Weighted Differential Gradient Method for Filling Pits in Light Detection and Ranging (LiDAR) Canopy Height Model
Abstract
1. Introduction
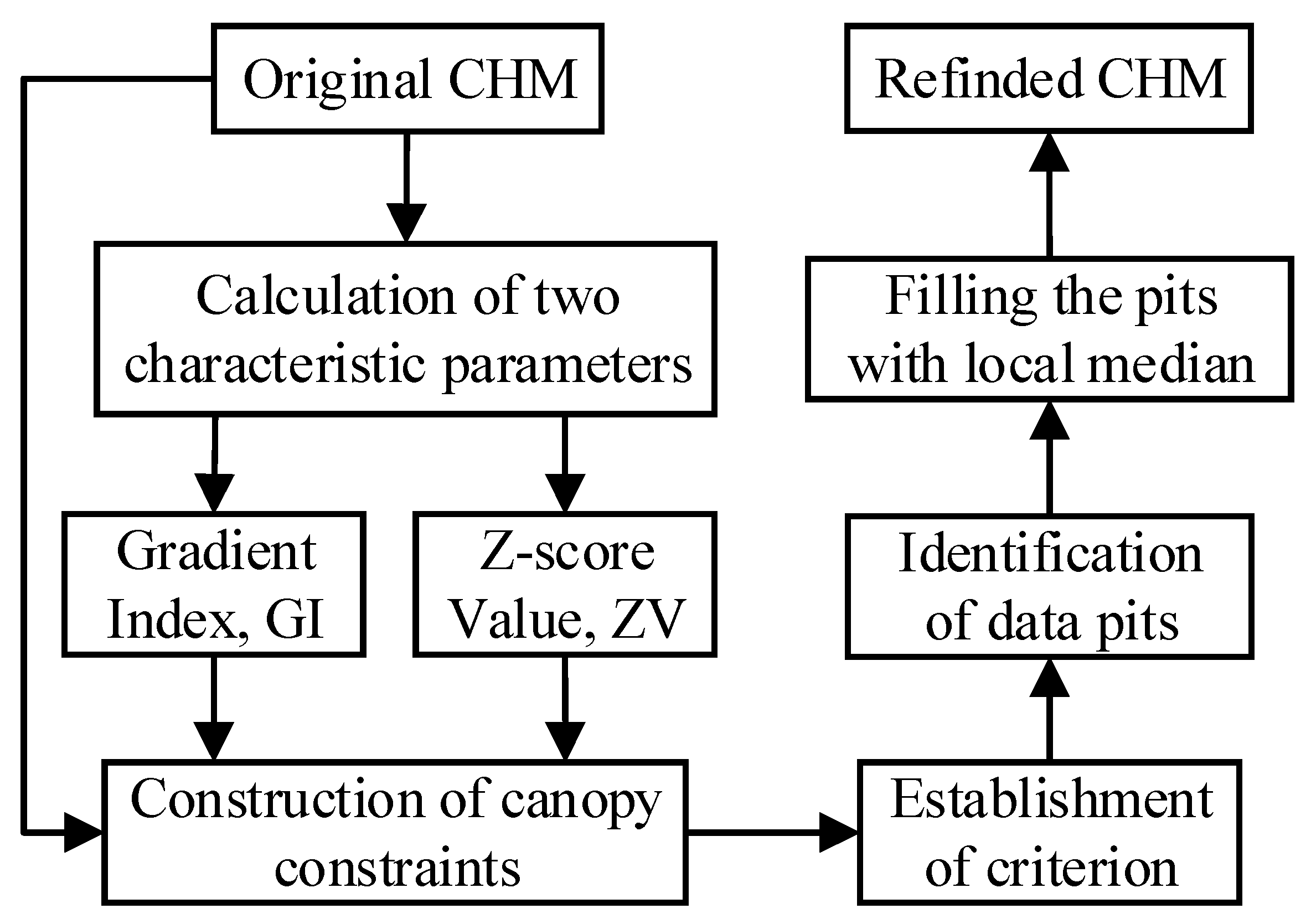
2. Development of a Weighted Differential Gradient Method
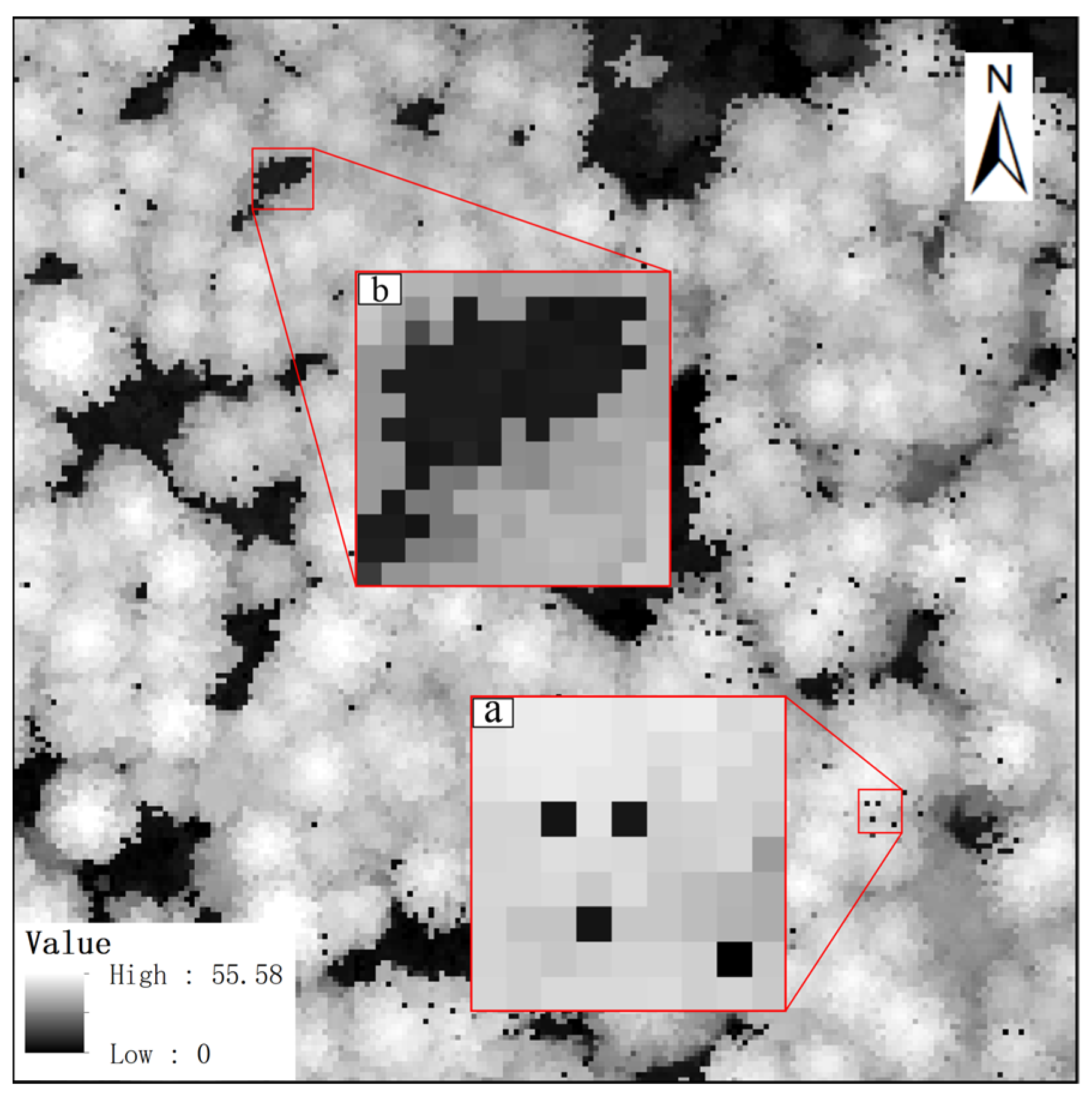
