Associations of ADIPOQ and LEP Gene Variants with Energy Intake: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Information Sources and Search Strategy
2.4. Study Selection
2.5. Data Collection Process and Data Items
2.6. Risk of Bias in Individual Studies
2.7. Summary Measures
3. Results
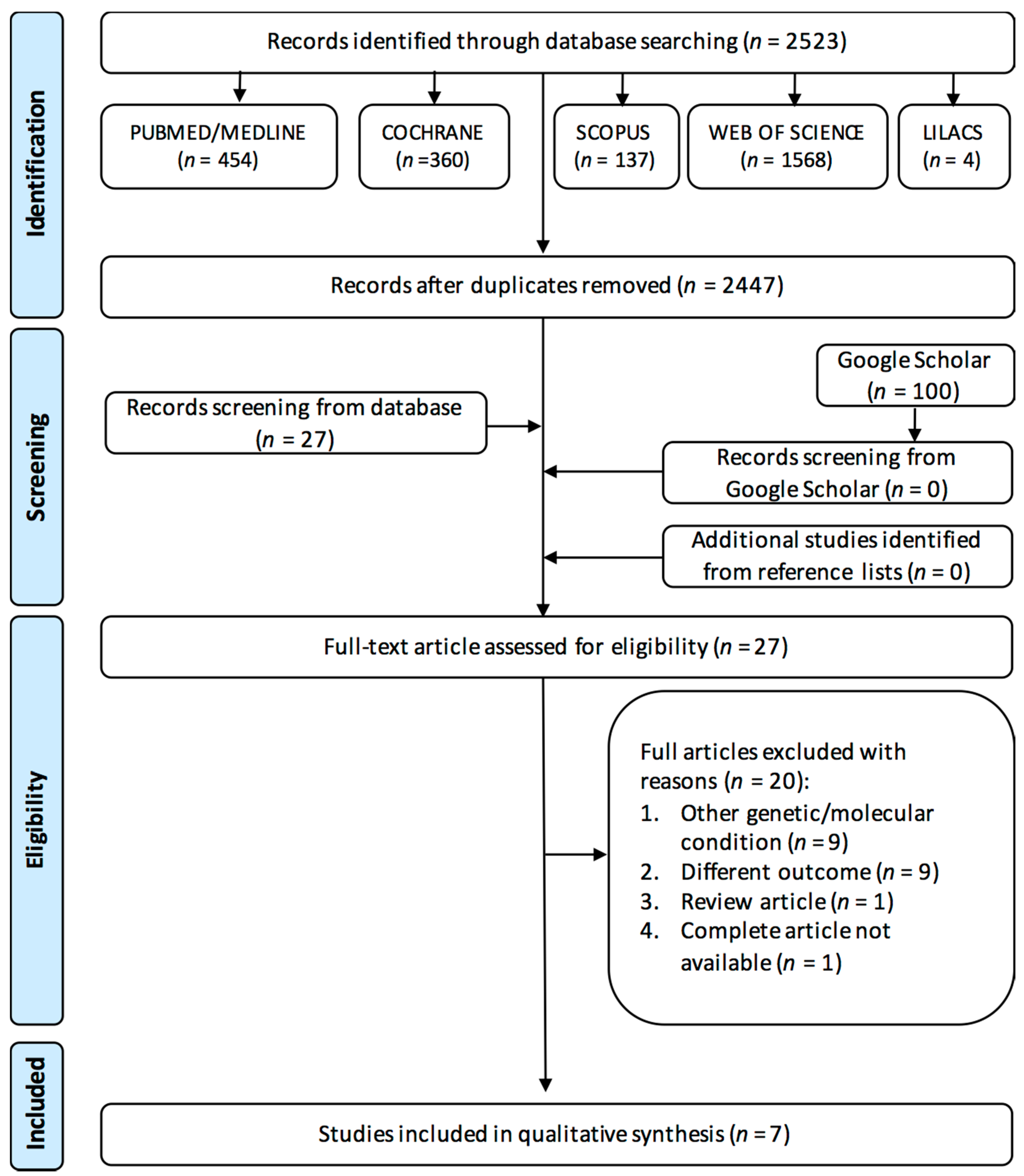
3.1. Study Selection
3.2. Study Characteristics
3.3. Risk of Bias within Studies
3.4. Synthesis of Results
3.4.1. LEP-rs7799039
3.4.2. LEP-rs2167270
3.4.3. ADIPOQ-rs2241766
3.4.4. ADIPOQ-rs17300539
3.4.5. ADIPOQ gene marker D3S1262
4. Discussion
4.1. Summary of Evidence
4.2. Limitations
Author Contributions
Funding
Conflicts of Interest
Appendix A. Search Strategy
| Database | Search |
|---|---|
| PubMed/MEDLINE (8 October 2018) | Search ((((((((((ADIPOQ gene)[Title/Abstract] OR adiponectin gene)[Title/Abstract] OR ACDC gene)[Title/Abstract] OR ADPN gene)[Title/Abstract] OR AMP1 gene)[Title/Abstract] OR APM-1 gene)[Title/Abstract] OR GBP28 gene)[Title/Abstract] OR ACRP30 gene [Title/Abstract])) OR (((((LEP gene)[Title/Abstract] OR leptin gene)[Title/Abstract] OR OB gene)[Title/Abstract] OR OBS gene)[Title/Abstract] OR LEPD gene [Title/Abstract])) AND energy intake Title/Abstract] Sort by: Best Match Filters: Humans |
| Scopus a,b (8 October 2018) | (TITLE-ABS-KEY (lep AND gene OR leptin AND gene OR ob AND gene OR obs AND gene OR lepd AND gene) OR TITLE-ABS-KEY (adipoq AND gene OR adiponectin AND gene OR acdc AND gene OR adpn AND gene OR apm1 AND gene OR apm-1 AND gene OR gbp28 AND gene OR acrp30 AND gene) AND TITLE-ABS-KEY (energy AND intake)) |
| Web of Science b (8 October 2018) | TITLE: (LEP gene OR leptin gene OR OB gene OR OBS gene OR LEPD gene) OR TITLE: (ADIPOQ gene OR adiponectin gene OR ACDC gene OR ADPN gene OR APM1 gene OR APM-1 gene OR GBP28 gene OR ACRP30 gene) AND TITLE: (energy intake) |
| Cochrane a (8 October 2018) | LEP gene OR leptin gene OR OB gene OR OBS gene OR LEPD gene in Title Abstract Keyword OR ADIPOQ gene OR adiponectin gene OR ACDC gene OR ADPN gene OR APM1 gene OR APM-1 gene OR GBP28 gene OR ACRP30 gene in Title Abstract Keyword AND energy intake in Title Abstract Keyword - (Word variations were searched) |
| LILACS (8 October 2018) | (tw: (LEP gene OR leptin gene OR OB gene OR OBS gene OR LEPD gene)) OR (tw: (ADIPOQ gene OR adiponectin gene OR ACDC gene OR ADPN gene OR APM1 gene OR APM-1 gene OR GBP28 gene OR ACRP30 gene)) AND (tw: (energy intake OR consumo energético OR ingestão calórica)) |
| Google Scholar (17 January 2017) | ADIPOQ gene OR LEP gene and energy intake (without patents or citations) |
Appendix B. List of Excluded Articles
| Article | Reason |
|---|---|
| Mariman et al. (2015) [52] | 1 |
| Brunkwall et al. (2016) [53] | 1 |
| Alharbi et al. (2014) [54] | 1 |
| Alharbi et al. (2014) [54], Liu et al. (2003) [55] | 1 |
| Li et al. (2014) [56] | 2 |
| Feitosa et al. (2000) [57] | 1 |
| Collaku et al. (2004) [58] | 1 |
| Dubois et al. (2013) [59] | 1 |
| Celis-Morales et al. (2017) [60] | 1 |
| Mizuta et al. (2008) [61] | 2 |
| de Krom et al. (2007) [62] | 2 |
| Trayhum et al. (1996) [63] | 3 |
