Lupin Peptide T9 (GQEQSHQDEGVIVR) Modulates the Mutant PCSK9D374Y Pathway: in vitro Characterization of its Dual Hypocholesterolemic Behavior
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. HMGCoAR Activity Assay
2.3. Cell Culture Conditions and Transfection
2.4. Western Blot Analysis
2.5. In Cell-Western
2.6. Assay for the Evaluation of Fluorescent LDL Uptake by HepG2 Cells
2.7. Statistical Analysis
3. Results
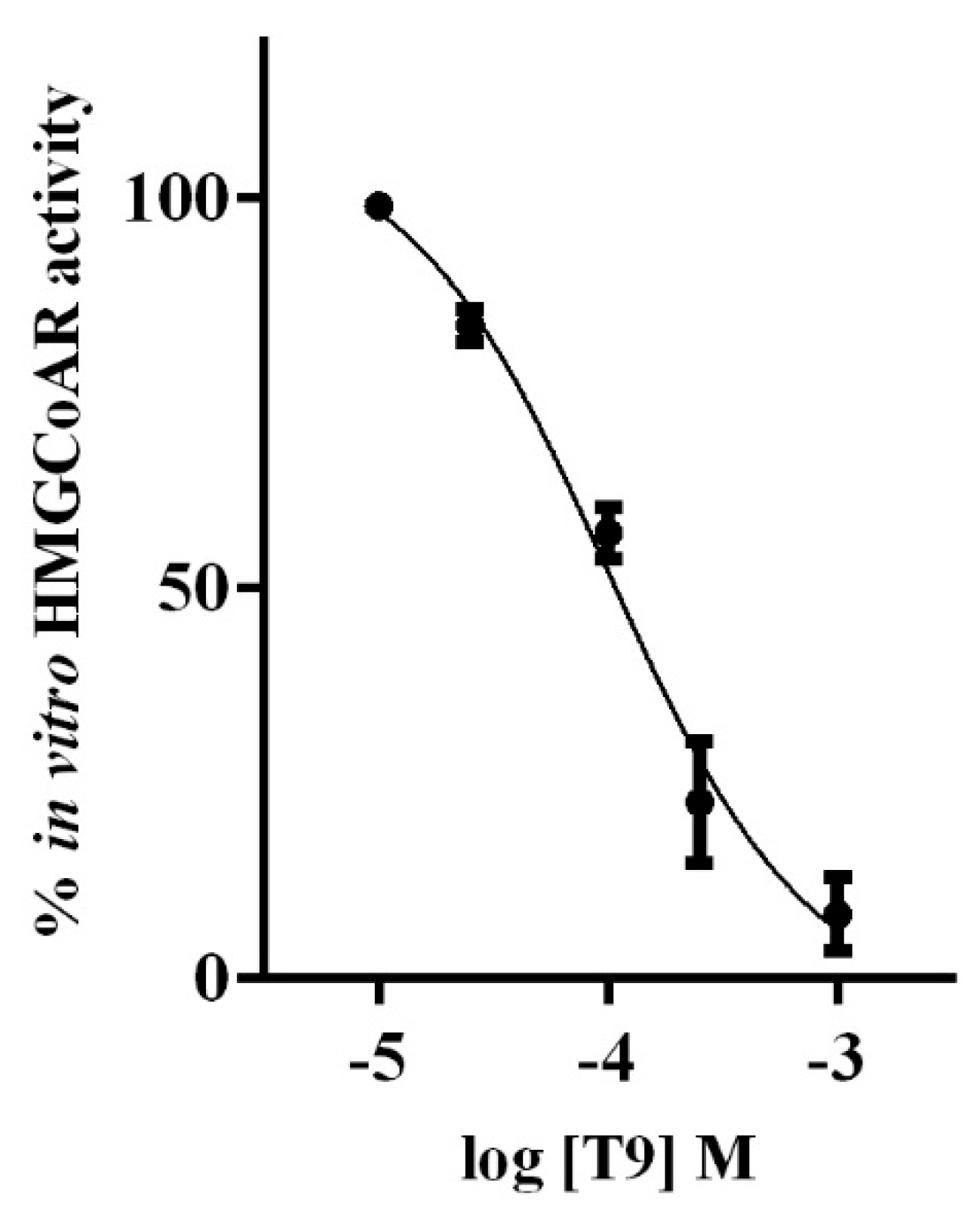
3.1. T9 Drops the HMGCOAR Activity In Vitro
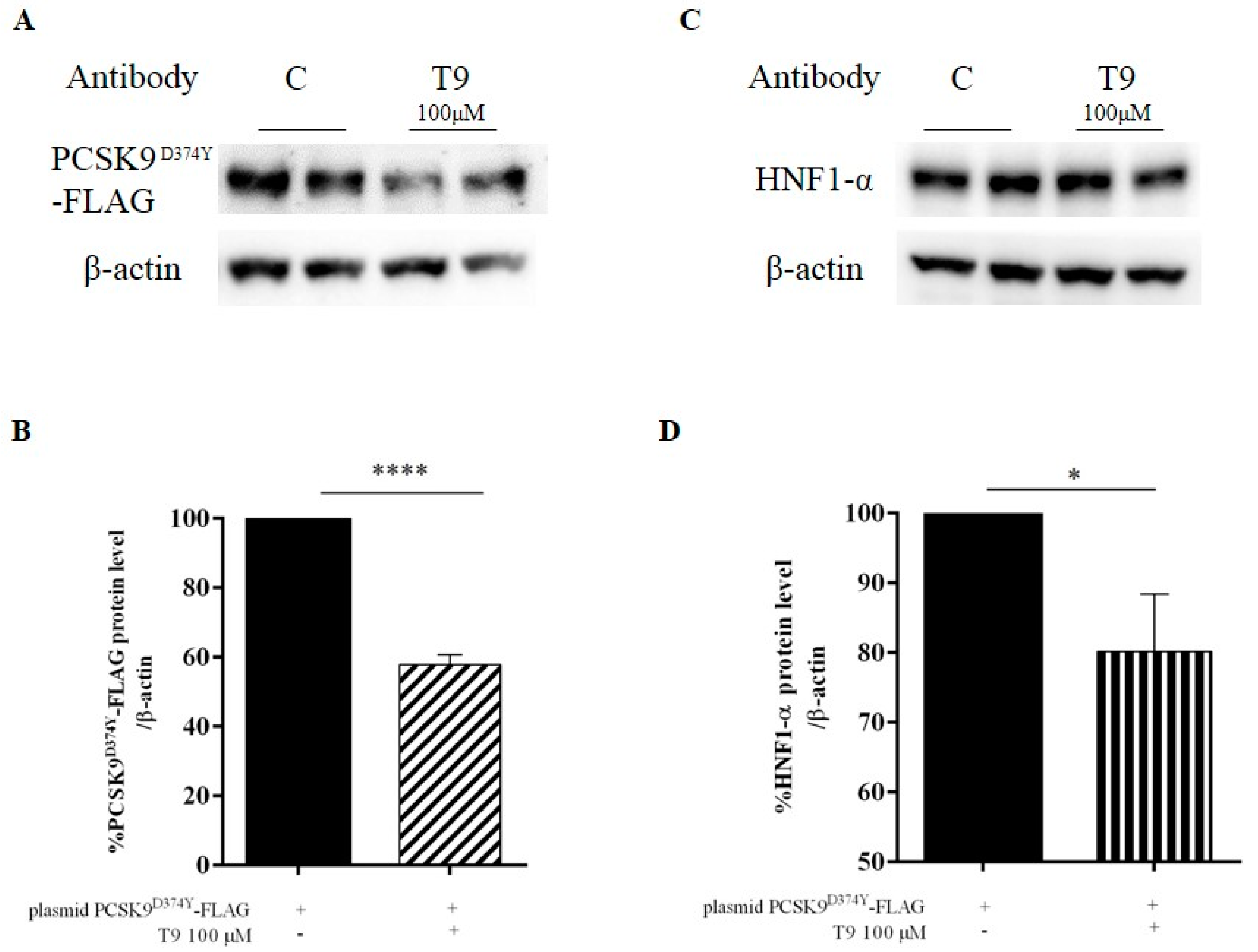
3.2. Peptide T9 Modulates the PCSK9D374Y Protein Level on HepG2 Cells
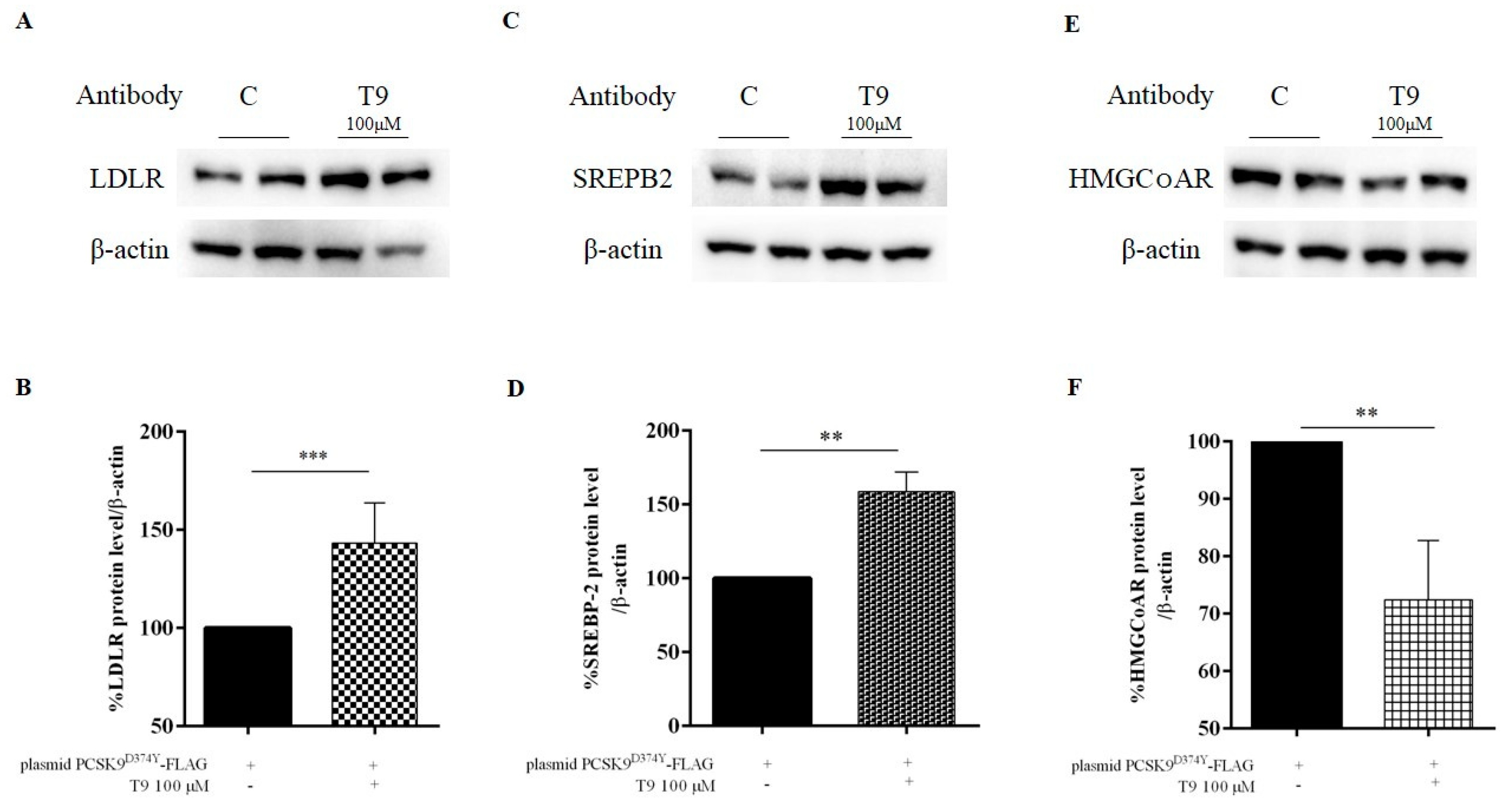
3.3. T9 Modulates the LDLR Pathway on HepG2 Cells Transfected with PCSK9D374Y
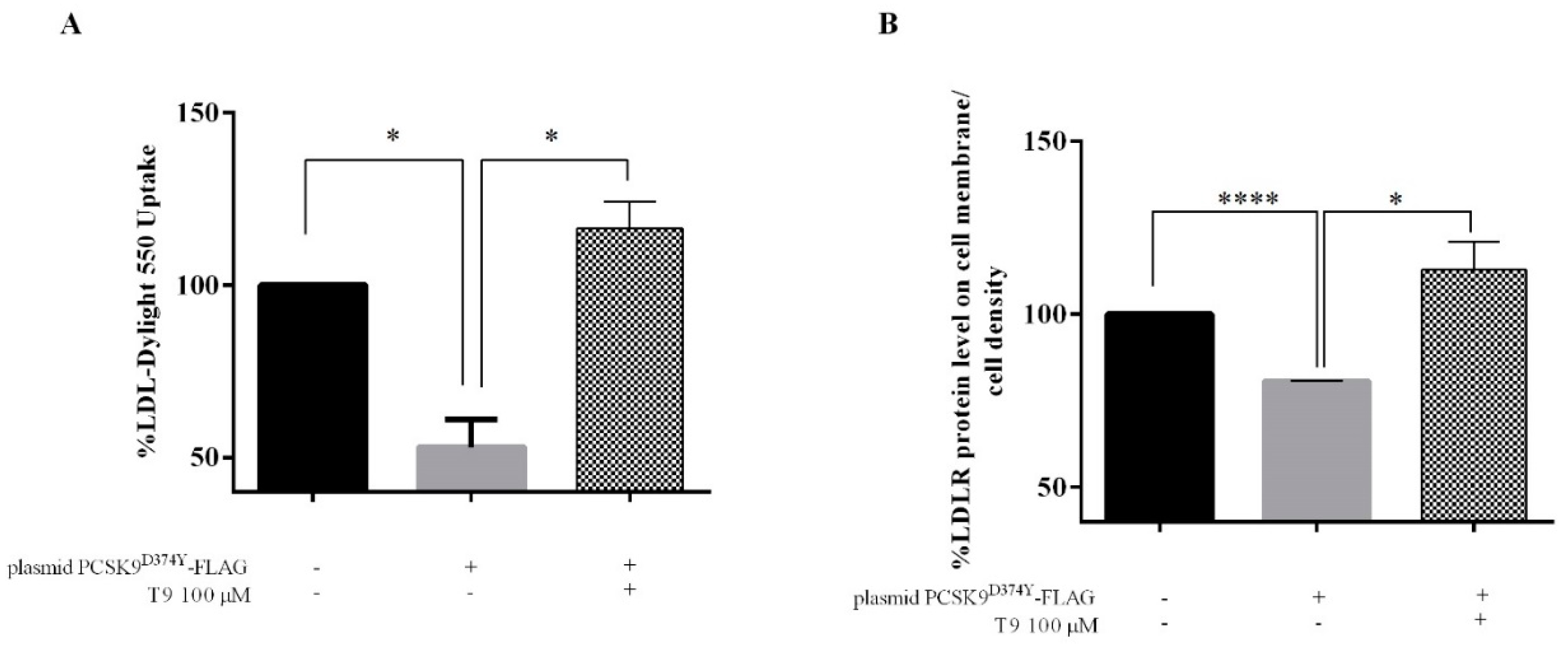
3.4. T9 Increases the Ability of Human Hepatic Cells Transfected with PCSK9D374Y to Uptake Extracellular LDL
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Piepoli, M.F.; Hoes, A.W.; Agewall, S.; Albus, C.; Brotons, C.; Catapano, A.L.; Cooney, M.T.; Corrà, U.; Cosyns, B.; Deaton, C.; et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts) Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Atherosclerosis 