The Timing Effects of Soy Protein Intake on Mice Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
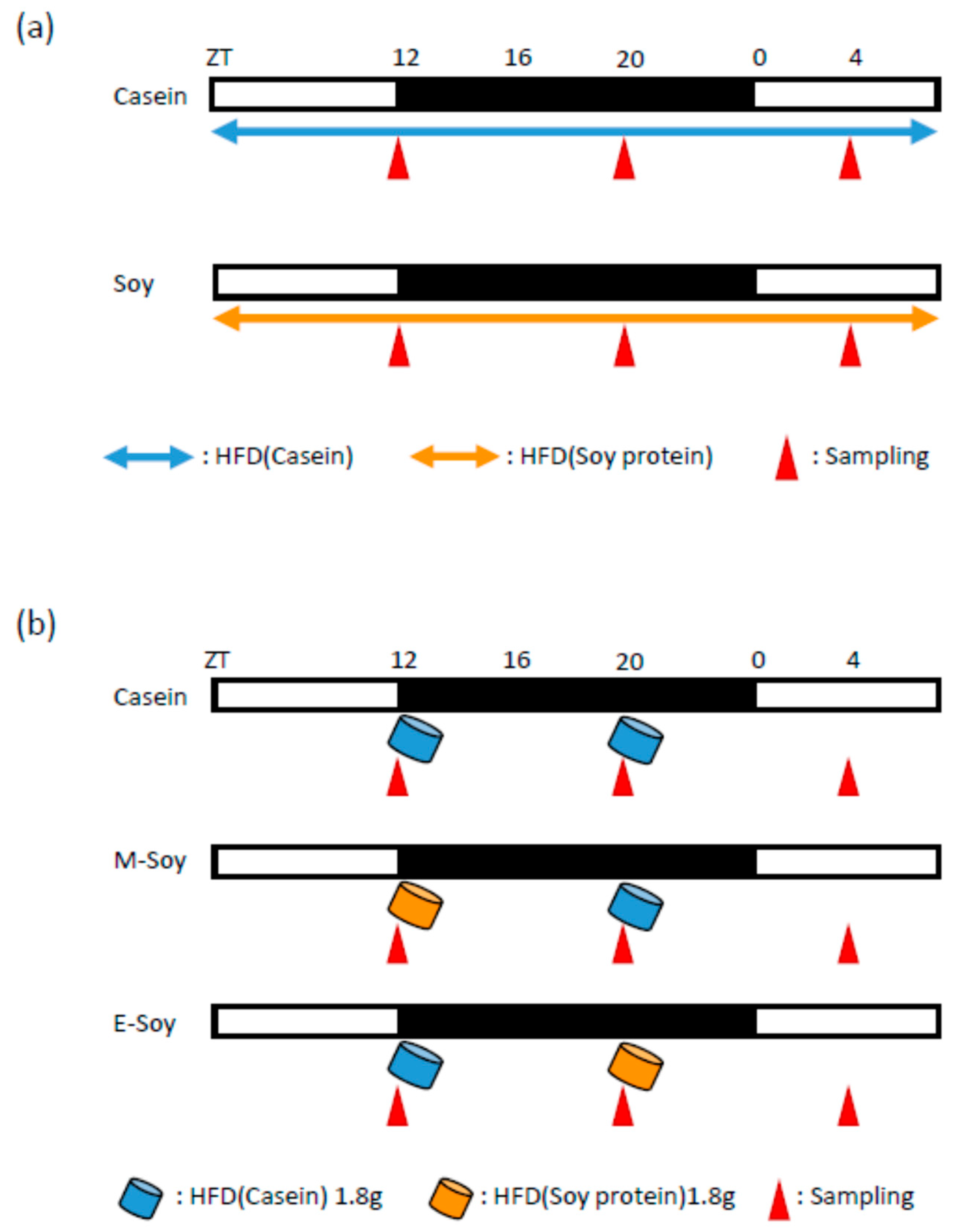
2.2. Experimental Design
2.3. Cholesterol and Triglyceride Measurement
2.4. Real-Time RT-PCR
2.5. Cecal pH Measurement
2.6. Short-Chain Fatty Acid (SCFA) Measurement
2.7. Fecal DNA Extraction
2.8. 16S rRNA Gene Sequencing
2.9. Analysis of 16S rRNA Gene Sequences
2.10. Metagenome Prediction
2.11. Statistical Analysis
3. Results
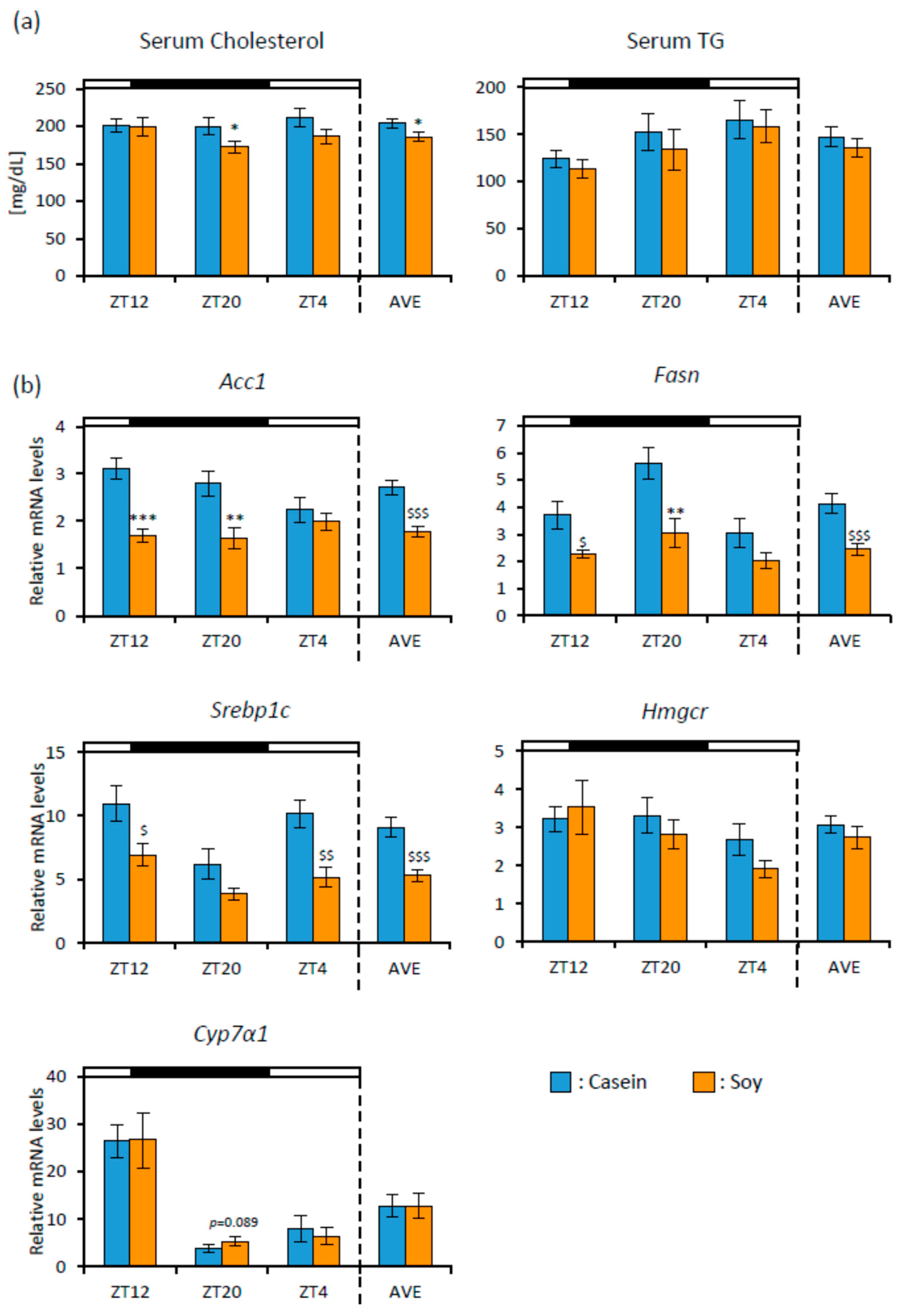
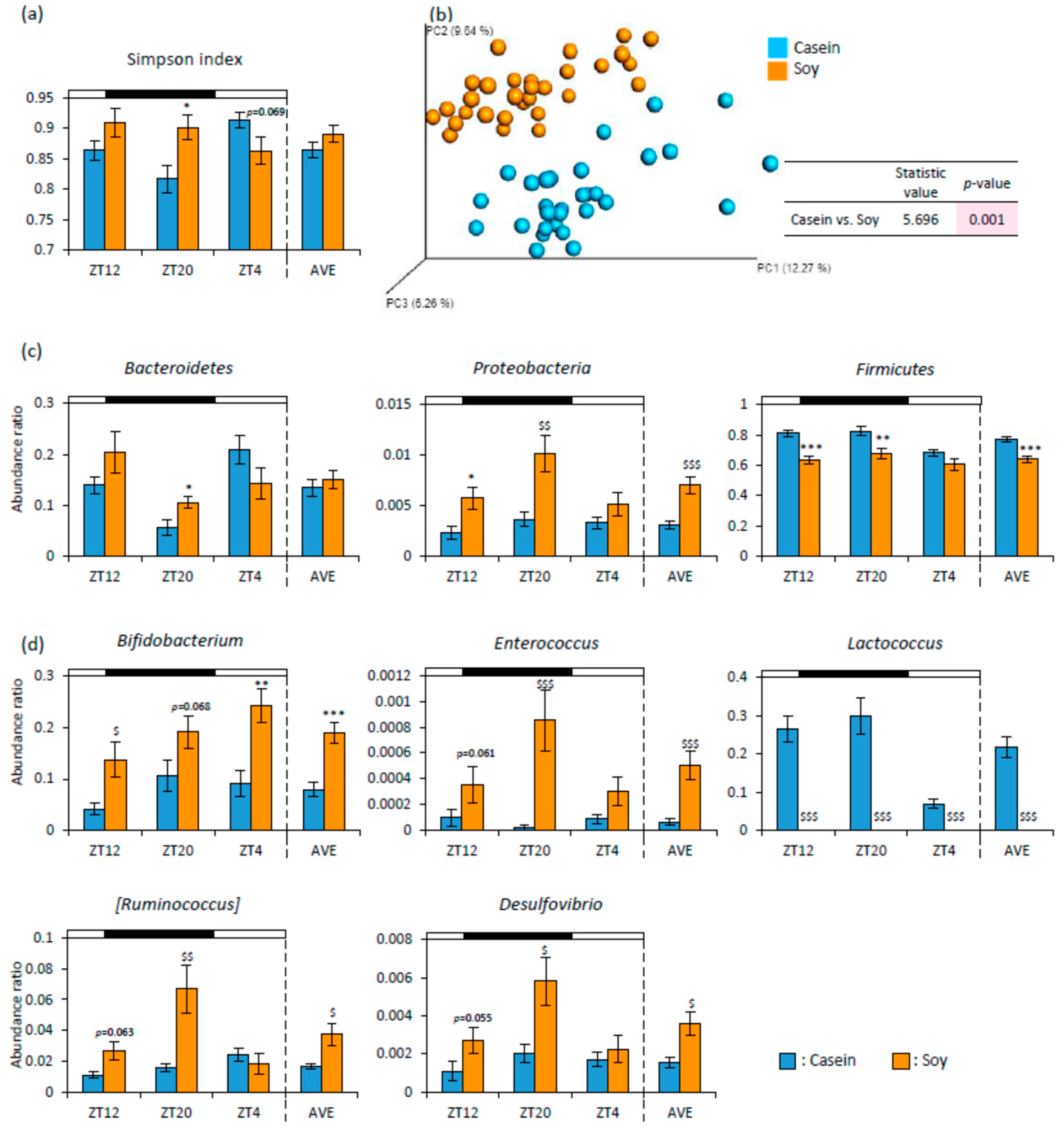
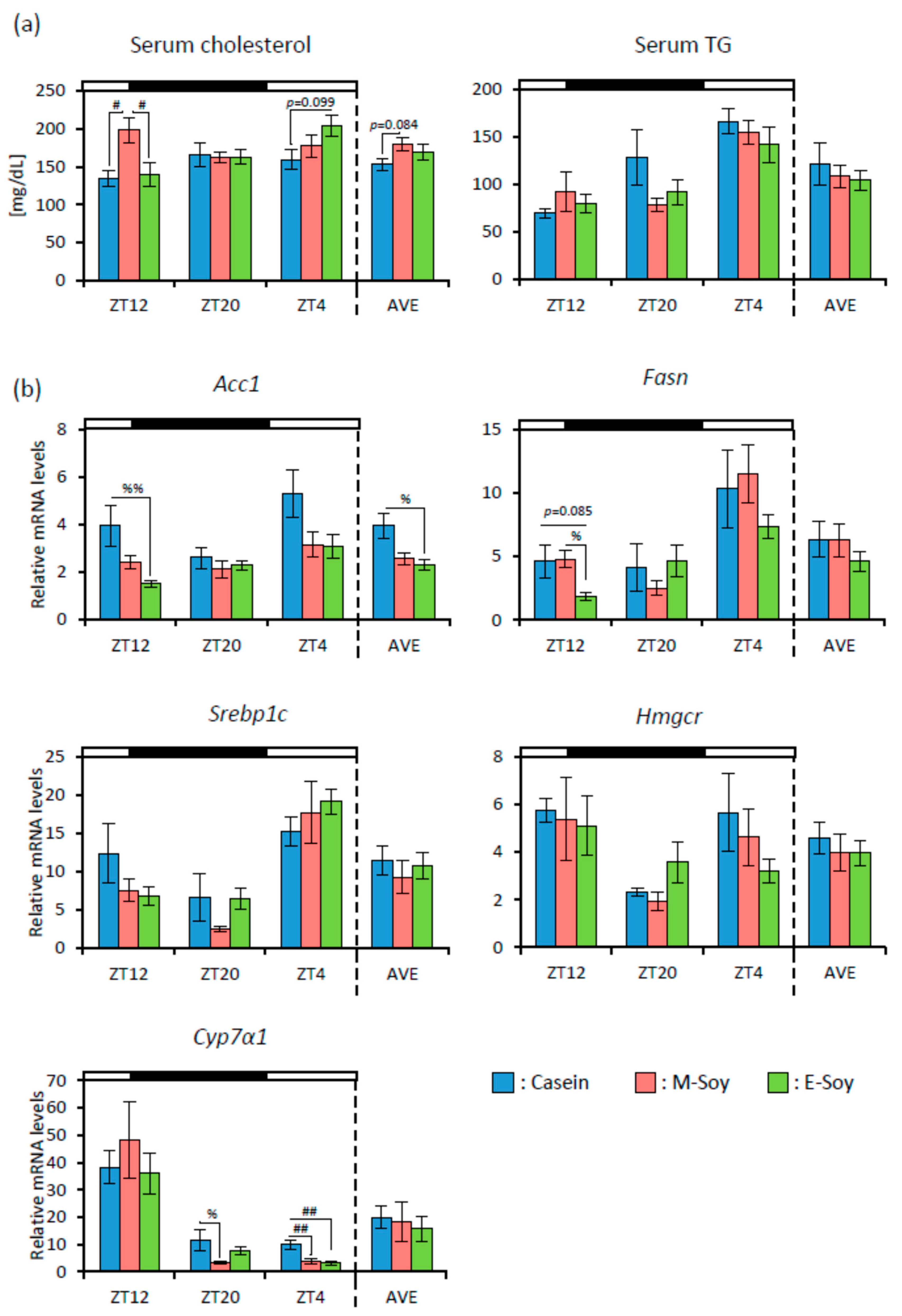
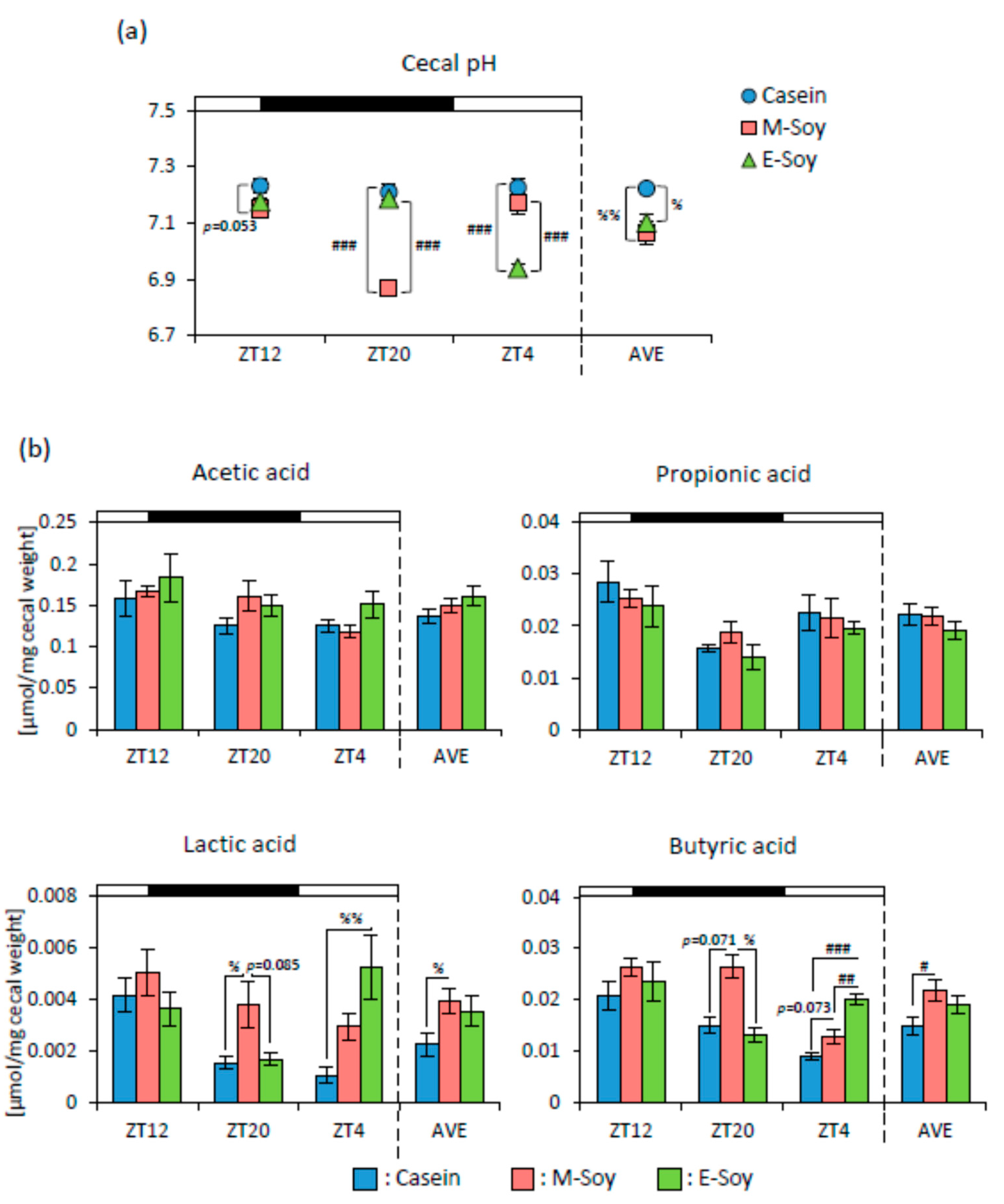
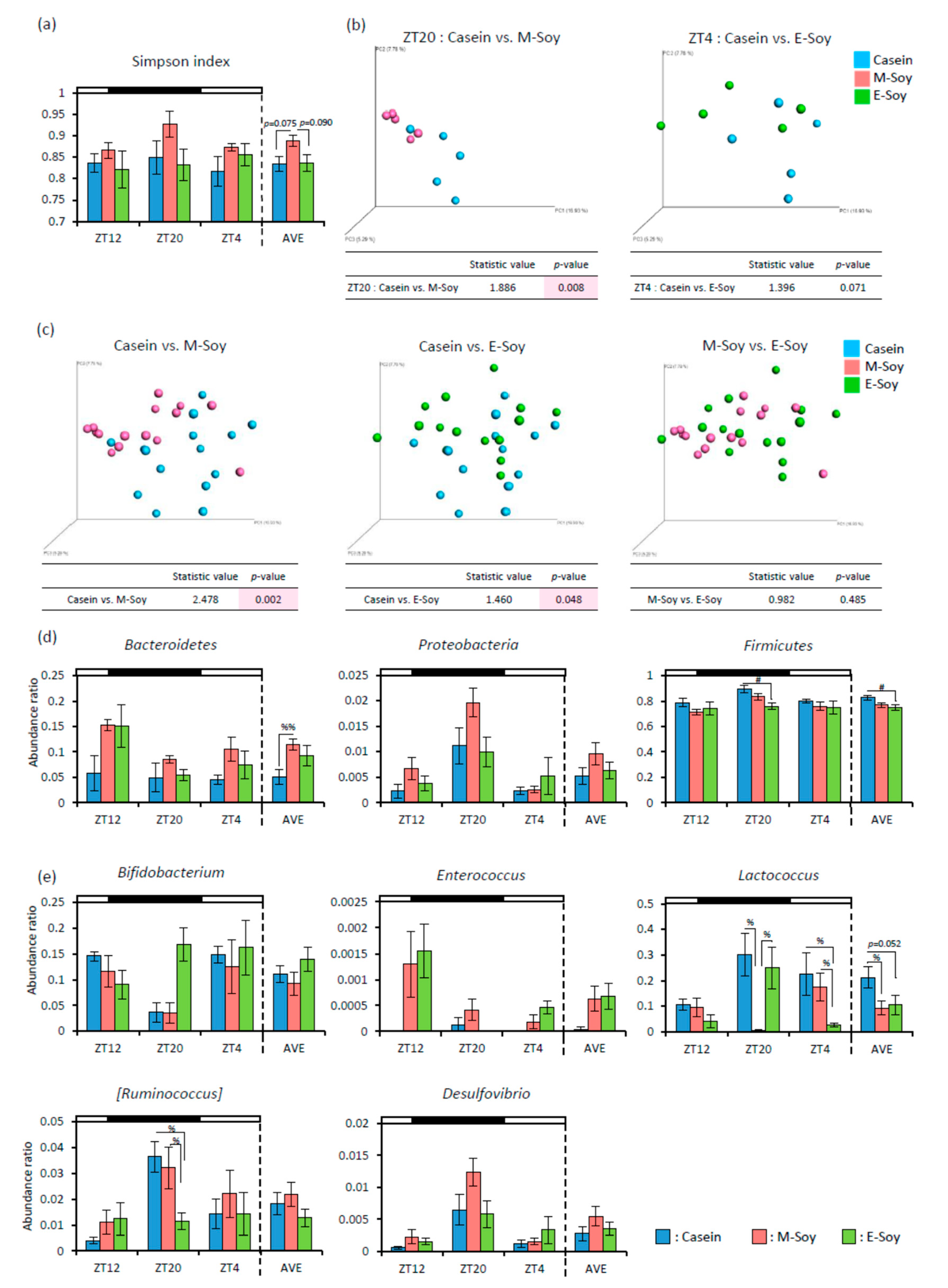
3.1. Soy Protein Intake Affected Lipid Metabolism and the Gut Microbiota
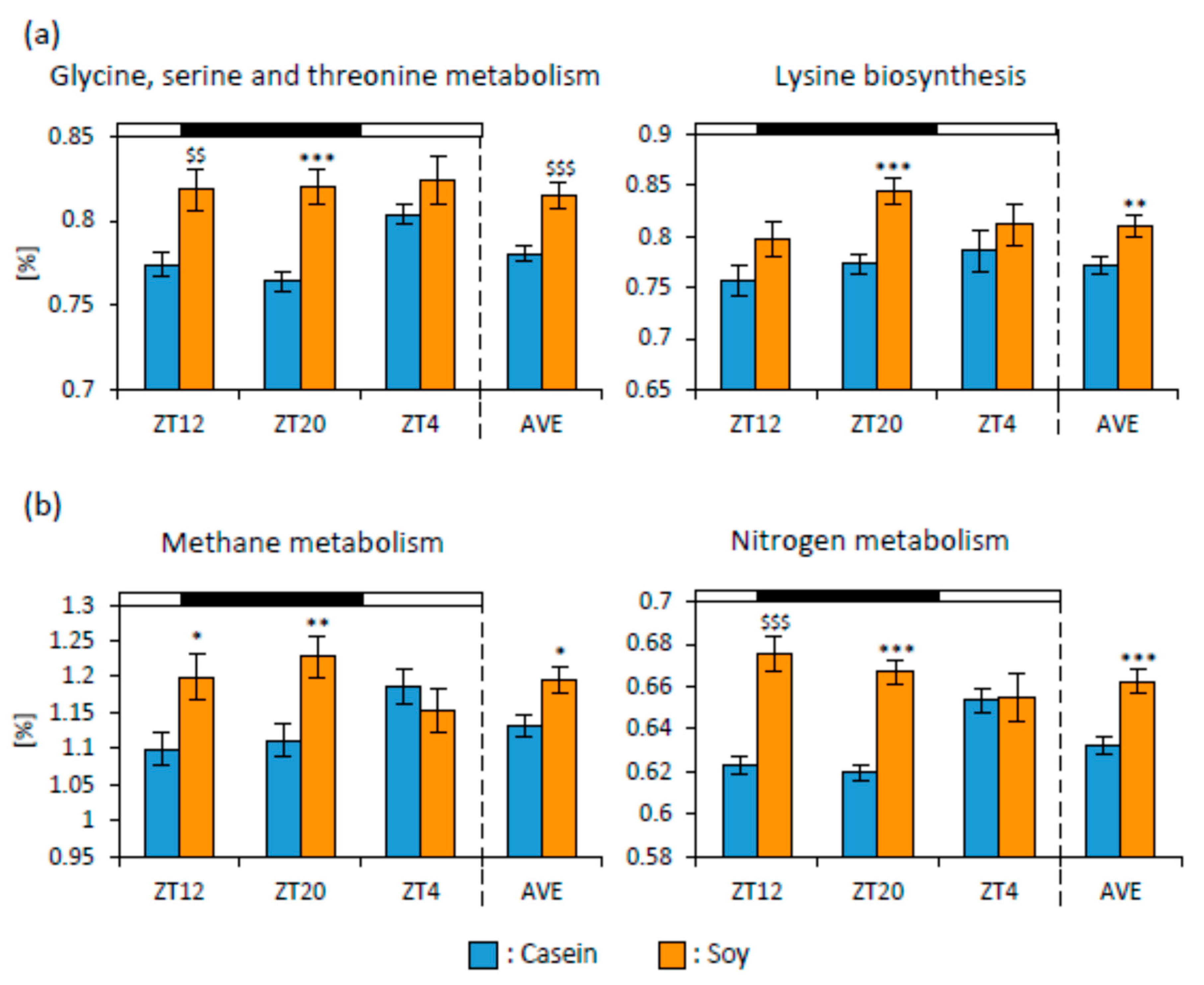
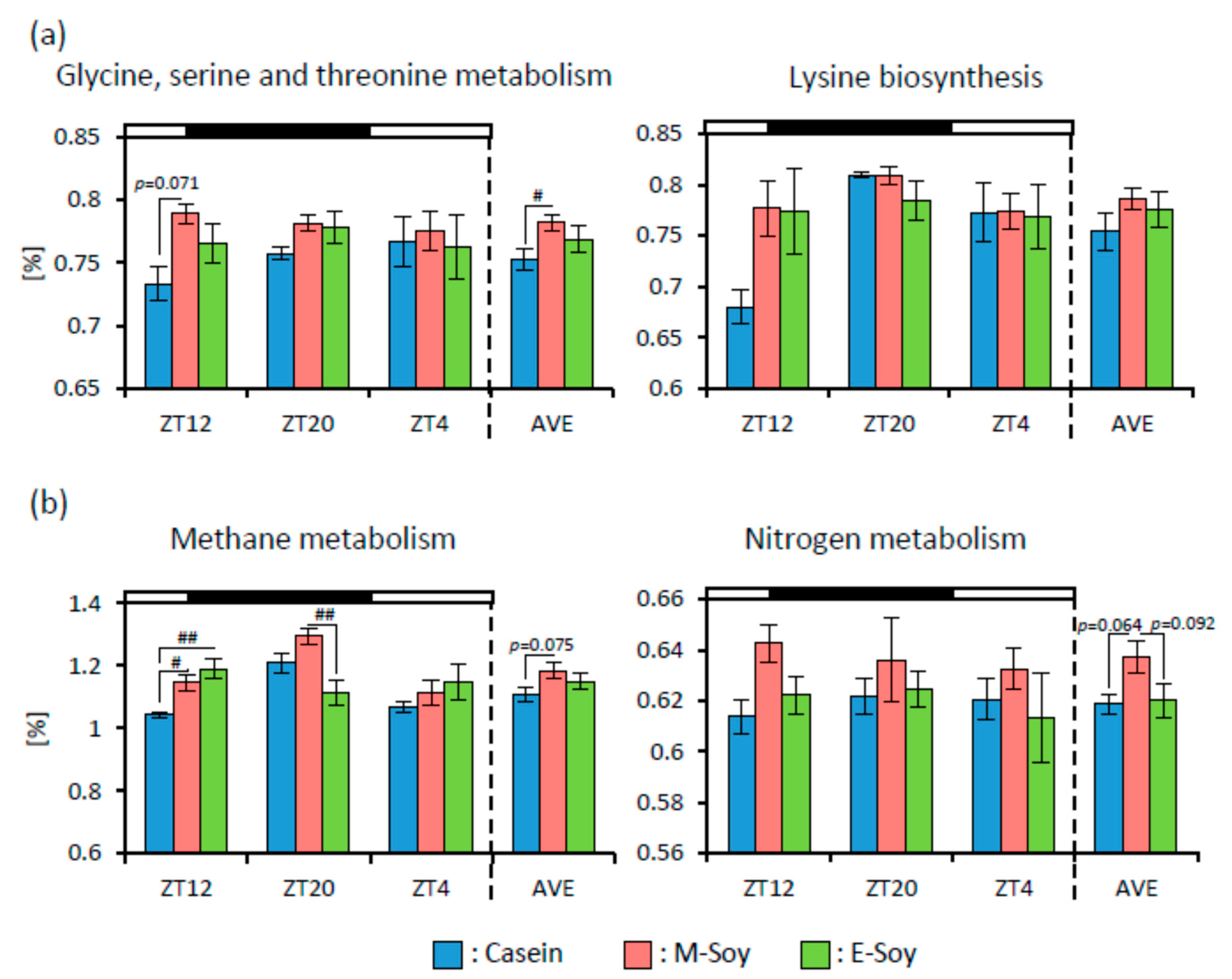
3.2. Soy Protein Intake in the Morning Affected the Gut Microbiota More Than That in the Evening
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Morita, T.; Kasaoka, S.; Oh-hashi, A.; Ikai, M.; Numasaki, Y.; Kiriyama, S. Resistant proteins alter cecal short-chain fatty acid profiles in rats fed high amylose cornstarch. J. Nutr. 1998, 128, 1156–1164. [Google Scholar] [CrossRef]
- Cook, S. Review article: Short chain fatty acids in health and disease. Aliment. Pharmacol. Ther. 1998, 12, 499–507. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The role of ph in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Davila, A.-M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.-H.; Sanz, Y.; Tomé, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107. [Google Scholar] [CrossRef]