| Valladares et al. (2014) [64] | 2 |
| de Luis et al. (2018) [65] | 2 |
| Gajewska et al. (2016) [66] | 2 |
| Licinio et al. (2007) [67] | 1 |
| Mammes et al. (1998) [68] | 2 |
| Park et al. (2014) [69] | 2 |
| Faith et al. (1999) [70] | 4 |
References
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- Palou, A.; Bonet, M.L. Challenges in obesity research. Nutr. Hosp. 2013, 28 (Suppl. 5), 144–153. [Google Scholar]
- Gonzalez Jimenez, E. Obesity: Etiologic and pathophysiological analysis. Endocrinol. Nutr. 2013, 60, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wadden, T.A.; Webb, V.L.; Moran, C.H.; Bailer, B.A. Lifestyle modification for obesity: New developments in diet, physical activity, and behavior therapy. Circulation 2012, 125, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Willett, W.C.; Hu, F.B. Global obesity: Trends, risk factors and policy implications. Nat. Rev. Endocrinol. 2013, 9, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Church, T.; Martin, C.K. The Obesity Epidemic: A Consequence of Reduced Energy Expenditure and the Uncoupling of Energy Intake? Obesity 2018, 26, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Penney, K.L.; Giovannucci, E.; Kraft, P.; Wilson, K.M. A genome-wide association study of energy intake and expenditure. PLoS ONE 2018, 13, e0201555. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, K.M.; Celis-Morales, C.; Lara, J.; Ashor, A.W.; Lovegrove, J.A.; Martinez, J.A.; Saris, W.H.; Gibney, M.; Manios, Y.; Traczyk, I.; et al. Associations between FTO genotype and total energy and macronutrient intake in adults: A systematic review and meta-analysis. Obes. Rev. 2015, 16, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Hasselbalch, A.L. Genetics of dietary habits and obesity—A twin study. Dan. Med. Bull. 2010, 57, B4182. [Google Scholar] [PubMed]
- Giralt, M.; Cereijo, R.; Villarroya, F. Adipokines and the Endocrine Role of Adipose Tissues. Handb. Exp. Pharmacol. 2016, 233, 265–282. [Google Scholar]
- Lehr, S.; Hartwig, S.; Lamers, D.; Famulla, S.; Muller, S.; Hanisch, F.G.; Cuvelier, C.; Ruige, J.; Eckardt, K.; Ouwens, D.M.; et al. Identification and validation of novel adipokines released from primary human adipocytes. Mol. Cell. Proteom. 2012, 11, M111.010504. [Google Scholar] [CrossRef] [PubMed]
- Rankinen, T.; Zuberi, A.; Chagnon, Y.C.; Weisnagel, S.J.; Argyropoulos, G.; Walts, B.; Perusse, L.; Bouchard, C. The human obesity gene map: The 2005 update. Obesity 2006, 14, 529–644. [Google Scholar] [CrossRef] [PubMed]
- Turcot, V.; Lu, Y.; Highland, H.M.; Schurmann, C.; Justice, A.E.; Fine, R.S.; Bradfield, J.P.; Esko, T.; Giri, A.; Graff, M.; et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018, 50, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Laursen, T.L.; Zak, R.B.; Shute, R.J.; Heesch, M.W.S.; Dinan, N.E.; Bubak, M.P.; la Salle, D.T.; Slivka, D.R. Leptin, adiponectin, and ghrelin responses to endurance exercise in different ambient conditions. Temperature 2017, 4, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Arita, Y.; Kihara, S.; Ouchi, N.; Takahashi, M.; Maeda, K.; Miyagawa, J.; Hotta, K.; Shimomura, I.; Nakamura, T.; Miyaoka, K.; et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999, 257, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Ghalandari, H.; Hosseini-Esfahani, F.; Mirmiran, P. The Association of Polymorphisms in Leptin/Leptin Receptor Genes and Ghrelin/Ghrelin Receptor Genes with Overweight/Obesity and the Related Metabolic Disturbances: A Review. Int. J. Endocrinol. Metab. 2015, 13, e19073. [Google Scholar] [CrossRef] [PubMed]
