Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Measurement of Appendicular Lean Mass by Dual X-ray Absorptiometry (DXA)
2.3. Blood Sample and Stool Collection
2.4. Measurement of Circulating Inflammatory Mediators
2.5. Determination of Circulating Amino Acids
2.6. Gut Microbiota DNA Extraction, 16S rRNA Amplification, and Sequencing
2.7. Statistical Analysis
- CovSel algorithm is used to select relevant variables and calculate a regression model between the first block and the responses
- The second block is orthogonalized with respect to the variables selected in the first block
- CovSel algorithm is used to select relevant variables and calculate a regression model between the orthogonalized second block and the residuals from the first fit
- The third block is orthogonalized with respect to the variables selected in the first and second blockswhere
- CovSel algorithm is used to select relevant variables and calculate a regression model between the orthogonalized third block and the residuals from the second fit
- An overall prediction model is built aswhere the predicted response is calculated as
3. Results
3.1. Characteristics of the Study Population
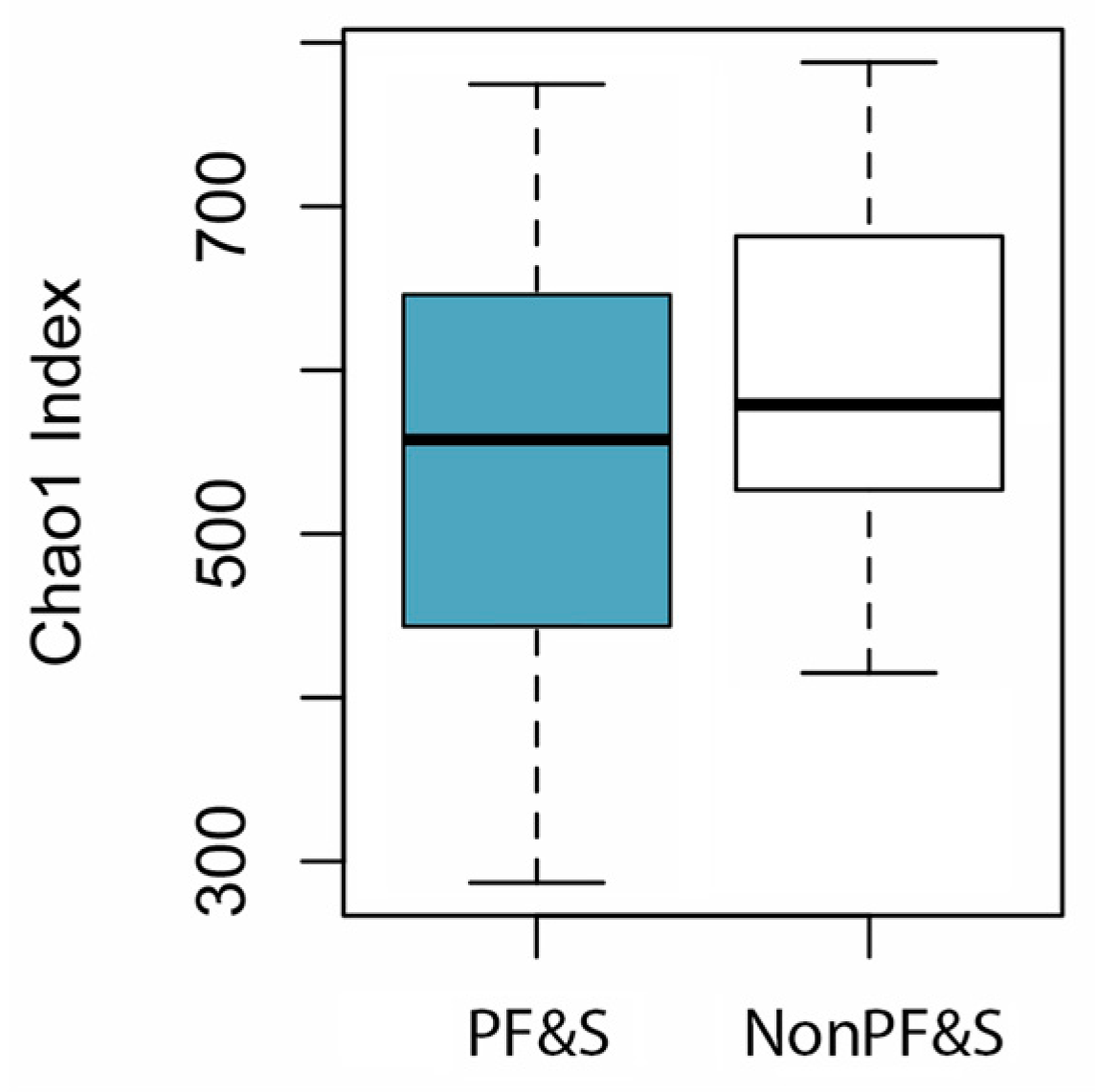
3.2. Features of Gut Microbiota According to the Presence of PF&S
3.3. SO-CovSel Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hirani, V.; Blyth, F.; Naganathan, V.; Le Couteur, D.G.; Seibel, M.J.; Waite, L.M.; Handelsman, D.J.; Cumming, R.G. Sarcopenia is associated with incident disability, institutionalization, and mortality in community-dwelling older men: The Concord Health and Ageing in Men Project. J. Am. Med. Dir. Assoc. 2015, 16, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Marzetti, E.; Calvani, R.; Tosato, M.; Cesari, M.; Di Bari, M.; Cherubini, A.; Collamati, A.; D’Angelo, E.; Pahor, M.; Bernabei, R.; et al. SPRINTT Consortium. Sarcopenia: An overview. Aging Clin. Exp. Res. 2017, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Calvani, R.; Marzetti, E. Frailty in older persons. Clin. Geriatr. Med. 2017, 33, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Calvani, R.; Cesari, M.; Tosato, M.; Martone, A.M.; Bernabei, R.; Onder, G.; Marzetti, E. Sarcopenia as the biological substrate of physical frailty. Clin. Geriatr. Med. 2015, 31, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Landi, F.; Vellas, B.; Bernabei, R.; Marzetti, E. Sarcopenia and physical frailty: Two sides of the same coin. Front. Aging Neurosci. 2014, 6, 192. [Google Scholar] [CrossRef]
- Cesari, M.; Landi, F.; Calvani, R.; Cherubini, A.; Di Bari, M.; Kortebein, P.; Del Signore, S.; Le Lain, R.; Vellas, B.; Pahor, M.; et al. SPRINTT Consortium. Rationale for a preliminary operational definition of physical frailty and sarcopenia in the SPRINTT trial. Aging Clin. Exp. Res. 2017, 29, 81–88. [Google Scholar] [CrossRef]