- Hamer, H.M.; Preter, V.D.; Windey, K.; Verbeke, K. Functional analysis of colonic bacterial metabolism: Relevant to health? Am. J. Physiol.—Gastrointest. Liver Physiol. 2012, 302, G1–G9. [Google Scholar] [CrossRef] [PubMed]
- Butteiger, D.N.; Hibberd, A.A.; McGraw, N.J.; Napawan, N.; Hall-Porter, J.M.; Krul, E.S. Soy protein compared with milk protein in a western diet increases gut microbial diversity and reduces serum lipids in golden syrian hamsters. J. Nutr. 2016, 146, 697–705. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Kuda, T.; Yazaki, T.; Takahashi, H.; Kimura, B. Caecal fermentation, putrefaction and microbiotas in rats fed milk casein, soy protein or fish meal. Appl. Microbiol. Biotechnol. 2014, 98, 2779–2787. [Google Scholar] [CrossRef] [PubMed]
- Gachon, F.; Nagoshi, E.; Brown, S.A.; Ripperger, J.; Schibler, U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma 2004, 113, 103–112. [Google Scholar] [CrossRef]
- Narishige, S.; Kuwahara, M.; Shinozaki, A.; Okada, S.; Ikeda, Y.; Kamagata, M.; Tahara, Y.; Shibata, S. Effects of caffeine on circadian phase, amplitude and period evaluated in cells in vitro and peripheral organs in vivo in per2::Luciferase mice. Br. J. Pharmacol. 2014, 171, 5858–5869. [Google Scholar] [CrossRef]
- Tahara, Y.; Shibata, S. Chrono-biology, chrono-pharmacology, and chrono-nutrition. J. Pharmacol. Sci. 2014, 124, 320–335. [Google Scholar] [CrossRef] [PubMed]
- Furutani, A.; Ikeda, Y.; Itokawa, M.; Nagahama, H.; Ohtsu, T.; Furutani, N.; Kamagata, M.; Yang, Z.-H.; Hirasawa, A.; Tahara, Y.; et al. Fish oil accelerates diet-induced entrainment of the mouse peripheral clock via gpr120. PLoS ONE 2015, 10, e0132472. [Google Scholar] [CrossRef]
