Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Samples
- -
- Four grades of pea protein (PeaP1, PeaP2, PeaP3, and PeaP4);
- -
- Hydrolyzed pea protein (HPeaP);
- -
- Two grades of wheat protein (WP1 and WP2);
- -
- Potato protein (PP);
- -
- Fava bean protein (FBP);
- -
- Oat protein (OP).
2.2. Materials
2.3. In Vitro Simulated Gastrointestinal Digestion (SGID)
2.4. Solubility Tests
2.5. Size Exclusion Chromatography
2.6. Bioactivity Tests
2.6.1. CCK and GLP-1 Secretion Study
2.6.2. DPP-IV Inhibition Assay
2.6.3. Opioid Receptor Binding Assay
2.6.4. ACE Inhibition Assay
2.6.5. IL-8 Secretion Assay
2.6.6. Antioxidant Assays
Superoxide Anion Assay
Hydrogen Peroxide Assay
Hydroxyl Radical Assay
3. Results and Discussion
3.1. Good Reproducibility of SGID Process and Peptide Generation
3.2. Dose-Dependent Increase in CCK and GLP-1 Secretion in Response to Intestinal Digestions (after SGID)
3.3. Good Inhibition of DPP-IV Activity by Intestinal Digestions (after SGID)
3.4. Good Potential to Bind Opioid Receptors by Protein Samples
3.5. Good Inhibition of ACE Activity by Intestinal Digestions (after SGID)
3.6. No Effect on Pro-Inflammatory Cytokine Secretion by Protein Samples
3.7. Antioxidant Properties of Protein Samples (after SGID)
3.7.1. Superoxide Anion Assay
3.7.2. Hydrogen Peroxide Assay
3.7.3. Hydroxyl Radical Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lonnie, M.; Hooker, E.; Brunstrom, J.M.; Corfe, B.M.; Green, M.A.; Watson, A.W.; Williams, E.A.; Stevenson, E.J.; Penson, S.; Johnstone, A.M. Protein for Life: Review of Optimal Protein Intake, Sustainable Dietary Sources and the Effect on Appetite in Ageing Adults. Nutrients 2018, 10, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pihlanto, A.; Mattila, P.; Mäkinen, S.; Pajari, A.-M. Bioactivities of alternative protein sources and their potential health benefits. Food Funct. 2017, 8, 3443–3458. [Google Scholar] [CrossRef] [PubMed]
- González, A.D.; Frostell, B.; Carlsson-Kanyama, A. Protein efficiency per unit energy and per unit greenhouse gas emissions: Potential contribution of diet choices to climate change mitigation. Food Policy 2011, 36, 562–570. [Google Scholar] [CrossRef]
- Lefranc-Millot, C.; Teichman-Dubois, V. Protein from Vegetable Sources: A Focus on Pea Protein. In Novel Proteins for Food, Pharmaceuticals and Agriculture; Hayes, D., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2018; pp. 197–216. [Google Scholar]