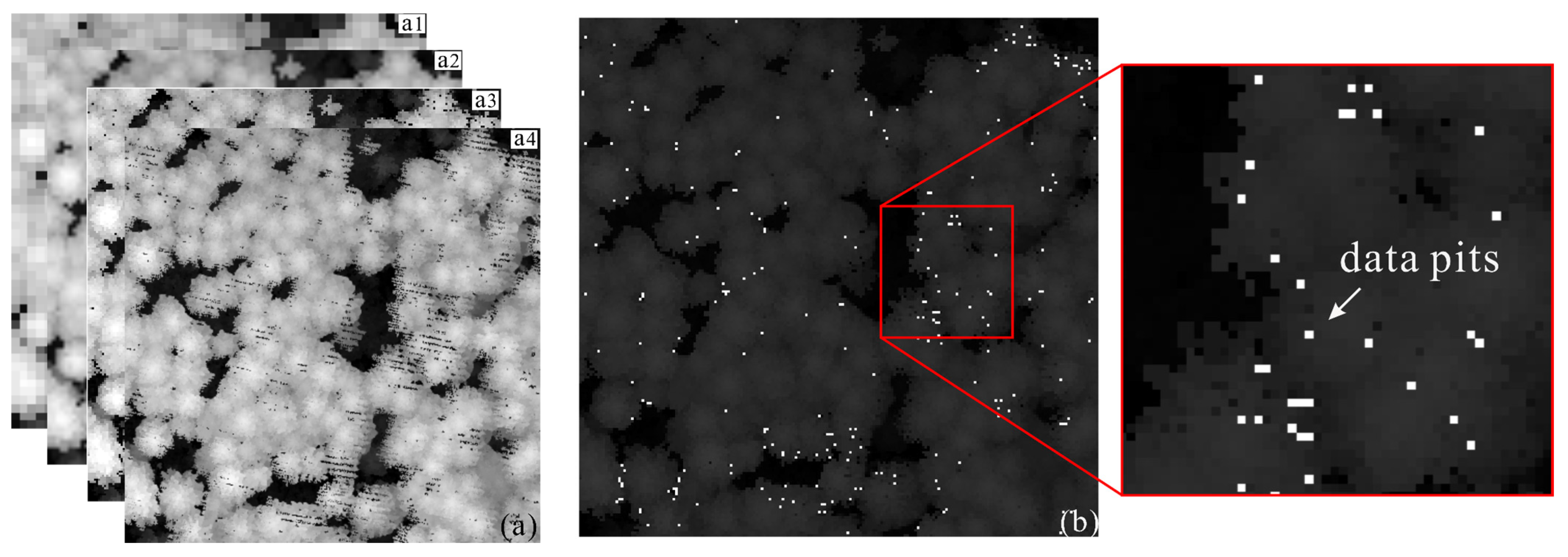
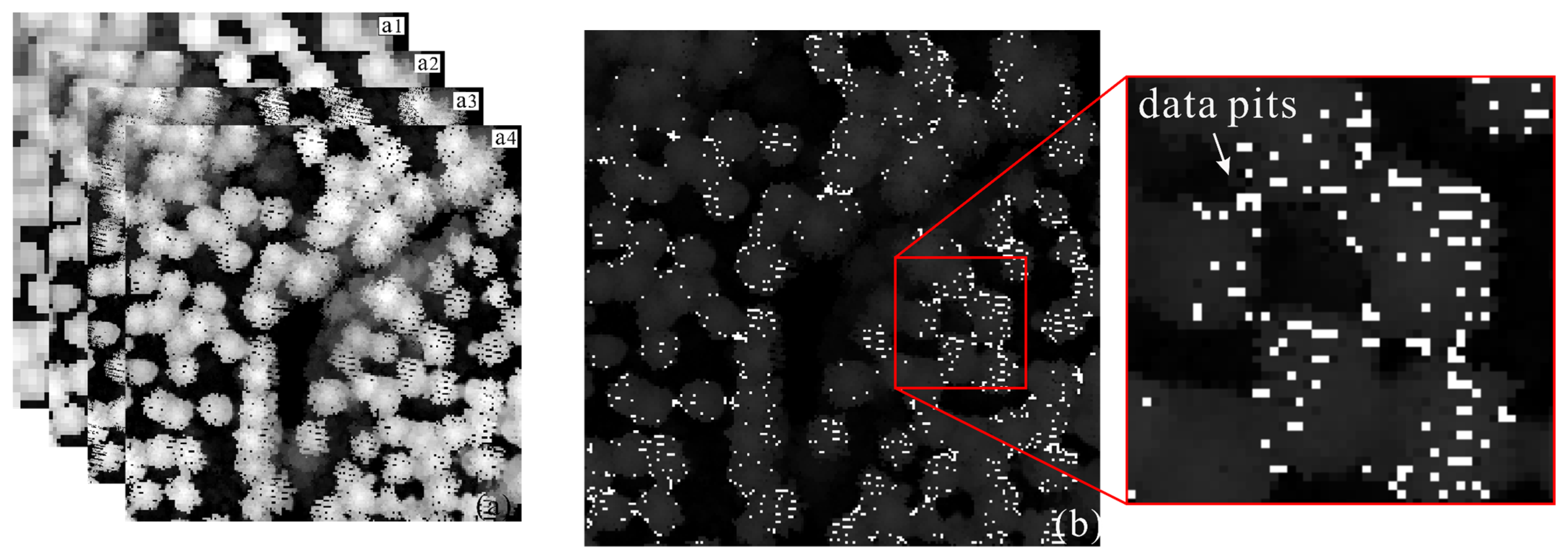
2.1. Characteristic Parameters for Canopy Pits Identification