- Tahara, Y.; Shiraishi, T.; Kikuchi, Y.; Haraguchi, A.; Kuriki, D.; Sasaki, H.; Motohashi, H.; Sakai, T.; Shibata, S. Entrainment of the mouse circadian clock by sub-acute physical and psychological stress. Sci. Rep. 2015, 5, 11417. [Google Scholar] [CrossRef]
- Tahara, Y.; Shibata, S. Circadian rhythms of liver physiology and disease: Experimental and clinical evidence. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci. Rep. 2018, 8, 1395. [Google Scholar] [CrossRef]
- Thaiss Christoph, A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler Anouk, C.; Abramson, L.; Katz Meirav, N.; Korem, T.; Zmora, N.; et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Takahashi, M.; Ozaki, M.; Miyashita, M.; Fukazawa, M.; Nakaoka, T.; Wakisaka, T.; Matsui, Y.; Hibi, M.; Osaki, N.; Shibata, S. Effects of timing of acute catechin-rich green tea ingestion on postprandial glucose metabolism in healthy men. J. Nutr. Biochem. 2019, 73, 108221. [Google Scholar] [CrossRef]
- Oishi, K.; Konishi, T.; Hashimoto, C.; Yamamoto, S.; Takahashi, Y.; Shiina, Y. Dietary fish oil differentially ameliorates high-fructose diet-induced hepatic steatosis and hyperlipidemia in mice depending on time of feeding. J. Nutr. Biochem. 2018, 52, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Miyakawa, H.; Watanabe, A.; Nakayama, Y.; Lyu, Y.; Hama, K.; Shibata, S. Mice microbiota composition changes by inulin feeding with a long fasting period under a two-meals-per-day schedule. Nutrients 2019, 11, 2802. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Fukui, K.; Takamatsu, K.; Hashimoto, Y.; Yamamoto, T. Soy protein isolate and its hydrolysate reduce body fat of dietary obese rats and genetically obese mice (yellow kk). Nutrition 2000, 16, 349–354. [Google Scholar] [CrossRef]