- Jequier, E. Leptin signaling, adiposity, and energy balance. Ann. N. Y. Acad. Sci. 2002, 967, 379–388. [Google Scholar] [CrossRef]
- Friedman, J.M. Leptin and the regulation of body weigh. Keio J. Med. 2011, 60, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.D.; Kang, S.H.; Ju, G.; Han, J.W.; Kim, T.H.; Lee, C.S.; Kim, K.W.; Yoon, I.Y. The prevalence of and risk factors for sleep-disordered breathing in an elderly Korean population. Respiration 2014, 87, 372–378. [Google Scholar] [CrossRef]
- Wedellova, Z.; Dietrich, J.; Siklova-Vitkova, M.; Kolostova, K.; Kovacikova, M.; Duskova, M.; Broz, J.; Vedral, T.; Stich, V.; Polak, J. Adiponectin inhibits spontaneous and catecholamine-induced lipolysis in human adipocytes of non-obese subjects through AMPK-dependent mechanisms. Physiol. Res. 2011, 60, 139–148. [Google Scholar]
- Lee, B.; Shao, J. Adiponectin and energy homeostasis. Rev. Endocr. Metab. Disord. 2014, 15, 149–156. [Google Scholar] [CrossRef]
- Boumaiza, I.; Omezzine, A.; Rejeb, J.; Rebhi, L.; Ouedrani, A.; Rejeb, N.B.; Nabli, N.; Abdelaziz, A.B.; Bouslama, A. Relationship between leptin G2548A and leptin receptor Q223R gene polymorphisms and obesity and metabolic syndrome risk in Tunisian volunteers. Genet. Test. Mol. Biomark. 2012, 16, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Hasnain, S. Leptin promoter variant G2548A is associated with serum leptin and HDL-C levels in a case control observational study in association with obesity in a Pakistani cohort. J. Biosci. 2016, 41, 251–255. [Google Scholar]
- Hoffstedt, J.; Eriksson, P.; Mottagui-Tabar, S.; Arner, P. A polymorphism in the leptin promoter region (-2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm. Metab. Res. 2002, 34, 355–359. [Google Scholar] [CrossRef]
- Mammes, O.; Betoulle, D.; Aubert, R.; Herbeth, B.; Siest, G.; Fumeron, F. Association of the G-2548A polymorphism in the 5′ region of the LEP gene with overweight. Ann. Hum. Genet. 2000, 64, 391–394. [Google Scholar] [CrossRef]
- Hinuy, H.M.; Hirata, M.H.; Forti, N.; Diament, J.; Sampaio, M.F.; Armaganijan, D.; Salazar, L.A.; Hirata, R.D. Leptin G-2548A promoter polymorphism is associated with increased plasma leptin and BMI in Brazilian women. Archiv. Endocrinol. Metabol. 2008, 52, 611–616. [Google Scholar] [CrossRef]
- Sahin, D.S.; Tumer, C.; Demir, C.; Celik, M.M.; Celik, M.; Ucar, E.; Gunesacar, R. Association with leptin gene C.-2548 G>A polymorphism, serum leptin levels, and body mass index in Turkish obese patients. Cell Biochem. Biophys. 2013, 65, 243–247. [Google Scholar]
- Lu, J.F.; Zhou, Y.; Huang, G.H.; Jiang, H.X.; Hu, B.L.; Qin, S.Y. Association of ADIPOQ polymorphisms with obesity risk: A meta-analysis. Hum. Immunol. 2014, 75, 1062–1068. [Google Scholar] [CrossRef]