- Rutherfurd-Markwick, K.J. Food proteins as a source of bioactive peptides with diverse functions. Br. J. Nutr. 2012, 108, S149–S157. [Google Scholar] [CrossRef] [Green Version]
- Anderson, G.H.; Aziz, A. Multifunctional roles of dietary proteins in the regulation of metabolism and food intake: Application to feeding infants. J. Pediatr. 2006, 149, S74–S79. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [Green Version]
- Moughan, P.J.; Rutherfurd, S.M.; Montoya, C.A.; Dave, L.A. Food-derived bioactive peptides—A new paradigm. Nutr. Res. Rev. 2014, 27, 16–20. [Google Scholar] [CrossRef] [Green Version]
- Manzanares, P.; Gandía, M.; Garrigues, S.; Marcos, J.F. Improving Health-Promoting Effects of Food-Derived Bioactive Peptides through Rational Design and Oral Delivery Strategies. Nutrients 2019, 11, 2545. [Google Scholar] [CrossRef] [Green Version]
- Möller, N.P.; Scholz-Ahrens, K.E.; Roos, N.; Schrezenmeir, J. Bioactive peptides and proteins from foods: Indication for health effects. Eur. J. Nutr. 2008, 47, 171–182. [Google Scholar] [CrossRef]
- Caron, J.; Domenger, D.; Dhulster, P.; Ravallec, R.; Cudennec, B. Protein Digestion-Derived Peptides and the Peripheral Regulation of Food Intake. Front. Endocrinol. 2017, 8, 85. [Google Scholar] [CrossRef]
- Janssen, S.; Depoortere, I. Nutrient sensing in the gut: New roads to therapeutics? Trends Endocrinol. Metab. 2013, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Rønnestad, I.; Akiba, Y.; Kaji, I.; Kaunitz, J.D. Duodenal luminal nutrient sensing. Curr. Opin. Pharmacol. 2014, 19, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadkarni, P.; Chepurny, O.G.; Holz, G.G. Regulation of Glucose Homeostasis by GLP-1. Prog. Mol. Biol. Transl. Sci. 2014, 121, 23–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juillerat-Jeanneret, L. Dipeptidyl Peptidase IV and Its Inhibitors: Therapeutics for Type 2 Diabetes and What Else? J. Med. Chem. 2014, 57, 2197–2212. [Google Scholar] [CrossRef]
- Drucker, D.J.; Nauck, M.A. The incretin system: Glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 2006, 368, 1696–1705. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [Green Version]