- Panasevich, M.R.; Schuster, C.M.; Phillips, K.E.; Meers, G.M.; Chintapalli, S.V.; Wankhade, U.D.; Shankar, K.; Butteiger, D.N.; Krul, E.S.; Thyfault, J.P.; et al. Soy compared with milk protein in a western diet changes fecal microbiota and decreases hepatic steatosis in obese oletf rats. J. Nutr. Biochem. 2017, 46, 125–136. [Google Scholar] [CrossRef]
- Watanabe, K.; Igarashi, M.; Li, X.; Nakatani, A.; Miyamoto, J.; Inaba, Y.; Sutou, A.; Saito, T.; Sato, T.; Tachibana, N.; et al. Dietary soybean protein ameliorates high-fat diet-induced obesity by modifying the gut microbiota-dependent biotransformation of bile acids. PLoS ONE 2018, 13, e0202083. [Google Scholar] [CrossRef]
- Kohno, M.; Hirotsuka, M.; Kito, M.; Matsuzawa, Y. Decreases in serum triacylglycerol and visceral fat mediated by dietary soybean & beta-conglycinin. J. Atheroscler. Thromb. 2006, 13, 247–255. [Google Scholar]
- Yamazaki, T.; Kishimoto, K.; Miura, S.; Ezaki, O. Dietary β-conglycinin prevents fatty liver induced by a high-fat diet by a decrease in peroxisome proliferator-activated receptor γ2 protein. J. Nutr. Biochem. 2012, 23, 123–132. [Google Scholar] [CrossRef]
- Feillet, C.A.; Ripperger, J.A.; Magnone, M.C.; Dulloo, A.; Albrecht, U.; Challet, E. Lack of food anticipation in per2 mutant mice. Curr. Biol. 2006, 16, 2016–2022. [Google Scholar] [CrossRef]