- Loos, R.J.; Ruchat, S.; Rankinen, T.; Tremblay, A.; Perusse, L.; Bouchard, C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am. J. Clin. Nutr. 2007, 85, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Z.; Meng, K.; Zhang, L. Association of adiponectin gene (ADIPOQ) rs2241766 polymorphism with obesity in adults: A meta-analysis. PLoS ONE 2014, 9, e95270. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Farace, D.J.; Frantzen, J. Fourth International Conference on Grey Literature: New Frontiers in Grey Literature; GreyNet, Grey Literature Network Service: Washington, DC, USA, 1999. [Google Scholar]
- Downs, S.H.; Black, N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J. Epidemiol. Community Health 1998, 52, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Bienertová-Vašků, J.; Bienert, P.; Forejt, M.; Tomandl, J.; Brázdová, Z.; Vašků, A. Genotype × nutrient association of common polymorphisms in obesity-related genes with food preferences and time structure of energy intake. Br. J. Nutr. 2010, 103, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Bienertova-Vasku, J.; Bienert, P.; Tomandl, J.; Forejt, M.; Vavrina, M.; Kudelkova, J.; Vasku, A. No association of defined variability in leptin, leptin receptor, adiponectin, proopiomelanocortin and ghrelin gene with food preferences in the Czech population. Nutr. Neurosci. 2008, 11, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Choquette, A.C.; Lemieux, S.; Tremblay, A.; Chagnon, Y.C.; Bouchard, C.; Vohl, M.C.; Perusse, L. Evidence of a quantitative trait locus for energy and macronutrient intakes on chromosome 3q27.3: The Quebec Family Study. Am. J. Clin. Nutr. 2008, 88, 1142–1148. [Google Scholar] [PubMed]
- Dougkas, A.; Yaqoob, P.; Givens, D.I.; Reynolds, C.K.; Minihane, A.M. The impact of obesity-related SNP on appetite and energy intake. Br. J. Nutr. 2013, 110, 1151–1156. [Google Scholar] [CrossRef]
- Martins, M.C.; Trujillo, J.; Freitas-Vilela, A.A.; Farias, D.R.; Rosado, E.L.; Struchiner, C.J.; Kac, G. Associations between obesity candidate gene polymorphisms (fat mass and obesity-associated (FTO), melanocortin-4 receptor (MC4R), leptin (LEP) and leptin receptor (LEPR)) and dietary intake in pregnant women. Br. J. Nutr. 2018, 120, 454–463. [Google Scholar] [CrossRef]
- Zandona, M.R.; Rodrigues, R.O.; Albiero, G.; Campagnolo, P.D.; Vitolo, M.R.; Almeida, S.; Mattevi, V.S. Polymorphisms in LEPR, PPARG and APM1 genes: Associations with energy intake and metabolic traits in young children. Arq. Bras. Endocrinol. Metabol. 2013, 57, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.W.; Woods, S.C.; Porte, D., Jr.; Seeley, R.J.; Baskin, D.G. Central nervous system control of food intake. Nature 2000, 404, 661–771. [Google Scholar] [CrossRef]
- Feng, H.; Zheng, L.; Feng, Z.; Zhao, Y.; Zhang, N. The role of leptin in obesity and the potential for leptin replacement therapy. Endocrine 2013, 44, 33–39. [Google Scholar] [CrossRef]