- Duraffourd, C.; De Vadder, F.; Goncalves, D.; Delaere, F.; Penhoat, A.; Brusset, B.; Rajas, F.; Chassard, D.; Duchampt, A.; Stefanutti, A.; et al. Mu-Opioid Receptors and Dietary Protein Stimulate a Gut-Brain Neural Circuitry Limiting Food Intake. Cell 2012, 150, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Mithieux, G.; Misery, P.; Magnan, C.; Pillot, B.; Gautier-Stein, A.; Bernard, C.; Rajas, F.; Zitoun, C. Portal sensing of intestinal gluconeogenesis is a mechanistic link in the diminution of food intake induced by diet protein. Cell Metab. 2005, 2, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, A.; Sharma, K.; Majumder, K. Food-derived bioactive peptides and their role in ameliorating hypertension and associated cardiovascular diseases. Adv. Food Nutr. Res. 2019, 89, 165–207. [Google Scholar] [CrossRef]
- Fernández-Tomé, S.; Hernández-Ledesma, B.; Chaparro, M.; Indiano-Romacho, P.; Bernardo, D.; Gisbert, J.P. Role of food proteins and bioactive peptides in inflammatory bowel disease. Trends Food Sci. Technol. 2019, 88, 194–206. [Google Scholar] [CrossRef]
- Caron, J.; Cudennec, B.; Domenger, D.; Belguesmia, Y.; Flahaut, C.; Kouach, M.; Lesage, J.; Goossens, J.-F.; Dhulster, P.; Ravallec, R. Simulated GI digestion of dietary protein: Release of new bioactive peptides involved in gut hormone secretion. Food Res. Int. 2016, 89, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Dipeptidyl peptidase-IV inhibitory activity of dairy protein hydrolysates. Int. Dairy J. 2012, 25, 97–102. [Google Scholar] [CrossRef]
- Garreau, I.; Zhao, Q.; Pejoan, C.; Cupo, A.; Piot, J.M. VV-hemorphin-7 and LVV-hemorphin-7 released during in vitro peptic hemoglobin hydrolysis are morphinomimetic peptides. Neuropeptides 1995, 28, 243–250. [Google Scholar] [CrossRef]
- Sentandreu, M.A.; Toldrá, F. A fluorescence-based protocol for quantifying angiotensin-converting enzyme activity. Nat Protoc 2006, 1, 2423–2427. [Google Scholar] [CrossRef]
- Aruoma, O.I.; Halliwell, B.; Hoey, B.M.; Butler, J. The antioxidant action of N-acetylcysteine: Its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic. Biol. Med. 1989, 6, 593–597. [Google Scholar] [CrossRef]
- Thurman, R.G.; Ley, H.G.; Scholz, R. Hepatic microsomal ethanol oxidation. Hydrogen peroxide formation and the role of catalase. Eur. J. Biochem. 1972, 25, 420–430. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C.; Aruoma, O.I. The deoxyribose method: A simple “test-tube” assay for determination of rate constants for reactions of hydroxyl radicals. Anal. Biochem. 1987, 165, 215–219. [Google Scholar] [CrossRef]