- Hirao, A.; Nagahama, H.; Tsuboi, T.; Hirao, M.; Tahara, Y.; Shibata, S. Combination of starvation interval and food volume determines the phase of liver circadian rhythm in per2: Luc knock-in mice under two meals per day feeding. Am. J. Physiol. -Gastrointest. Liver Physiol. 2010, 299, G1045–G1053. [Google Scholar] [CrossRef] [PubMed]
- Huazano, A.; López, M.G. Metabolism of short chain fatty acids in colon and faeces of mice after a supplementation of diets with agave fructans. Lipid Metab. 2013, 8, 163–182. [Google Scholar]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.-W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy japanese and its microbial and functional uniqueness. DNA Res. 2016, 23, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- den Besten, G.; Gerding, A.; van Dijk, T.H.; Ciapaite, J.; Bleeker, A.; van Eunen, K.; Havinga, R.; Groen, A.K.; Reijngoud, D.-J.; Bakker, B.M. Protection against the metabolic syndrome by guar gum-derived short-chain fatty acids depends on peroxisome proliferator-activated receptor γ and glucagon-like peptide-1. PLoS ONE 2015, 10, e0136364. [Google Scholar] [CrossRef]
- Ding, X.; Saxena, N.K.; Lin, S.; Gupta, N.A.; Anania, F.A. Exendin-4, a glucagon-like protein-1 (glp-1) receptor agonist, reverses hepatic steatosis in ob/ob mice. Hepatology 2006, 43, 173–181. [Google Scholar] [CrossRef]
- ZHANG, J.; Sun, Y.s.; Zhao, L.; Chen, T.; Fan, M.; Jiao, H.C.; Zhao, J.; Wang, X.; Li, F.c.; Li, H.; et al. Scfas-induced glp-1 secretion links the regulation of gut microbiome on hepatic lipogenesis in chickens. BioRxiv 2019. [Google Scholar] [CrossRef]
- Huang, H.; Krishnan, H.B.; Pham, Q.; Yu, L.L.; Wang, T.T.Y. Soy and gut microbiota: Interaction and implication for human health. J. Agric. Food Chem. 2016, 64, 8695–8709. [Google Scholar] [CrossRef]