- Kroll, C.; Mastroeni, S.S.; Veugelers, P.J.; Mastroeni, M.F. Association of ADIPOQ, LEP, and FTO gene polymorphisms with large for gestational age infants. Am. J. Hum. Biol. 2017, 29, e22893. [Google Scholar] [CrossRef]
- Dasgupta, S.; Salman, M.; Siddalingaiah, L.B.; Lakshmi, G.L.; Xaviour, D.; Sreenath, J. Genetic variants in leptin: Determinants of obesity and leptin levels in South Indian population. Adipocyte 2015, 4, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, T.; Maleki, M.; Saidijam, M.; Paoli, M. Association between leptin gene G-2548A polymorphism with metabolic syndrome. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2013, 18, 668–673. [Google Scholar]
- Ahima, R.S. Metabolic actions of adipocyte hormones: Focus on adiponectin. Obesity 2006, 14 (Suppl. 1), 9s–15s. [Google Scholar] [CrossRef] [PubMed]
- Scherer, P.E.; Williams, S.; Fogliano, M.; Baldini, G.; Lodish, H.F. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 1995, 270, 26746–26749. [Google Scholar] [CrossRef] [PubMed]
- Bouatia-Naji, N.; Meyre, D.; Lobbens, S.; Seron, K.; Fumeron, F.; Balkau, B.; Heude, B.; Jouret, B.; Scherer, P.E.; Dina, C.; et al. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes 2006, 55, 545–550. [Google Scholar] [CrossRef]
- Warodomwichit, D.; Shen, J.; Arnett, D.K.; Tsai, M.Y.; Kabagambe, E.K.; Peacock, J.M.; Hixson, J.E.; Straka, R.J.; Province, M.A.; An, P.; et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: The GOLDN study. Obesity 2009, 17, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.B.; Nasrullah, A.; Haq, S.; Akhtar, A.; Ghazanfar, H.; Nasir, A.; Afzal, R.M.; Bukhari, M.M.; Chaudhary, A.Y.; Naqvi, S.W. The Interplay of Genetics and Environmental Factors in the Development of Obesity. Cureus 2017, 9, e1435. [Google Scholar] [CrossRef] [PubMed]
- Lyon, H.N.; Hirschhorn, J.N. Genetics of common forms of obesity: A brief overview. Am. J. Clin. Nutr. 2005, 82, 215s–217s. [Google Scholar] [CrossRef]
- Mariman, E.C.; Bouwman, F.G.; Aller, E.E.; van Baak, M.A.; Wang, P. Extreme obesity is associated with variation in genes related to the circadian rhythm of food intake and hypothalamic signaling. Physiol. Genom. 2015, 47, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Brunkwall, L.; Chen, Y.; Hindy, G.; Rukh, G.; Ericson, U.; Barroso, I.; Johansson, I.; Franks, P.W.; Orho-Melander, M.; Renstrom, F. Sugar-sweetened beverage consumption and genetic predisposition to obesity in 2 Swedish cohorts. Am. J. Clin. Nutr. 2016, 104, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, K.K.; Richardson, T.G.; Khan, I.A.; Syed, R.; Mohammed, A.K.; Boustred, C.R.; Gaunt, T.R.; Tamimi, W.; Al-Daghri, N.M.; Day, I.N. Influence of adiposity-related genetic markers in a population of saudi arabians where other variables influencing obesity may be reduced. Dis. Mark. 2014, 2014, 758232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Lacorte, J.M.; Viguerie, N.; Poitou, C.; Pelloux, V.; Guy-Grand, B.; Coussieu, C.; Langin, D.; Basdevant, A.; Clement, K. Adiponectin gene expression in subcutaneous adipose tissue of obese women in response to short-term very low calorie diet and refeeding. J. Clin. Endocrinol. Metab. 2003, 88, 5881–5886. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Tiwari, H.K.; Lin, W.Y.; Allison, D.B.; Chung, W.K.; Leibel, R.L.; Yi, N.; Liu, N. Genetic association analysis of 30 genes related to obesity in a European American population. Int. J. Obes. (Lond.) 2014, 38, 724–729. [Google Scholar] [CrossRef]