2016, 252, 207–274. [Google Scholar] [PubMed]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Endo, A. The discovery and development of HMG-CoA reductase inhibitors. J. Lipid Res. 1992, 33, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Lammi, C.; Zanoni, C.; Scigliuolo, G.M.; D’Amato, A.; Arnoldi, A. Lupin Peptides Lower Low-Density Lipoprotein (LDL) Cholesterol through an Up-regulation of the LDL Receptor/Sterol Regulatory Element Binding Protein 2 (SREBP2) Pathway at HepG2 Cell Line. J. Agric. Food Chem. 2014, 62, 7151–7159. [Google Scholar] [CrossRef]
- Lammi, C.; Arnoldi, A.; Aiello, G. Soybean Peptides Exert Multifunctional Bioactivity Modulating 3-Hydroxy-3-Methylglutaryl-CoA Reductase and Dipeptidyl Peptidase-IV Targets in Vitro. J. Agric. Food Chem. 2019, 67, 4824–4830. [Google Scholar] [CrossRef]
- Soares, R.A.; Mendonça, S.; De Castro, L.; Menezes, A.C.; Arêas, J.A. Ívini Major Peptides from Amaranth (Amaranthus cruentus) Protein Inhibit HMG-CoA Reductase Activity. Int. J. Mol. Sci. 2015, 16, 4150–4160. [Google Scholar] [CrossRef]