- Giromini, C.; Cheli, F.; Rebucci, R.; Baldi, A. Invited review: Dairy proteins and bioactive peptides: Modeling digestion and the intestinal barrier. J. Dairy Sci. 2019, 102, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.W.; Nam, M.S. Bioactive Peptides in Milk and Dairy Products: A Review. Korean J. Food Sci. Anim. Resour. 2015, 35, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Santos-Hernández, M.; Tomé, D.; Gaudichon, C.; Recio, I. Stimulation of CCK and GLP-1 secretion and expression in STC-1 cells by human jejunal contents and in vitro gastrointestinal digests from casein and whey proteins. Food Funct. 2018, 9, 4702–4713. [Google Scholar] [CrossRef]
- Geraedts, M.C.P.; Troost, F.J.; Fischer, M.A.J.G.; Edens, L.; Saris, W.H.M. Direct induction of CCK and GLP-1 release from murine endocrine cells by intact dietary proteins. Mol. Nutr. Food Res. 2011, 55, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Power-Grant, O.; Bruen, C.; Brennan, L.; Giblin, L.; Jakeman, P.; FitzGerald, R.J. In vitro bioactive properties of intact and enzymatically hydrolysed whey protein: Targeting the enteroinsular axis. Food Funct. 2015, 6, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Cudennec, B.; Ravallec-Plé, R.; Courois, E.; Fouchereau-Peron, M. Peptides from fish and crustacean by-products hydrolysates stimulate cholecystokinin release in STC-1 cells. Food Chem. 2008, 111, 970–975. [Google Scholar] [CrossRef]
- Cudennec, B.; Fouchereau-Peron, M.; Ferry, F.; Duclos, E.; Ravallec, R. In Vitro and in vivo evidence for a satiating effect of fish protein hydrolysate obtained from blue whiting (Micromesistius poutassou) muscle. J. Funct. Foods 2012, 4, 271–277. [Google Scholar] [CrossRef]
- Sufian, K.N.B.; Hira, T.; Nakamori, T.; Furuta, H.; Asano, K.; Hara, H. Soybean β-Conglycinin Bromelain Hydrolysate Stimulates Cholecystokinin Secretion by Enteroendocrine STC-1 Cells to Suppress the Appetite of Rats under Meal-Feeding Conditions. Biosci. Biotechnol. Biochem. 2011, 75, 848–853. [Google Scholar] [CrossRef]
- Foltz, M.; Ansems, P.; Schwarz, J.; Tasker, M.C.; Lourbakos, A.; Gerhardt, C.C. Protein Hydrolysates Induce CCK Release from Enteroendocrine Cells and Act as Partial Agonists of the CCK-1-Receptor. J. Agric. Food Chem. 2008, 56, 837–843. [Google Scholar] [CrossRef]
- Geraedts, M.C.P.; Troost, F.J.; Tinnemans, R.; Söderholm, J.D.; Brummer, R.-J.; Saris, W.H.M. Release of Satiety Hormones in Response to Specific Dietary Proteins Is Different between Human and Murine Small Intestinal Mucosa. Ann Nutr Metab 2010, 56, 308–313. [Google Scholar] [CrossRef]
