Gemma E. Walton. 2020. "The Impact of Low-Level Iron Supplements on the Faecal Microbiota of Irritable Bowel Syndrome and Healthy Donors Using In Vitro Batch Cultures" Nutrients 12, no. 12: 3819. https://doi.org/10.3390/nu12123819
APA StylePoveda, C., Pereira, D. I. A., Lewis, M. C., & Walton, G. E. (2020). The Impact of Low-Level Iron Supplements on the Faecal Microbiota of Irritable Bowel Syndrome and Healthy Donors Using In Vitro Batch Cultures. Nutrients, 12(12), 3819. https://doi.org/10.3390/nu12123819