- Marzetti, E.; Calvani, R.; Landi, F.; Hoogendijk, E.O.; Fougère, B.; Vellas, B.; Pahor, M.; Bernabei, R.; Cesari, M. SPRINTT Consortium. Innovative Medicines Initiative: The SPRINTT project. J. Frailty Aging 2015, 4, 207–208. [Google Scholar] [CrossRef]
- Calvani, R.; Marini, F.; Cesari, M.; Tosato, M.; Anker, S.D.; von Haehling, S.; Miller, R.R.; Bernabei, R.; Landi, F.; Marzetti, E. SPRINTT consortium. Biomarkers for physical frailty and sarcopenia: State of the science and future developments. J. Cachexia Sarcopenia Muscle 2015, 6, 278–286. [Google Scholar] [CrossRef]
- Picca, A.; Calvani, R.; Bossola, M.; Allocca, E.; Menghi, A.; Pesce, V.; Lezza, A.M.S.; Bernabei, R.; Landi, F.; Marzetti, E. Update on mitochondria and muscle aging: All wrong roads lead to sarcopenia. Biol. Chem. 2018, 399, 421–436. [Google Scholar] [CrossRef]
- Calvani, R.; Picca, A.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Coelho-Junior, H.J.; Bossola, M.; Urbani, A.; et al. A distinct pattern of circulating amino acids characterizes older persons with physical frailty and sarcopenia: Results from the BIOSPHERE study. Nutrients 2018, 10, 1691. [Google Scholar] [CrossRef]
- Marzetti, E.; Picca, A.; Marini, F.; Biancolillo, A.; Coelho-Junior, H.J.; Gervasoni, J.; Bossola, M.; Cesari, M.; Onder, G.; Landi, F.; et al. Inflammatory signatures in older persons with physical frailty and sarcopenia: The frailty “cytokinome” at its core. Exp. Gerontol. 2019, 122, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Calvani, R.; Tosato, M.; Martone, A.M.; Ortolani, E.; Savera, G.; Sisto, A.; Marzetti, E. Anorexia of aging: Risk factors, consequences, and potential treatments. Nutrients 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Calvani, R.; Tosato, M.; Martone, A.M.; Ortolani, E.; Savera, G.; D’Angelo, E.; Sisto, A.; Marzetti, E. Protein intake and muscle health in old age: From biological plausibility to clinical evidence. Nutrients 2016, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Cesari, M.; Calvani, R.; Cherubini, A.; Di Bari, M.; Bejuit, R.; Mshid, J.; Andrieu, S.; Sinclair, A.J.; Sieber, C.C.; et al. SPRINTT Consortium. The “Sarcopenia and Physical fRailty IN older people: Multi-componenT Treatment strategies” (SPRINTT) randomized controlled trial: Design and methods. Aging Clin. Exp. Res. 2017, 29, 89–100. [Google Scholar] [CrossRef]
- Chen, H.; Rejeski, W.J.; Gill, T.M.; Guralnik, J.; King, A.C.; Newman, A.; Blair, S.N.; Conroy, D.; Liu, C.; Manini, T.M.; et al. LIFE Study. A comparison of self-report indices of major mobility disability to failure on the 400-m walk test: The LIFE study. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 513–518. [Google Scholar] [CrossRef]
- Roubenoff, R. Sarcopenic obesity: Does muscle loss cause fat gain? Lessons from rheumatoid arthritis and osteoarthritis. Ann. N. Y. Acad. Sci. 2006, 904, 553–557. [Google Scholar] [CrossRef]