- Chen, W.; Hira, T.; Nakajima, S.; Hara, H. Wheat gluten hydrolysate potently stimulates peptide-YY secretion and suppresses food intake in rats. Biosci. Biotechnol. Biochem. 2018, 82, 1992–1999. [Google Scholar] [CrossRef]
- Caron, J.; Domenger, D.; Dhulster, P.; Ravallec, R.; Cudennec, B. Using Caco-2 cells as novel identification tool for food-derived DPP-IV inhibitors. Food Res. Int. 2017, 92, 113–118. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Lalmahomed, M.; Paolella, S.; FitzGerald, R.J. Milk protein isolate (MPI) as a source of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2017, 231, 202–211. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Investigation of the Potential of Hemp, Pea, Rice and Soy Protein Hydrolysates as a Source of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides. Food Dig. Res. Curr. Opin. 2015, 6, 19–29. [Google Scholar]
- Wang, F.; Yu, G.; Zhang, Y.; Zhang, B.; Fan, J. Dipeptidyl Peptidase IV Inhibitory Peptides Derived from Oat (Avena sativa L.), Buckwheat (Fagopyrum esculentum), and Highland Barley (Hordeum vulgare trifurcatum (L.) Trofim) Proteins. J. Agric. Food Chem. 2015, 63, 9543–9549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Udenigwe, C.C. Role of food-derived opioid peptides in the central nervous and gastrointestinal systems. J. Food Biochem. 2019, 43, e12629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domenger, D.; Cudennec, B.; Kouach, M.; Touche, V.; Landry, C.; Lesage, J.; Gosselet, F.; Lestavel, S.; Goossens, J.-F.; Dhulster, P.; et al. Food-Derived Hemorphins Cross Intestinal and Blood–Brain Barriers In Vitro. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef]
- Kim, S.-K.; Ngo, D.-H.; Vo, T.-S. Marine Fish-Derived Bioactive Peptides as Potential Antihypertensive Agents. Adv. Food Nutr. Res. 2012, 65, 249–260. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Safet. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Manoharan, S.; Shuib, A.S.; Abdullah, N. Structural Characteristics and Antihypertensive Effects of Angiotensin-I-Converting Enzyme Inhibitory Peptides in THE Renin-Angiotensin and Kallikrein Kinin Systems. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 383–406. [Google Scholar] [CrossRef]
- Boschin, G.; Scigliuolo, G.M.; Resta, D.; Arnoldi, A. ACE-inhibitory activity of enzymatic protein hydrolysates from lupin and other legumes. Food Chem. 2014, 145, 34–40. [Google Scholar] [CrossRef]