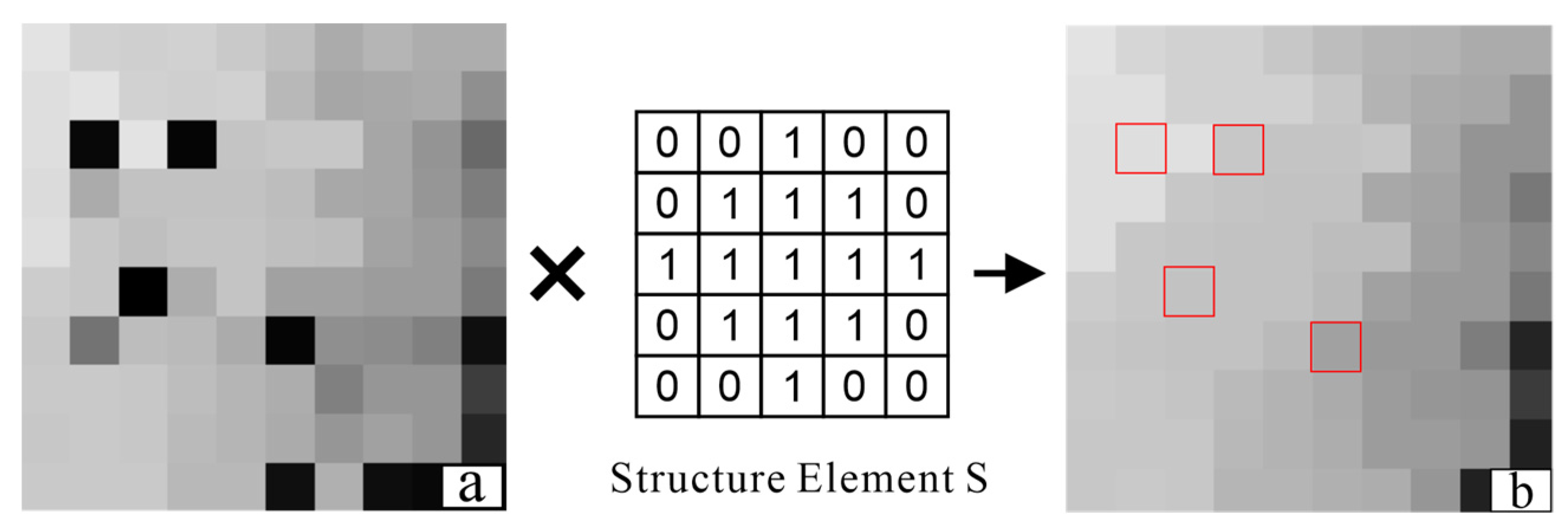
2.2. Establishment of Canopy Constraints for the Weighted Differential Gradient