- Lachenmeier, D.W.; Monakhova, Y.B.; Kuballa, T.; Löbell-Behrends, S.; Maixner, S.; Kohl-Himmelseher, M.; Waldner, A.; Steffen, C. NMR evaluation of total statin content and HMG-CoA reductase inhibition in red yeast rice (Monascus spp.) food supplements. Chin. Med. 2012, 7, 8. [Google Scholar] [CrossRef]
- Seidah, N.G.; Prat, A. The proprotein convertases are potential targets in the treatment of dyslipidemia. J. Mol. Med. 2007, 85, 685–696. [Google Scholar] [CrossRef]
- Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem. Sci. 2007, 32, 71–77. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 2006, 116, 2995–3005. [Google Scholar] [CrossRef]
- Deisenhofer, J.; Kwon, H.J.; A Lagace, T.; McNutt, M.C.; Horton, J.D. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA 2008, 105, 1820–1825. [Google Scholar] [Green Version]
- Chaudhary, R.; Garg, J.; Shah, N.; Sumner, A. PCSK9 inhibitors: A new era of lipid lowering therapy. World J. Cardiol. 2017, 9, 76–91. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Vistoli, G.; Zanoni, C.; Arnoldi, A.; Sambuy, Y.; Ferruzza, S.; Ranaldi, G. A multidisciplinary investigation on the bioavailability and activity of peptides from lupin protein. J. Funct. Foods 2016, 24, 297–306. [Google Scholar] [CrossRef]