- Binkley, N.; Krueger, D.; Buehring, B. What’s in a name revisited: Should osteoporosis and sarcopenia be considered components of “dysmobility syndrome?”. Osteoporos. Int. 2013, 24, 2955–2959. [Google Scholar] [CrossRef]
- Porter Starr, K.N.; McDonald, S.R.; Bales, C.W. Obesity and physical frailty in older adults: A scoping review of lifestyle intervention trials. J. Am. Med. Dir. Assoc. 2014, 15, 240–250. [Google Scholar] [CrossRef]
- Hulsegge, G.; Herber-Gast, G.-C.M.; Spijkerman, A.M.W.; Susan, H.; Picavet, J.; van der Schouw, Y.T.; Bakker, S.J.L.; Gansevoort, R.T.; Dollé, M.E.T.; Smit, H.A.; et al. Obesity and age-related changes in markers of oxidative stress and inflammation across four generations. Obesity 2016, 24, 1389–1396. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; Carlson, C.L.; Visser, M.; Kelley, D.E.; Scherzinger, A.; Harris, T.B.; Stamm, E.; Newman, A.B. Attenuation of skeletal muscle and strength in the elderly: The health ABC study. J. Appl. Physiol. 2001, 90, 2157–2165. [Google Scholar] [CrossRef]
- Visser, M.; Goodpaster, B.H.; Kritchevsky, S.B.; Newman, A.B.; Nevitt, M.; Rubin, S.M.; Simonsick, E.M.; Harris, T.B. Muscle mass, muscle strength, and muscle fat infiltration as predictors of incident mobility limitations in well-functioning older persons. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 324–333. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, E.M. The role of gut microbiota in nutritional status. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Fanelli, F.; Calvani, R.; Mulè, G.; Pesce, V.; Sisto, A.; Pantanelli, C.; Bernabei, R.; Landi, F.; Marzetti, E. Gut dysbiosis and muscle aging: Searching for novel targets against sarcopenia. Mediat. Inflamm. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Calvani, R.; Picca, A.; Marini, F.; Biancolillo, A.; Cesari, M.; Pesce, V.; Lezza, A.M.S.; Bossola, M.; Leeuwenburgh, C.; Bernabei, R.; et al. The “BIOmarkers associated with Sarcopenia and PHysical frailty in EldeRly pErsons” (BIOSPHERE) study: Rationale, design and methods. Eur. J. Intern. Med. 2018, 56, 19–25. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Putignani, L.; Paroni Sterbini, F.; Petito, V.; Picca, A.; Del Chierico, F.; Reddel, S.; Calvani, R.; Marzetti, E.; Sanguinetti, M.; et al. Influence of hepatitis C virus eradication with direct-acting antivirals on the gut microbiota in patients with cirrhosis. Aliment. Pharmacol. Ther. 2018, 48, 1301–1311. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef]
- Marzetti, E.; Cesari, M.; Calvani, R.; Msihid, J.; Tosato, M.; Rodriguez-Mañas, L.; Lattanzio, F.; Cherubini, A.; Bejuit, R.; Di Bari, M.; et al. SPRINTT Consortium. The “Sarcopenia and Physical fRailty IN older people: Multi-componenT Treatment strategies” (SPRINTT) randomized controlled trial: Case finding, screening and characteristics of eligible participants. Exp. Gerontol. 2018, 113, 48–57. [Google Scholar] [CrossRef]