2.3. Filling the Pits
3. Experiments and Analysis
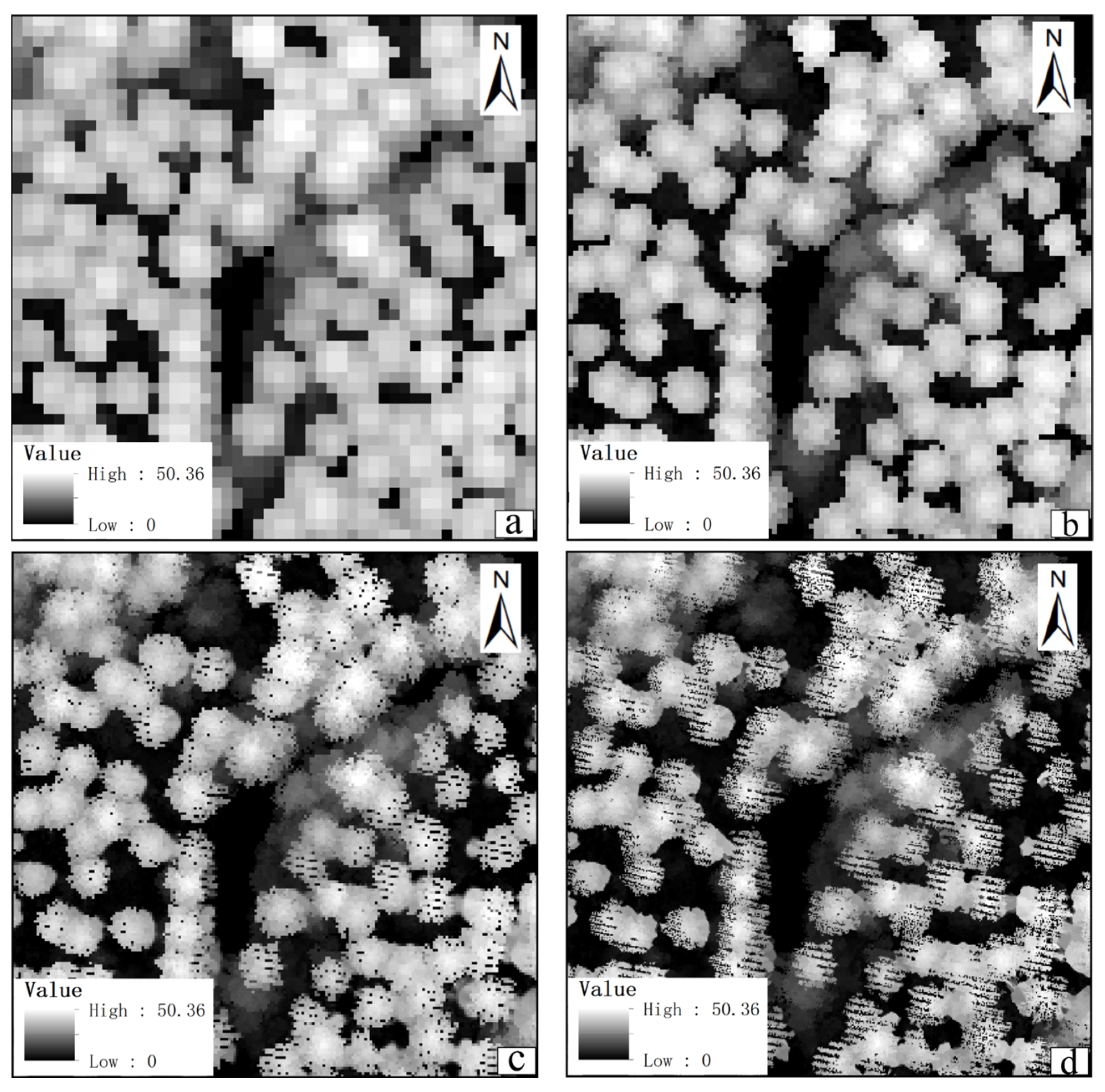
3.1. Experimental Data and Preprocessing