- Dong, B.; Li, H.; Singh, A.B.; Cao, A.; Liu, J. Inhibition of PCSK9 transcription by berberine involves down-regulation of hepatic HNF1α protein expression through the ubiquitin-proteasome degradation pathway. J. Biol. Chem. 2015, 290, 4047–4058. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Aiello, G.; Arnoldi, A.; Grazioso, G. Lupin Peptides Modulate the Protein-Protein Interaction of PCSK9 with the Low Density Lipoprotein Receptor in HepG2 Cells. Sci. Rep. 2016, 6, 29931. [Google Scholar] [CrossRef]
- Zanoni, C.; Aiello, G.; Arnoldi, A.; Lammi, C. Investigations on the hypocholesterolaemic activity of LILPKHSDAD and LTFPGSAED, two peptides from lupin beta-conglutin: Focus on LDLR and PCSK9 pathways. J Funct. Foods 2017, 32, 1–8. [Google Scholar] [CrossRef]
- Grazioso, G.; Bollati, C.; Sgrignani, J.; Arnoldi, A.; Lammi, C. The first food-derived peptide inhibitor of the protein-protein interaction between Gain-of-Function PCSK9D374Y and the LDL receptor. J. Agric. Food Chem. 2018, 66, 10552–10557. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A.; Aiello, G. YDFYPSSTKDQQS (P3), a peptide from lupin protein, absorbed by Caco-2 cells, modulates cholesterol metabolism in HepG2 cells via SREBP-1 activation. J. Food Biochem. 