- Lee, K.-Y.; So, J.-S.; Heo, T.-R. Thin layer chromatographic determination of organic acids for rapid identification of bifidobacteria at genus level. J. Microbiol. Methods 2001, 45, 1–6. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus enterococcus: Between probiotic potential and safety concerns-an update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shoji, H.; Sato, H.; Nagata, S.; Ohtsuka, Y.; Shimizu, T.; Yamashiro, Y. Effects of oral administration of bifidobacterium breve on fecal lactic acid and short-chain fatty acids in low birth weight infants. J. Pediatric Gastroenterol. Nutr. 2007, 44, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Casalta, E.; Montel, M.-C. Safety assessment of dairy microorganisms: The lactococcus genus. Int. J. Food Microbiol. 2008, 126, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, A.; Kilcoyne, M.; Clyne, M.; Joshi, L.; Carrington, S.; Martin, S.T.; Coffey, J.C.; Winter, D.C.; et al. A preliminary study examining the binding capacity of akkermansia muciniphila and desulfovibrio spp., to colonic mucin in health and ulcerative colitis. PLoS ONE 2015, 10, e0135280. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong Eric, A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick James, A.J.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef]
- Kuroda, H.; Tahara, Y.; Saito, K.; Ohnishi, N.; Kubo, Y.; Seo, Y.; Otsuka, M.; Fuse, Y.; Ohura, Y.; Hirao, A.; et al. Meal frequency patterns determine the phase of mouse peripheral circadian clocks. Sci. Rep. 2012, 2, 711. [Google Scholar] [CrossRef]