- Feitosa, M.F.; Rice, T.; Nirmala, A.; Rao, D.C. Major gene effect on body mass index: The role of energy intake and energy expenditure. Hum. Biol. 2000, 72, 781–799. [Google Scholar]
- Collaku, A.; Rankinen, T.; Rice, T.; Leon, A.S.; Rao, D.C.; Skinner, J.S.; Wilmore, J.H.; Bouchard, C. A genome-wide linkage scan for dietary energy and nutrient intakes: The Health, Risk Factors, Exercise Training, and Genetics (HERITAGE) Family Study. Am. J. Clin. Nutr. 2004, 79, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Dubois, L.; Diasparra, M.; Bedard, B.; Kaprio, J.; Fontaine-Bisson, B.; Perusse, D.; Tremblay, R.; Boivin, M. Gene-environment contributions to energy and macronutrient intakes in 9-year-old children: Results from the Quebec Newborn Twin Study. Physiol. Behav. 2013, 119, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Celis-Morales, C.A.; Lyall, D.M.; Gray, S.R.; Steell, L.; Anderson, J.; Iliodromiti, S.; Welsh, P.; Guo, Y.; Petermann, F.; Mackay, D.F.; et al. Dietary fat and total energy intake modifies the association of genetic profile risk score on obesity: Evidence from 48 170 UK Biobank participants. Int. J. Obes. (Lond.) 2017, 41, 1761–1768. [Google Scholar] [CrossRef] [PubMed]
- Mizuta, E.; Kokubo, Y.; Yamanaka, I.; Miyamoto, Y.; Okayama, A.; Yoshimasa, Y.; Tomoike, H.; Morisaki, H.; Morisaki, T. Leptin gene and leptin receptor gene polymorphisms are associated with sweet preference and obesity. Hypertens. Res. 2008, 31, 1069–1077. [Google Scholar] [CrossRef]
- De Krom, M.; van der Schouw, Y.T.; Hendriks, J.; Ophoff, R.A.; van Gils, C.H.; Stolk, R.P.; Grobbee, D.E.; Adan, R. Common genetic variations in CCK, leptin, and leptin receptor genes are associated with specific human eating patterns. Diabetes 2007, 56, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P.; Rayner, D.V. Hormones and the ob gene product (leptin) in the control of energy balance. Biochem. Soc. Trans. 1996, 24, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Valladares, M.; Obregon, A.M.; Weisstaub, G.; Burrows, R.; Patino, A.; Ho-Urriola, J.; Santos, J.L. Association between feeding behavior, and genetic polymorphism of leptin and its receptor in obese Chilean children. Nutr. Hosp. 2014, 31, 1044–1051. [Google Scholar] [PubMed]
- De Luis, D.A.; Izaola, O.; Primo, D.; Aller, R.; Ortola, A.; Gomez, E.; Lopez, J.J. The association of SNP276G>T at adiponectin gene with insulin resistance and circulating adiponectin in response to two different hypocaloric diets. Diabetes Res. Clin. Pract. 2018, 137, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Gajewska, J.; Kurylowicz, A.; Ambroszkiewicz, J.; Mierzejewska, E.; Chelchowska, M.; Szamotulska, K.; Weker, H.; Puzianowska-Kuznicka, M. ADIPOQ-11377C>G Polymorphism Increases the Risk of Adipokine Abnormalities and Child Obesity Regardless of Dietary Intake. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Licinio, J.; Milane, M.; Thakur, S.; Whelan, F.; Yildlz, B.O.; Delibasi, T.; de Miranda, P.B.; Ozata, M.; Bolu, E.; DePaoli, A.; et al. Effects of leptin on intake of specific micro- and macronutrients in a woman with leptin gene deficiency studied off and on leptin at stable body weight. Appetite 2007, 49, 594–599. [Google Scholar] [CrossRef]