- Guralnik, J.M.; Simonsick, E.M.; Ferrucci, L.; Glynn, R.J.; Berkman, L.F.; Blazer, D.G.; Scherr, P.A.; Wallace, R.B. A short physical performance battery assessing lower extremity function: Association with self-reported disability and prediction of mortality and nursing home admission. J. Gerontol. 1994, 49, M85–M94. [Google Scholar] [CrossRef]
- Studenski, S.A.; Peters, K.W.; Alley, D.E.; Cawthon, P.M.; McLean, R.R.; Harris, T.B.; Ferrucci, L.; Guralnik, J.M.; Fragala, M.S.; Kenny, A.M.; et al. The FNIH sarcopenia project: Rationale, study description, conference recommendations, and final estimates. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 547–558. [Google Scholar] [CrossRef]
- Newman, A.B.; Simonsick, E.M.; Naydeck, B.L.; Boudreau, R.M.; Kritchevsky, S.B.; Nevitt, M.C.; Pahor, M.; Satterfield, S.; Brach, J.S.; Studenski, S.A.; et al. Association of long-distance corridor walk performance with mortality, cardiovascular disease, mobility limitation, and disability. JAMA 2006, 295, 2018–2026. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Bucci, C.; Lo Monaco, M.R.; Bentivoglio, A.R.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial-derived vesicles as candidate biomarkers in Parkinson’s disease: Rationale, design and methods of the EXosomes in PArkiNson Disease (EXPAND) study. Int. J. Mol. Sci. 2019, 20, 2373. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Marini, F.; Roger, J. SO-CovSel: A novel method for variable selection in a multiblock framework. J. Chemom. 2019, e3120. [Google Scholar] [CrossRef]
- Roger, J.M.; Palagos, B.; Bertrand, D.; Fernandez-Ahumada, E. CovSel: Variable selection for highly multivariate and multi-response calibration. Application to IR spectroscopy. Chemom. Intell. Lab. Syst. 2011, 106, 216–223. [Google Scholar] [CrossRef]
- Naes, T.; Tomic, O.; Mevik, B.-H.; Martens, H. Path modelling by sequential PLS regression. J. Chemom. 2011, 25, 28–40. [Google Scholar] [CrossRef]
- Biancolillo, A.; Naes, T. The Sequential and Orthogonalized PLS Regression for Multiblock Regression: Theory, Examples, and Extensions. In Data Fusion Methodology and Applications; Cocchi, M., Ed.; Elsevier: Oxford, UK, 2019; Volume 31, pp. 157–177. [Google Scholar]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Jackson, M.A.; Jackson, M.; Jeffery, I.B.; Beaumont, M.; Bell, J.T.; Clark, A.G.; Ley, R.E.; O’Toole, P.W.; Spector, T.D.; Steves, C.J. Signatures of early frailty in the gut microbiota. Genome Med. 2016, 8, 8. [Google Scholar] [CrossRef]
- Maffei, V.J.; Kim, S.; Blanchard, E.; Luo, M.; Jazwinski, S.M.; Taylor, C.M.; Welsh, D.A. Biological Aging and the Human Gut Microbiota. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1474–1482. [Google Scholar] [CrossRef]