2018, 42, e12524. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A. IAVPGEVA, IAVPTGVA, and LPYP, three peptides from soy glycinin, modulate cholesterol metabolism in HepG2 cells through the activation of the LDLR-SREBP2 pathway. J. Funct. Foods 2015, 14, 469–478. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Vistoli, G.; Arnoldi, A. Two Peptides from Soy β-Conglycinin Induce a Hypocholesterolemic Effect in HepG2 Cells by a Statin-Like Mechanism: Comparative in Vitro and in Silico Modeling Studies. J. Agric. Food Chem. 2015, 63, 7945–7951. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A. Three Peptides from Soy Glycinin Modulate Glucose Metabolism in Human Hepatic HepG2 Cells. Int. J. Mol. Sci. 2015, 16, 27362–27370. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.; Goldstein, J.; Brown, M. SREBPs: Transcriptional Mediators of Lipid Homeostasis. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 491–498. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Boschin, G.; Arnoldi, A. Multifunctional peptides for the prevention of cardiovascular disease: A new concept in the area of bioactive food-derived peptides. J. Funct. Foods 2019, 55, 135–145. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lammi, C.; Bollati, C.; Lecca, D.; Abbracchio, M.P.; Arnoldi, A. Lupin Peptide T9 (GQEQSHQDEGVIVR) Modulates the Mutant PCSK9D374Y Pathway: in vitro Characterization of its Dual Hypocholesterolemic Behavior. Nutrients 2019, 11, 1665. https://doi.org/10.3390/nu11071665
Lammi C, Bollati C, Lecca D, Abbracchio MP, Arnoldi A. Lupin Peptide T9 (GQEQSHQDEGVIVR) Modulates the Mutant PCSK9D374Y Pathway: in vitro Characterization of its Dual Hypocholesterolemic Behavior. Nutrients. 2019; 11(7):1665. https://doi.org/10.3390/nu11071665
Chicago/Turabian StyleLammi, Carmen, Carlotta Bollati, Davide Lecca, Maria Pia Abbracchio, and Anna Arnoldi. 2019. "Lupin Peptide T9 (GQEQSHQDEGVIVR) Modulates the Mutant PCSK9D374Y Pathway: in vitro Characterization of its Dual Hypocholesterolemic Behavior" Nutrients 11, no. 7: 1665. https://doi.org/10.3390/nu11071665
APA StyleLammi, C., Bollati, C., Lecca, D., Abbracchio, M. P., & Arnoldi, A. (2019). Lupin Peptide T9 (GQEQSHQDEGVIVR) Modulates the Mutant PCSK9D374Y Pathway: in vitro Characterization of its Dual Hypocholesterolemic Behavior. Nutrients, 11(7), 1665. https://doi.org/10.3390/nu11071665