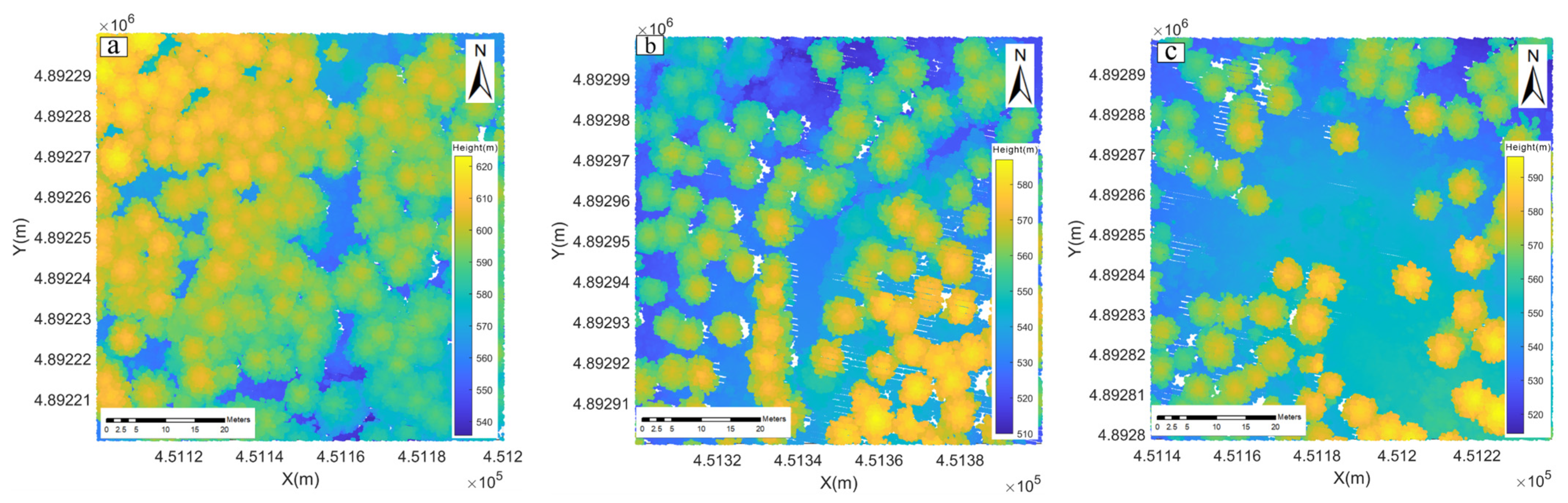
3.1.1. LiDAR Data
3.1.2. Data Preprocessing
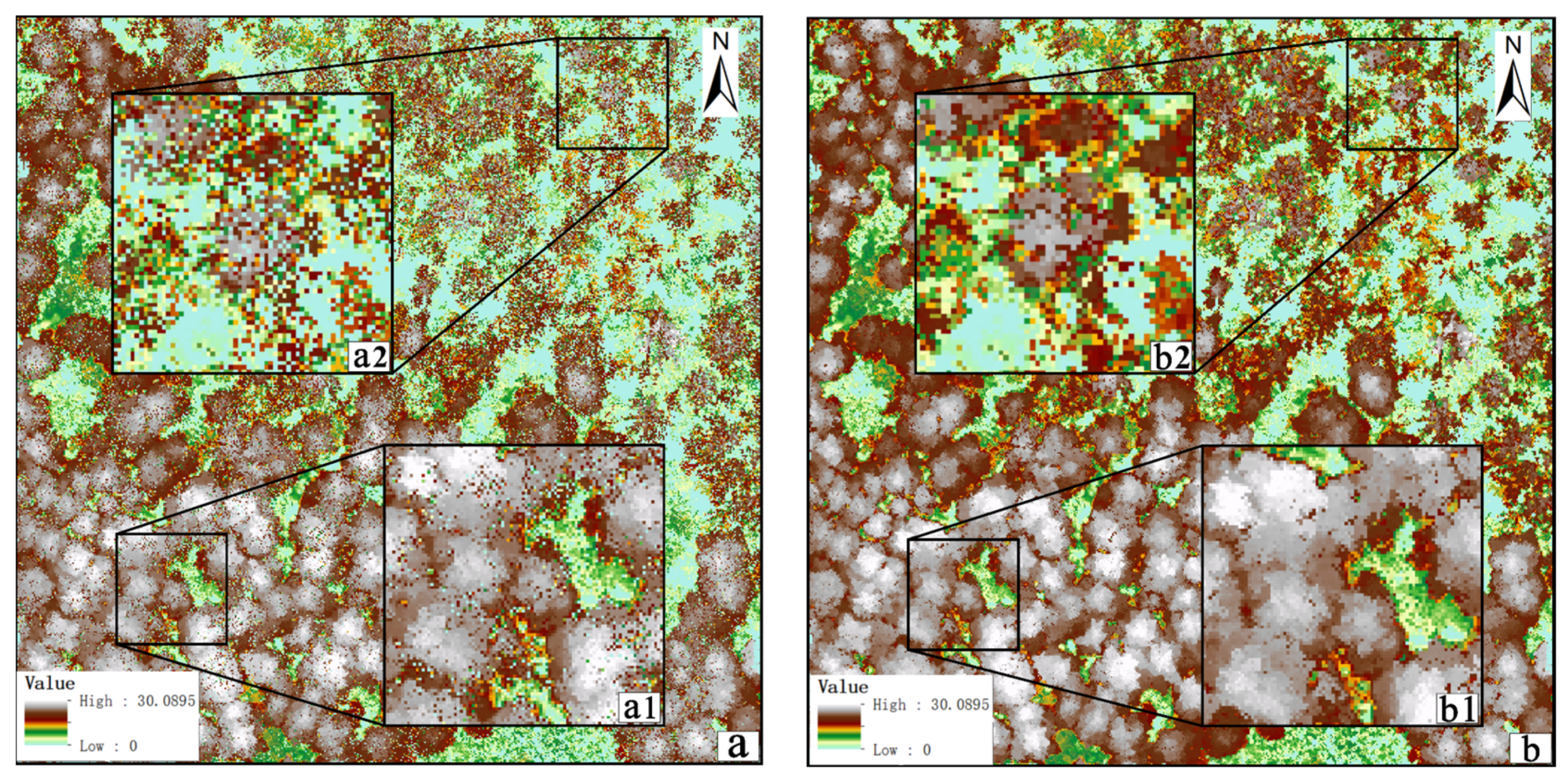