- Higaki, N.; Sato, K.; Suda, H.; Suzuka, T.; Komori, T.; Saeki, T.; Nakamura, Y.; Ohtsuki, K.; Iwami, K.; Kanamoto, R. Evidence for the existence of a soybean resistant protein that captures bile acid and stimulates its fecal excretion. Biosci. Biotechnol. Biochem. 2006, 70, 2844–2852. [Google Scholar] [CrossRef]
- Worthmann, A.; John, C.; Rühlemann, M.C.; Baguhl, M.; Heinsen, F.-A.; Schaltenberg, N.; Heine, M.; Schlein, C.; Evangelakos, I.; Mineo, C.; et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat. Med. 2017, 23, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gøbel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef] [PubMed]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.D.; Bailey, M.T. Impact of stressor exposure on the interplay between commensal microbiota and host inflammation. Gut Microbes 2014, 5, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, N.; Tahara, Y.; Kuriki, D.; Haraguchi, A.; Shibata, S. Warm water bath stimulates phase-shifts of the peripheral circadian clocks in per2: Luciferase mouse. PLoS ONE 2014, 9, e100272. [Google Scholar] [CrossRef]
- Hoogerwerf, W.A. Role of clock genes in gastrointestinal motility. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G549–G555. [Google Scholar] [CrossRef]
- Pilorz, V.; Helfrich-Förster, C.; Oster, H. The role of the circadian clock system in physiology. Pflügers Arch. Eur. J. Physiol. 2018, 470, 227–239. [Google Scholar] [CrossRef]
- Gomez, D.E.; Arroyo, L.G.; Costa, M.C.; Viel, L.; Weese, J.S. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J. Vet. Intern. Med. 2017, 31, 928–939. [Google Scholar] [CrossRef]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef]