- Rudolph, S.; Lunow, D.; Kaiser, S.; Henle, T. Identification and quantification of ACE-inhibiting peptides in enzymatic hydrolysates of plant proteins. Food Chem. 2017, 224, 19–25. [Google Scholar] [CrossRef]
- Guha, S.; Majumder, K. Structural-features of food-derived bioactive peptides with anti-inflammatory activity: A brief review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Zhang, H.; Kovacs-Nolan, J.; Kodera, T.; Eto, Y.; Mine, Y. γ-Glutamyl cysteine and γ-glutamyl valine inhibit TNF-α signaling in intestinal epithelial cells and reduce inflammation in a mouse model of colitis via allosteric activation of the calcium-sensing receptor. Biochim. Biophys. Acta 2015, 1852, 792–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Walle, J.; Hendrickx, A.; Romier, B.; Larondelle, Y.; Schneider, Y.-J. Inflammatory parameters in Caco-2 cells: Effect of stimuli nature, concentration, combination and cell differentiation. Toxicol. In Vitro 2010, 24, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
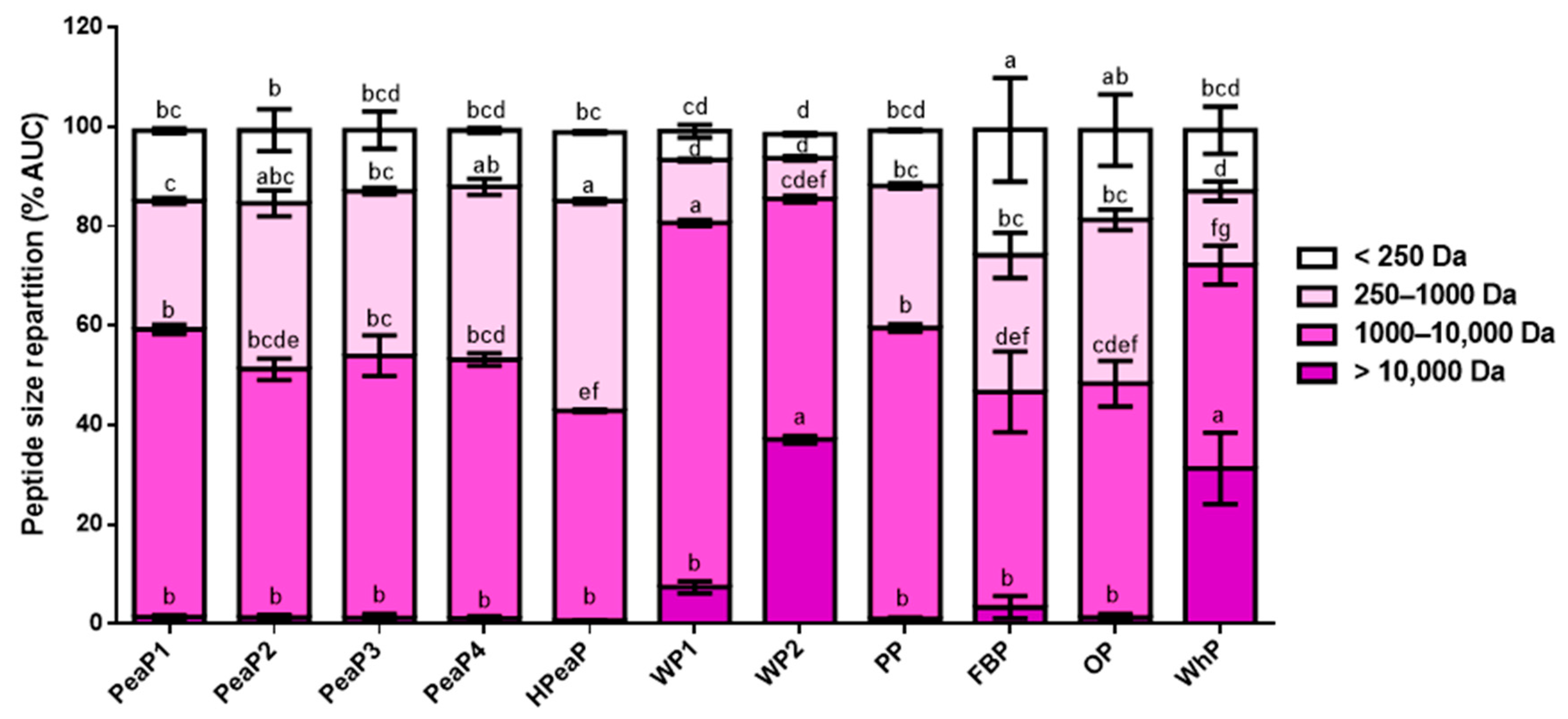
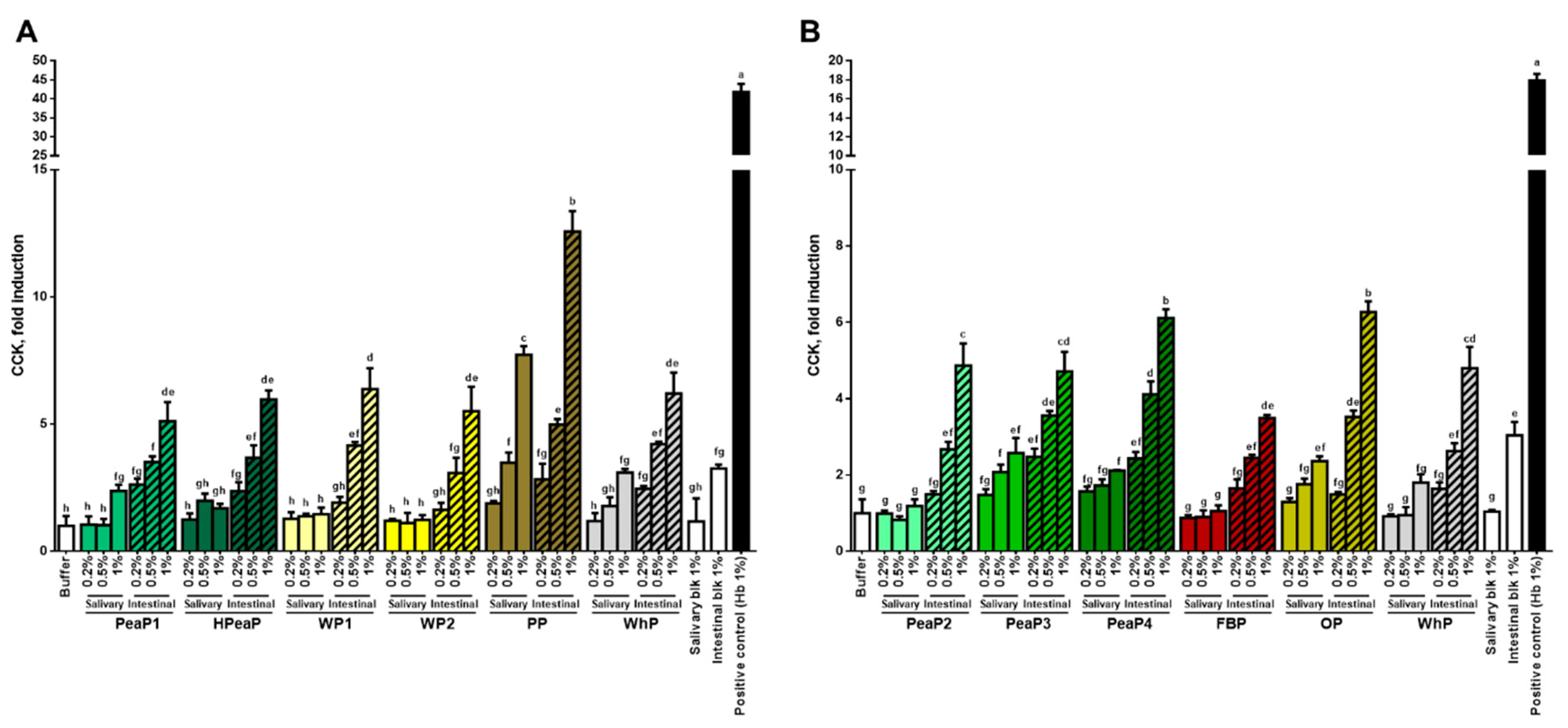
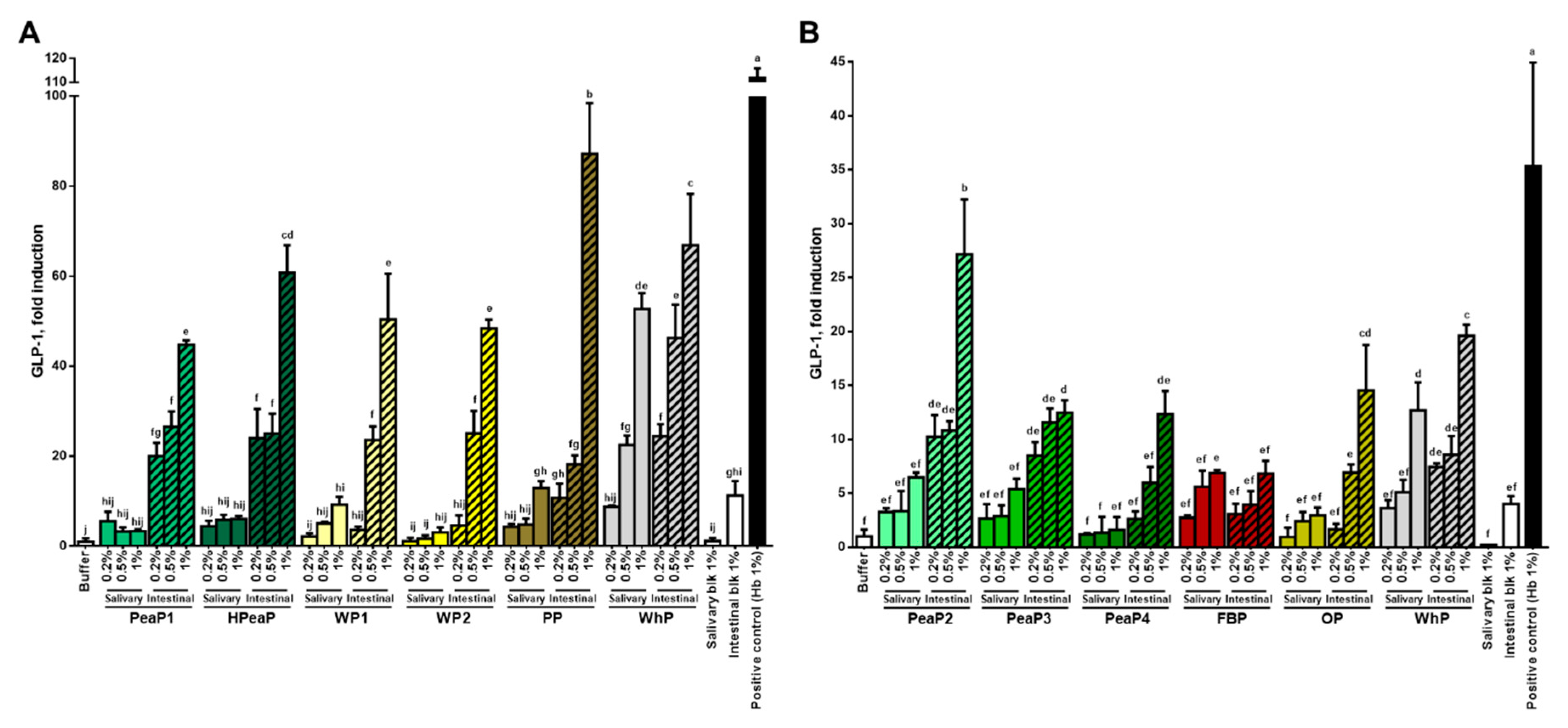

| Name | Description | % Dry Matter | % Protein |
|---|---|---|---|
| PeaP1 | Pea protein—grade 1 | 93.0 | 79.1 |
| PeaP2 | Pea protein—grade 2 | 95.2 | 79.7 |
| PeaP3 | Pea protein—grade 3 | 94.4 | 80.8 |
| PeaP4 | Pea protein—grade 4 | 95.6 | 81.5 |
| HPeaP | Hydrolyzed pea protein | 94.6 | 78.3 |
| WP1 | Wheat protein—grade 1 | 94.8 | 83.7 |
| WP2 | Wheat protein—grade 2 | 93.2 | 93.2 |
| PP | Potato protein | 93.8 | 79.1 |
| FBP | Fava bean protein | 96.0 | 89.8 |
| OP | Oat protein | 96.0 | 88.3 |
| WhP | Whey protein | 95.0 | 85.5 |
| Characteristics | Saliva | Gastric Juice | Duodenal Juice | Bile Juice |
|---|---|---|---|---|
| Chemical Composition | KCl (12 mM) | KCl (11 mM) | KCl (7.6 mM) | KCl (5 mM) |
| KSCN (2 mM) | NaH2PO4 (2.2 mM) | KH2PO4 (0.6 mM) | NaCl (90 mM) | |
| NaH2PO4 (7.4 mM) | NH4Cl (5.7 mM) | NaCl (120 mM) | NaHCO3 (69 mM) | |
| Na2SO4 (4 mM) | NaCl (47 mM) | NaHCO3 (40 mM) | HCl (1.5 mM) | |
| NaCl (5 mM) | HCl (6.5 mM) | HCl (1.8 mM) | CO(NH2)2 (4 mM) | |
| NaHCO3 (20 mM) | CaCl2 (2.7 mM) | MgCl2 (0.5 mM) | ||
| CO(NH2)2 (3.3 mM) | CO(NH2)2 (1.4 mM) | CO(NH2)2 (1.7 mM) | ||
| Proteases | Pepsin 1/40 (w/w) | Pancreatin 1/50 (w/w) | ||
| pH | 6.8 ± 0.2 | 1.3 ± 0.2 | 8.1 ± 0.2 | 8.2 ± 0.2 |
| Name | Salivary Sample | Gastric Digestion | Intestinal Digestion |
|---|---|---|---|
| PeaP1 | 20.53 | 61.65 | 85.97 |
| PeaP2 | 43.55 | 58.67 | 83.94 |
| PeaP3 | 47.27 | 69.47 | 86.32 |