- Mammes, O.; Betoulle, D.; Aubert, R.; Giraud, V.; Tuzet, S.; Petiet, A.; Colas-Linhart, N.; Fumeron, F. Novel polymorphisms in the 5′ region of the LEP gene: Association with leptin levels and response to low-calorie diet in human obesity. Diabetes 1998, 47, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, H.J.; Jang, H.B.; Hwang, J.Y.; Kang, J.H.; Han, B.G.; Lee, J.Y.; Song, J. Interactions between ADIPOQ gene variants and dietary monounsaturated: Saturated fatty acid ratio on serum lipid levels in Korean children. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Faith, M.S.; Rha, S.S.; Neale, M.C.; Allison, D.B. Evidence for genetic influences on human energy intake: Results from a twin study using measured observations. Behav. Genet. 1999, 29, 145–154. [Google Scholar] [CrossRef]
| Author, Year | Country, Ethnicity | Study Design | Sample Size, n | Sample Characteristics | LEP/ADIPOQ Gene Variants | Energy Intake | Main Conclusion | Risk of Bias * |
|---|---|---|---|---|---|---|---|---|
| Bienertova-Vasku, Bienert et al., 2010 [35] | Czech Republic, Caucasian | Case-control study | 409 | Obese (n = 252) and normal-weight (n = 157) groups | LEP-rs2167270 (LEP + 19A/G); ADIPOQ-rs2241766 (ADIPOQ 45T/G, ADIPOQ 94T/G) | Standardized 7-day food records | None of the examined polymorphisms served as an independent predictor for the percentage of daily energy intake. The ADIPOQ-rs2241766 polymorphism was associated with the energy value of breakfast, defined as the first meal during the day (β = 0.15; p = 0.02). Moreover, the LEP-rs2167270 polymorphism was correlated with the energy value of supper (β = 0.13; p = 0.05), with AG heterozygotes expressing a tendency toward the highest energy intake in their supper. | 72.7 |
| Bienertova-Vasku, Bienert et al., 2008 [36] | Czech Republic, Caucasian | Case-control study | 185 | Obese (n = 125) and normal-weight (n = 60) groups | LEP-rs7799039 (LEP-2548 G/A); ADIPOQ-rs2241766 (APM1 T94G) | 7-day food records, total energy | None of the examined polymorphisms were associated with total energy intake. | 72.7 |
| Boumaiza, Omezzine et al., 2012 [23] | Tunisia | Case-control | 329 | Obese (n = 160) and non-obese (n = 169) groups | LEP-rs7799039 (LEP G2548A) | 3-day dietary record: 2 weekdays and 1 weekend day | The AA genotype had significantly higher daily energy intake. GG: 2853 ± 1215 kcal; GA: 2889 ± 1277 kcal; AA: 3431 ± 1609 kcal; p = 0.048 | 77.3 |
| Choquette, Lemieux et al., 2008 [37] | French-Canadian subjects | Cohort | 836 | Random sampling and ascertainment through obese probands | Marker D3S1262 on chromosome 3q27.3—region harboring the ADIPOQ gene | 3-day dietary record: 2 weekdays and 1 weekend day | A significant linkage was observed on chromosome 3q27.3 with marker D3S1262. LOD score: 2.24, p < 0.0001 | 68.4 |