- Ticinesi, A.; Milani, C.; Lauretani, F.; Nouvenne, A.; Mancabelli, L.; Lugli, G.A.; Turroni, F.; Duranti, S.; Mangifesta, M.; Viappiani, A.; et al. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci. Rep. 2017, 7, 11102. [Google Scholar] [CrossRef]
- Haran, J.P.; Bucci, V.; Dutta, P.; Ward, D.; McCormick, B. The nursing home elder microbiome stability and associations with age, frailty, nutrition and physical location. J. Med. Microbiol. 2018, 67, 40–51. [Google Scholar] [CrossRef]
- Verdi, S.; Jackson, M.A.; Beaumont, M.; Bowyer, R.C.E.; Bell, J.T.; Spector, T.D.; Steves, C.J. An investigation into physical frailty as a link between the gut microbiome and cognitive health. Front. Aging Neurosci. 2018, 10, 398. [Google Scholar] [CrossRef]
- Ogawa, T.; Hirose, Y.; Honda-Ogawa, M.; Sugimoto, M.; Sasaki, S.; Kibi, M.; Kawabata, S.; Ikebe, K.; Maeda, Y. Composition of salivary microbiota in elderly subjects. Sci. Rep. 2018, 8, 414. [Google Scholar] [CrossRef] [PubMed]
- Siddharth, J.; Chakrabarti, A.; Pannérec, A.; Karaz, S.; Morin-Rivron, D.; Masoodi, M.; Feige, J.N.; Parkinson, S.J. Aging and sarcopenia associate with specific interactions between gut microbes, serum biomarkers and host physiology in rats. Aging 2017, 9, 1698–1720. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; Konikoff, T.; Nielsen, H.B. Oscillospira and related bacteria—From metagenomic species to metabolic features. Environ. Microbiol. 2017, 19, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Konikoff, T.; Gophna, U. Oscillospira: A central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.A.; Jackson, J.; Stanton, M.; Rojas-Triana, A.; Bober, L.; Laverty, M.; Yang, X.; Zhu, F.; Liu, J.; Wang, S.; et al. Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E2 and cytokines. World J. Gastroenterol. 2009, 15, 5549–5557. [Google Scholar] [CrossRef]
- Saint-Georges-Chaumet, Y.; Edeas, M. Microbiota-mitochondria inter-talk: Consequence for microbiota-host interaction. Pathog. Dis. 2016, 74, ftv096. [Google Scholar] [CrossRef]
- Hénique, C.; Mansouri, A.; Vavrova, E.; Lenoir, V.; Ferry, A.; Esnous, C.; Ramond, E.; Girard, J.; Bouillaud, F.; Prip-Buus, C.; et al. Increasing mitochondrial muscle fatty acid oxidation induces skeletal muscle remodeling toward an oxidative phenotype. FASEB J. 2015, 29, 2473–2483. [Google Scholar] [CrossRef]
- Gumucio, J.P.; Qasawa, A.H.; Ferrara, P.J.; Malik, A.N.; Funai, K.; McDonagh, B.; Mendias, C.L. Reduced mitochondrial lipid oxidation leads to fat accumulation in myosteatosis. FASEB J. 2019, 33, 7863–7881. [Google Scholar] [CrossRef]
- Poggiogalle, E.; Lubrano, C.; Gnessi, L.; Mariani, S.; Di Martino, M.; Catalano, C.; Lenzi, A.; Donini, L.M. The decline in muscle strength and muscle quality in relation to metabolic derangements in adult women with obesity. Clin. Nutr. 2019, 38, 2430–2435. [Google Scholar] [CrossRef]