3.2. Automatically Identifying and Filling Pits Pixels for CHM
- (1)
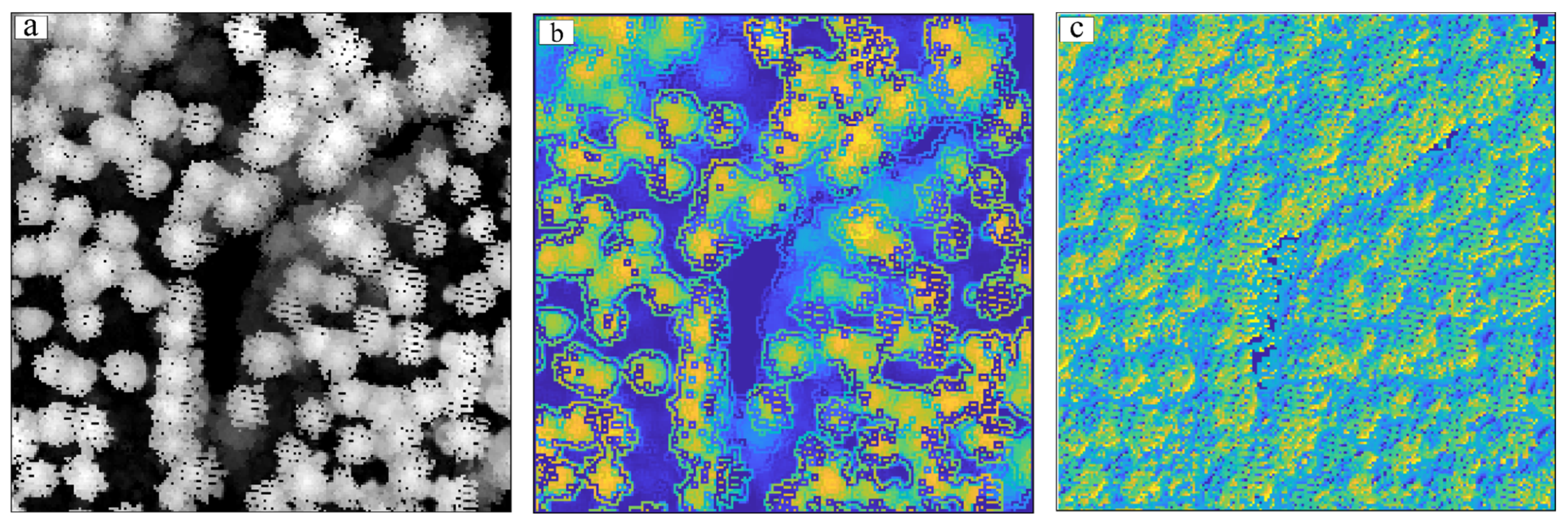
- Experiment with Test Area 1