| PeaP4 | 100.00 | 82.81 | 86.96 |
| HPeaP | 81.16 | 82.39 | 95.61 |
| WP1 | 63.33 | 75.56 | 93.40 |
| WP2 | 90.08 | 100.00 | 100.00 |
| PP | 9.13 | 51.79 | 69.77 |
| FBP | 39.41 | 56.72 | 90.34 |
| OP | 18.33 | 69.35 | 83.58 |
| WhP | 91.97 | 69.97 | 100.00 |
| DPP-IV-IC50 (mg mL−1) | Salivary Sample | Gastric Digestion | Intestinal Digestion |
|---|---|---|---|
| PeaP1 | ND | 1.13 ± 0.05 CD | 1.07 ± 0.06 bcde |
| PeaP2 | 3.87 ± 1.01 | 0.85 ± 0.19 CD | 0.98 ± 0.55 cde |
| PeaP3 | ND | 1.45 ± 0.15 BCD | 1.19 ± 0.05 bcd |
| PeaP4 | ND | 2.34 ± 1.18 AB | 1.46 ± 0.17 bc |
| HPeaP | 1.19 ± 0.12 | 0.86 ± 0.08 CD | 0.79 ± 0.08 de |
| WP1 | ND | 2.96 ± 0.34 A | 1.64 ± 0.07 ab |
| WP2 | ND | 2.96 ± 0.51 A | 2.03 ± 0.30 a |
| PP | ND | 1.88 ± 0.21 ABC | 1.07 ± 0.12 bcde |
| FBP | ND | 0.65 ± 0.07 D | 0.54 ± 0.03 e |
| OP | ND | 0.75 ± 0.07 CD | 0.83 ± 0.10 cde |
| WhP | ND | 1.70 ± 0.10 BCD | 1.02 ± 0.14 bcde |
| Opioid R-ED50 (mg mL−1) | Salivary Sample | Intestinal Digestion |
|---|---|---|
| PeaP1 | 26.80 | 14.90 |
| PeaP2 | 4.98 | 4.23 |
| PeaP3 | 4.63 | 2.44 |
| PeaP4 | 24.98 | 3.17 |
| HPeaP | 8.60 | 10.27 |
| WP1 | 39.50 | 26.73 |
| WP2 | 80.30 | 17.90 |
| PP | 14.55 | 15.40 |
| FBP | 2.83 | 12.51 |
| OP | 31.03 | 10.25 |
| WhP | 78.21 | 22.70 |
| ACE-IC50 (µg mL−1) | Salivary Sample | Gastric Digestion | Intestinal Digestion |
|---|---|---|---|
| PeaP1 | ND | 90.70 ± 25.80 CD | 101.19 ± 23.15 cd |
| PeaP2 | 407.78 ± 28.10 | 37.37 ± 19.83 D | 221.79 ± 45.57 ab |
| PeaP3 | ND | 47.38 ± 44.33 CD | 185.54 ± 9.97 abc |
| PeaP4 | ND | 187.97 ± 45.38 AB | 29.54 ± 34.98 d |
| HPeaP | 148.97 ± 40.79 | 93.37 ± 24.74 CD | 113.64 ± 50.51 cd |
| WP1 | ND | 60.79 ± 12.22 CD | 63.82 ± 20.18 d |
| WP2 | ND | 186.78 ± 0.91 AB | 265.46 ± 22.88 a |
| PP | ND | 107.34 ± 40.79 BCD | 85.23 ± 19.24 d |
| FBP | ND | 133.50 ± 35.09 BC | 52.02 ± 43.06 d |
| OP | ND | 98.04 ± 32.21 CD | 99.46 ± 9.88 cd |
| WhP | ND | 126.38 ± 82.34 BCD | 127.50 ± 25.61 bcd |
| O2−-IC50 (mg mL−1) | Salivary Sample | Intestinal Digestion |
|---|---|---|
| PeaP1 | 0.70 | ND |
| PeaP2 | ND | ND |
| PeaP3 | ND | ND |
| PeaP4 | ND | ND |
| HPeaP | ND | ND |
| WP1 | ND | ND |
| WP2 | 1.30 | ND |
| PP | 2.00 | 1.80 |
| FBP | ND | ND |
| OP | 2.50 | ND |
| WhP | ND | ND |
| H2O2-IC50 (mg mL−1) | Salivary Sample | Intestinal Digestion |
|---|---|---|
| PeaP1 | 0.65 | 0.55 |
| PeaP2 | 0.65 | 1.40 |
| PeaP3 | 0.80 | 4.60 |
| PeaP4 | 1.05 | ND |
| HPeaP | 0.65 | 1.20 |
| WP1 | ND | ND |