- Sachs, S.; Zarini, S.; Kahn, D.E.; Harrison, K.A.; Perreault, L.; Phang, T.; Newsom, S.A.; Strauss, A.; Kerege, A.; Schoen, J.A.; et al. Intermuscular adipose tissue directly modulates skeletal muscle insulin sensitivity in humans. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E866–E879. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Yokoyama, H.; Imai, D.; Takeda, R.; Ota, A.; Kawai, E.; Hisada, T.; Emoto, M.; Suzuki, Y.; Okazaki, K. Aerobic exercise training with brisk walking increases intestinal bacteroides in healthy elderly women. Nutrients 2019, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Calvani, R.; Tosato, M.; Martone, A.M.; Picca, A.; Ortolani, E.; Savera, G.; Salini, S.; Ramaschi, M.; Bernabei, R.; et al. Animal-derived protein consumption is associated with muscle mass and strength in community-dwellers: Results from the Milan EXPO survey. J. Nutr. Health Aging 2017, 21, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Portune, K.J.; Steuer, N.; Lan, A.; Cerrudo, V.; Audebert, M.; Dumont, F.; Mancano, G.; Khodorova, N.; Andriamihaja, M.; et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 2017, 106, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Wagenmakers, A.J.M. Protein and amino acid metabolism in human muscle. Adv. Exp. Med. Biol. 1998, 441, 307–319. [Google Scholar] [CrossRef]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef]
- Lu, Y.; Karagounis, L.G.; Ng, T.P.; Carre, C.; Narang, V.; Wong, G.; Ying Tan, C.T.; Zin Nyunt, M.S.; Gao, Q.; Abel, B.; et al. Systemic and metabolic signature of sarcopenia in community-dwelling older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2019. [Google Scholar] [CrossRef]
- Adachi, Y.; Ono, N.; Imaizumi, A.; Muramatsu, T.; Andou, T.; Shimodaira, Y.; Nagao, K.; Kageyama, Y.; Mori, M.; Noguchi, Y.; et al. Plasma amino acid profile in severely frail elderly patients in Japan. Int. J. Gerontol. 2018, 12, 290–293. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.J.; Kiesswetter, E.; Drey, M.; Sieber, C.C. Nutrition, frailty, and sarcopenia. Aging Clin. Exp. Res. 2017, 29, 43–48. [Google Scholar] [CrossRef]
- Thevaranjan, N.; Puchta, A.; Schulz, C.; Naidoo, A.; Szamosi, J.C.; Verschoor, C.P.; Loukov, D.; Schenck, L.P.; Jury, J.; Foley, K.P.; et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 2017, 21, 455–466. [Google Scholar] [CrossRef]
- Grosicki, G.J.; Fielding, R.A.; Lustgarten, M.S. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: Biological basis for a gut-muscle axis. Calcif. Tissue Int. 2018, 102, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Ticinesi, A.; Lauretani, F.; Milani, C.; Nouvenne, A.; Tana, C.; Del Rio, D.; Maggio, M.; Ventura, M.; Meschi, T. Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: Is there a gut–muscle axis? Nutrients 2017, 9, 1303. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, L.; Fabbri, E. Inflammageing: Chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018, 15, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Sovran, B.; Hugenholtz, F.; Elderman, M.; Van Beek, A.A.; Graversen, K.; Huijskes, M.; Boekschoten, M.V.; Savelkoul, H.F.J.; De Vos, P.; Dekker, J.; et al. Age-associated impairment of the mucus barrier function is associated with profound changes in microbiota and immunity. Sci. Rep. 2019, 9, 1437. [Google Scholar] [CrossRef]
- Ticinesi, A.; Lauretani, F.; Tana, C.; Nouvenne, A.; Ridolo, E.; Meschi, T. Exercise and immune system as modulators of intestinal microbiome: Implications for the gut-muscle axis hypothesis. Exerc. Immunol. Rev. 2019, 25, 84–95. [Google Scholar]
- Ticinesi, A.; Nouvenne, A.; Cerundolo, N.; Catania, P.; Prati, B.; Tana, C.; Meschi, T. Gut microbiota, muscle mass and function in aging: A focus on physical frailty and sarcopenia. Nutrients 2019, 11, 1633. [Google Scholar] [CrossRef]

| Cytokines | IFNγ, IL1β, IL1Ra, IL2, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12, IL13, IL15, IL17, TNF-α |
| Chemokines | CCL5, CCL11, IP-10, MCP-1, MIP-1α, MIP-1β |
| Growth factors | FGF-β, G-CSF, GM-CSF, PDGF-BB |
| Data Block | Biological Pathway | Variables |
|---|---|---|
| Matrix 1 | Inflammation | CCL5, CCL11, IFN-γ, FGF-β, G-CSF, GM-CSF, IL1β, IL1ra, IL2, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12, IL13, IL15, IL17, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, TNF-α |
| Matrix 2 | Protein/amino acid metabolism | 1-methylhistidine, 3-methylhistidine, 4-hydroxyproline, α-aminobutyric acid, β-alanine, β-aminobutyric acid, γ-aminobutyric acid, alanine, aminoadipic acid, anserine, arginine, asparagine, aspartic acid, carnosine, citrulline, cystathionine, cystine, ethanolamine, glutamic acid, glycine, histidine, isoleucine, leucine, lysine, methionine, ornithine, phenylalanine, phosphoethanolamine, phosphoserine, proline, sarcosine, serine, taurine, threonine, tryptophan, tyrosine, valine |
| Matrix 3 | Gut microbiota | Actinobacteria, Adlercreutzia, Aerostipes, Aerotruncus, Akkermansia, Alcaligenaceae, Atopobium, Bacteroidaceae, Bacteroides, Bacteroidetes, Barnesiellaceae, Bifidobacteriaceae, Bifidobacterium, Bilophila, Blautia, Carnobacteriaceae, Christensenella, Christensenellaceae, Clostridiaceae, Collinsella, Coprococcus, Coriobacteriaceae, Cyanobacteria, Dehalobacteriaceae, Dehalobacterium, Desulfovibrionaceae, Dethiosulfovibrionaceae, Dialister, Dorea, Eggerthella, Enterobacteriaceae, Enterococcaceae, Enterococcus, Erysipelotrichaceae, EtOH8, Eubacterium, Euryarchaeota, Faecalibacterium, Firmicutes, Granulicatella, Haemophilus, Lachnobacterium, Lachnospira, Lachnospiraceae, Lactobacillaceae, Lactobacillus, Methanobacteriaceae, Methanobrevibacter, Mogibacteriacea, Oscillospira, Parabacteroides, Paraprevotella, Paraprevotellaceae, Pasteurellaceae, Peptostreptococcaceae, Phascolarctobacterium, Porphyromonadaceae, Prevotella, Prevotellaceae, Proteobacteria, Pyramidobacter, Rikenellaceae, Roseburia, Ruminococcaceae, Ruminococcus, Ruminococcus, S24-7, Slackia, Streptococcaceae, Streptococcus, Sutterella, Synergistetes, Tenericutes, TM7, Veillonella, Veillonellaceae, Verrucomicrobia, Verrucomicrobiaceae |