| Casein Diet | Soy Diet | |
|---|---|---|
| Casein | 22.86 | - |
| Soy protein | - | 23.78 |
| L-cysteine | 0.18 | 0.18 |
| βcorn starch | 13.71 | 12.79 |
| αcorn starch | 15.5 | 15.5 |
| Sucrose | 10 | 10 |
| Soybean oil | 4 | 4 |
| Lard | 24 | 24 |
| Cellulose | 5 | 5 |
| Mineral mixture | 3.5 | 3.5 |
| Vitamin mixture | 1 | 1 |
| Choline bitartrate | 0.25 | 0.25 |
| Total | 100 | 100 |
| Gene | Forward | Reverse |
|---|---|---|
| Acc1 | GCACTCCCGATTCATAATG | CCCAAATCAGAAAGTGTATC |
| Cyp7α1 | AGACCGCACATAAAGCCCGG | CTTTCATTGCTTCAGGGCTC |
| Fas | TGGGTTCTAGCCAGCAGAGT | ACCACCAGAGACCGTTATGC |
| Gapdh | TGGTGAAGGTCGGTGTGAAC | AATGAAGGGGTCGTTGATGG |
| Hmgcr | GATCATCCAGTTGGTCAATGC | GCAAGCTTTGTGGAGAGGAG |
| Srebp1c | CGCTACCGGTCTTCTATCAATG | CAAGAAGCGGATGTAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamura, K.; Sasaki, H.; Shiga, K.; Miyakawa, H.; Shibata, S. The Timing Effects of Soy Protein Intake on Mice Gut Microbiota. Nutrients 2020, 12, 87. https://doi.org/10.3390/nu12010087
Tamura K, Sasaki H, Shiga K, Miyakawa H, Shibata S. The Timing Effects of Soy Protein Intake on Mice Gut Microbiota. Nutrients. 2020; 12(1):87. https://doi.org/10.3390/nu12010087
Chicago/Turabian StyleTamura, Konomi, Hiroyuki Sasaki, Kazuto Shiga, Hiroki Miyakawa, and Shigenobu Shibata. 2020. "The Timing Effects of Soy Protein Intake on Mice Gut Microbiota" Nutrients 12, no. 1: 87. https://doi.org/10.3390/nu12010087
APA StyleTamura, K., Sasaki, H., Shiga, K., Miyakawa, H., & Shibata, S. (2020). The Timing Effects of Soy Protein Intake on Mice Gut Microbiota. Nutrients, 12(1), 87. https://doi.org/10.3390/nu12010087