| WP2 | 2.10 | ND |
| PP | 0.65 | 1.20 |
| FBP | 0.65 | 0.65 |
| OP | 0.75 | 3.05 |
| WhP | 4.45 | 2.08 |
| HO-IC50 (mg mL−1) | Salivary Sample | Intestinal Digestion |
|---|---|---|
| PeaP1 | 1.60 | 2.05 |
| PeaP2 | 1.25 | 1.00 |
| PeaP3 | 1.10 | 1.55 |
| PeaP4 | 1.60 | 1.35 |
| HPeaP | 1.45 | 2.50 |
| WP1 | 4.50 | 1.65 |
| WP2 | 1.35 | 0.85 |
| PP | 1.00 | 1.75 |
| FBP | 1.30 | 1.45 |
| OP | 1.50 | 1.70 |
| WhP | 2.95 | 1.33 |
| Protein | Metabolic Activity | Anti-Hypertensive | Anti-Inflammatory | Antioxidant | SCORE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CCK | GLP-1 | DPP-IV | OR | ACE | IL-8 | O2− | H2O2 | HO. | ||
| PeaP1 | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 2 | 1 | 10 |
| PeaP2 | 1 | 2 | 2 | 2 | 1 | 0 | 0 | 2 | 1 | 11 |
| PeaP3 | 1 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 8 |
| PeaP4 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 1 | 8 |
| HPeaP | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 2 | 1 | 10 |
| WP1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 7 |
| WP2 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 6 |
| PP | 2 | 2 | 2 | 1 | 2 | 0 | 2 | 2 | 1 | 14 |
| FBP | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 2 | 1 | 10 |
| OP | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 1 | 1 | 9 |
| WhP | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 1 | 1 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dugardin, C.; Cudennec, B.; Tourret, M.; Caron, J.; Guérin-Deremaux, L.; Behra-Miellet, J.; Lefranc-Millot, C.; Ravallec, R. Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion. Nutrients 2020, 12, 3746. https://doi.org/10.3390/nu12123746
Dugardin C, Cudennec B, Tourret M, Caron J, Guérin-Deremaux L, Behra-Miellet J, Lefranc-Millot C, Ravallec R. Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion. Nutrients. 2020; 12(12):3746. https://doi.org/10.3390/nu12123746
Chicago/Turabian StyleDugardin, Camille, Benoit Cudennec, Mélissa Tourret, Juliette Caron, Laetitia Guérin-Deremaux, Josette Behra-Miellet, Catherine Lefranc-Millot, and Rozenn Ravallec. 2020. "Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion" Nutrients 12, no. 12: 3746. https://doi.org/10.3390/nu12123746
APA StyleDugardin, C., Cudennec, B., Tourret, M., Caron, J., Guérin-Deremaux, L., Behra-Miellet, J., Lefranc-Millot, C., & Ravallec, R. (2020). Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion. Nutrients, 12(12), 3746. https://doi.org/10.3390/nu12123746