| PF&S (n = 18) | NonPF&S (n = 17) | p | |
|---|---|---|---|
| Age, years (mean ± SD) | 75.5 ± 3.9 | 73.9 ± 3.2 | 0.2204 |
| Gender (female), n (%) | 10 (56) | 5 (29) | 0.2223 |
| BMI, kg/m2 (mean ± SD) | 32.14 ± 6.02 | 26.27 ± 2.55 | 0.0008 |
| SPPB (mean ± SD) | 7.19 ± 1.22 | 11.24 ± 0.97 | <0.0001 |
| aLM, kg (mean ± SD) | 17.75 ± 3.17 | 22.50 ± 2.93 | <0.0001 |
| aLMBMI (mean ± SD) | 0.55 ± 0.11 | 0.87 ± 0.15 | <0.0001 |
| Number of disease conditions * (mean ± SD) | 3.2 ± 1.7 | 3.0 ± 2.1 | 0.8046 |
| Number of medications ** (mean ± SD) | 3.4 ± 1.2 | 2.9 ± 1.6 | 0.1034 |
| PF&S (n = 18) | nonPF&S (n = 17) | |
|---|---|---|
| MIP-1α (pg/mL) | 2.98 (11.04) | 10.64 (11.15) |
| Aspartic acid (µmol/L) | 26.95 (9.33) | 16.10 (9.28) |
| Threonine (µmol/L) | 109.90 (33.60) | 125.80 (55.60) |
| Barnesiellaceae (log2FC) | 0.0010 (0.007) | 0.0030 (0.003) |
| Christensenellaceae (log2FC) | 0.0004 (0.005) | 0.0023 (0.004) |
| Oscillospira (log2FC) | 0.0147 (0.227) | 0.0109 (0.009) |
| Ruminococcus (log2FC) | 0.0674 (0.091) | 0.0620 (0.058) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picca, A.; Ponziani, F.R.; Calvani, R.; Marini, F.; Biancolillo, A.; Coelho-Júnior, H.J.; Gervasoni, J.; Primiano, A.; Putignani, L.; Del Chierico, F.; et al. Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study. Nutrients 2020, 12, 65. https://doi.org/10.3390/nu12010065
Picca A, Ponziani FR, Calvani R, Marini F, Biancolillo A, Coelho-Júnior HJ, Gervasoni J, Primiano A, Putignani L, Del Chierico F, et al. Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study. Nutrients. 2020; 12(1):65. https://doi.org/10.3390/nu12010065
Chicago/Turabian StylePicca, Anna, Francesca Romana Ponziani, Riccardo Calvani, Federico Marini, Alessandra Biancolillo, Hélio José Coelho-Júnior, Jacopo Gervasoni, Aniello Primiano, Lorenza Putignani, Federica Del Chierico, and et al. 2020. "Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study" Nutrients 12, no. 1: 65. https://doi.org/10.3390/nu12010065
APA StylePicca, A., Ponziani, F. R., Calvani, R., Marini, F., Biancolillo, A., Coelho-Júnior, H. J., Gervasoni, J., Primiano, A., Putignani, L., Del Chierico, F., Reddel, S., Gasbarrini, A., Landi, F., Bernabei, R., & Marzetti, E. (2020). Gut Microbial, Inflammatory and Metabolic Signatures in Older People with Physical Frailty and Sarcopenia: Results from the BIOSPHERE Study. Nutrients, 12(1), 65. https://doi.org/10.3390/nu12010065