| Dougkas, Yaqoob et al., 2013 [38] | United Kingdom | Randomized crossover trial | 40 | Healthy, non-smoking, overweight men aged 18–50 years with a BMI of 25.0–29.9 kg/m2 | LEP-rs7799039 | Energy intake was assessed by an ad libitum lunch offered 90 min after dairy snacks or water control | rs7799039 did not have an effect on ad libitum energy intake at lunch. AA: 3744 ± 391 kJ; GG+GA: 4222 ± 285; p = 0.290 | 75.0 |
| Martins, Trujillo et al., 2018 [39] | Brazil | Cohort | 220 | Pregnant | LEP-rs7799039 | Semi-quantitative FFQ: consumption frequency of food items over the 6 months before the interview. Data collected at 5–13 weeks of gestation covered the pre-pregnancy period, and data collected at 30–36 weeks of gestation refers to dietary intake of women during pregnancy | There was a significant association between allele A of LEP-rs7799039 and change in dietary intake from pre-pregnancy to pregnancy, with individuals presenting lower total energy intake. GA+AA: 1964 kcal/day, 95%CI: 1684–2290 kcal/day; GG: 2192 kcal/day, 95%CI: 1890–2542 kcal/day; p = 0.04 | 100.0 |
| Zandona, Rodrigues et al., 2013 [40] | Brazil | Prospective cohort study | 325 | Children at 12–16 months and at 3–4 years of age | ADIPOQ-rs17300539 (APM1 -11391G>A); ADIPOQ- rs266729 (APM1-11377C>G); LEP-rs7799039 (LEP-2548G>A) | 12–16 months: 24-h diet recall to record the child’s food intake on the day before the home visit 3–4 years: two 24-h diet recalls were collected on two randomly selected and non-consecutive days | There was a strong linkage disequilibrium between ADIPOQ-rs17300539 and ADIPOQ-rs266729. Children carrying the ADIPOQ-rs17300539 A-allele had lower total energy intake/day than G/G homozygotes at 1 year. G: 952 ± 387 kcal; A: 841 ± 386 kcal; p = 0.045. The opposite effect was observed at 4 years of age: G-carriers had higher total energy intake/day than A-carriers, but without statistical significance. G: 1501 kcal; A: 1588 kcal; p = 0.149. Moreover, no associations were observed between LEP-rs7799039 and dietary parameters. | 95.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroll, C.; Mastroeni, S.S.B.S.; Veugelers, P.J.; Mastroeni, M.F. Associations of ADIPOQ and LEP Gene Variants with Energy Intake: A Systematic Review. Nutrients 2019, 11, 750. https://doi.org/10.3390/nu11040750
Kroll C, Mastroeni SSBS, Veugelers PJ, Mastroeni MF. Associations of ADIPOQ and LEP Gene Variants with Energy Intake: A Systematic Review. Nutrients. 2019; 11(4):750. https://doi.org/10.3390/nu11040750
Chicago/Turabian StyleKroll, Caroline, Silmara S.B.S. Mastroeni, Paul J. Veugelers, and Marco F Mastroeni. 2019. "Associations of ADIPOQ and LEP Gene Variants with Energy Intake: A Systematic Review" Nutrients 11, no. 4: 750. https://doi.org/10.3390/nu11040750
APA StyleKroll, C., Mastroeni, S. S. B. S., Veugelers, P. J., & Mastroeni, M. F. (2019). Associations of ADIPOQ and LEP Gene Variants with Energy Intake: A Systematic Review. Nutrients, 11(4), 750. https://doi.org/10.3390/nu11040750