- (2)
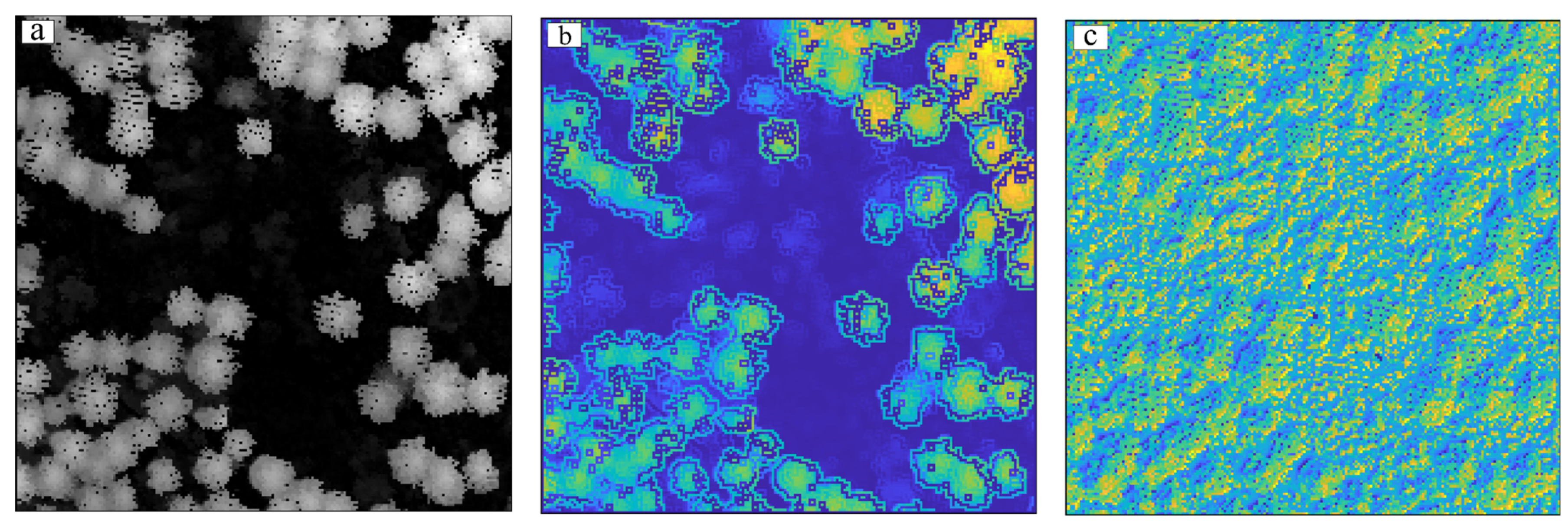
- Experiment with Test Area 2 and Test Area 3
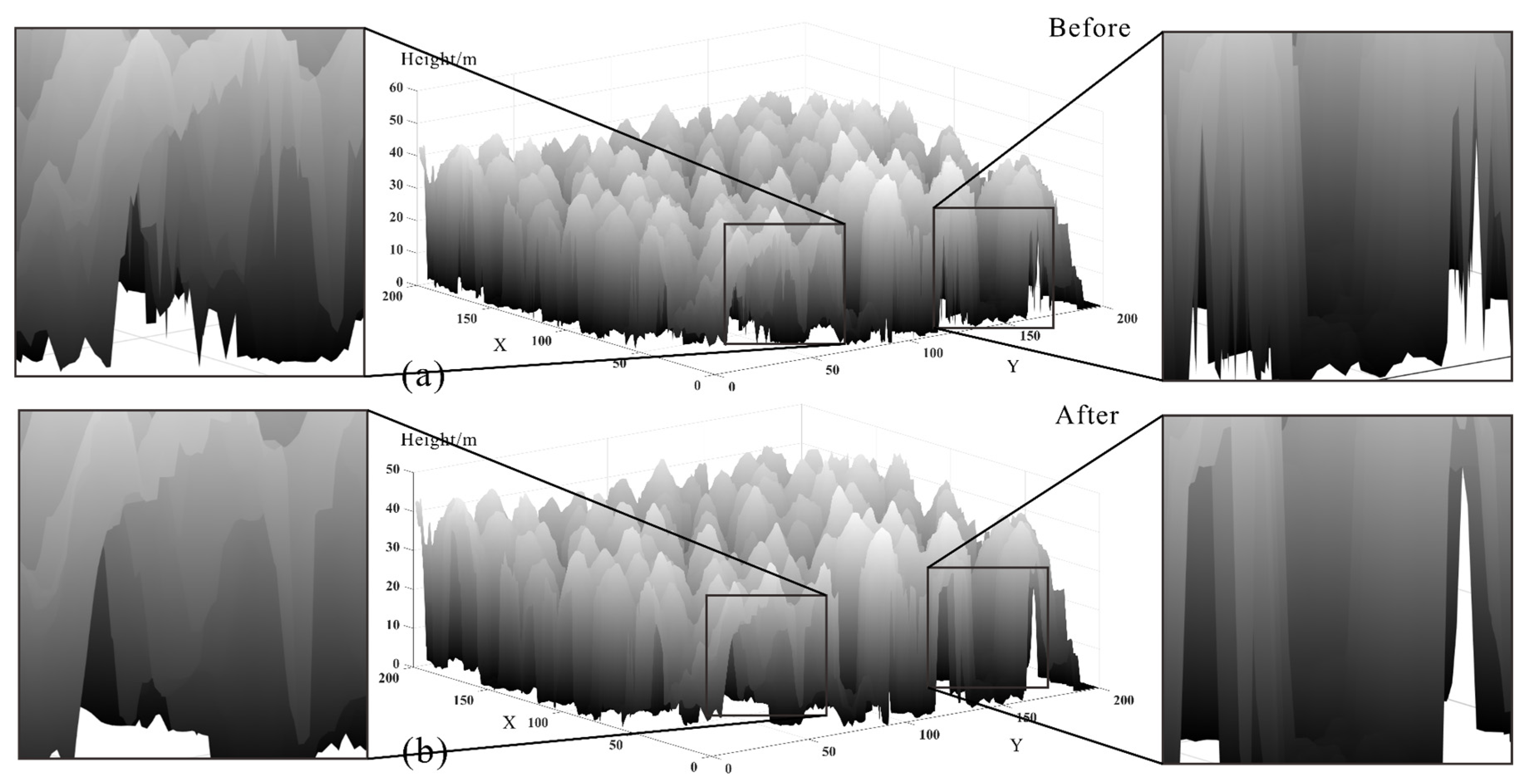
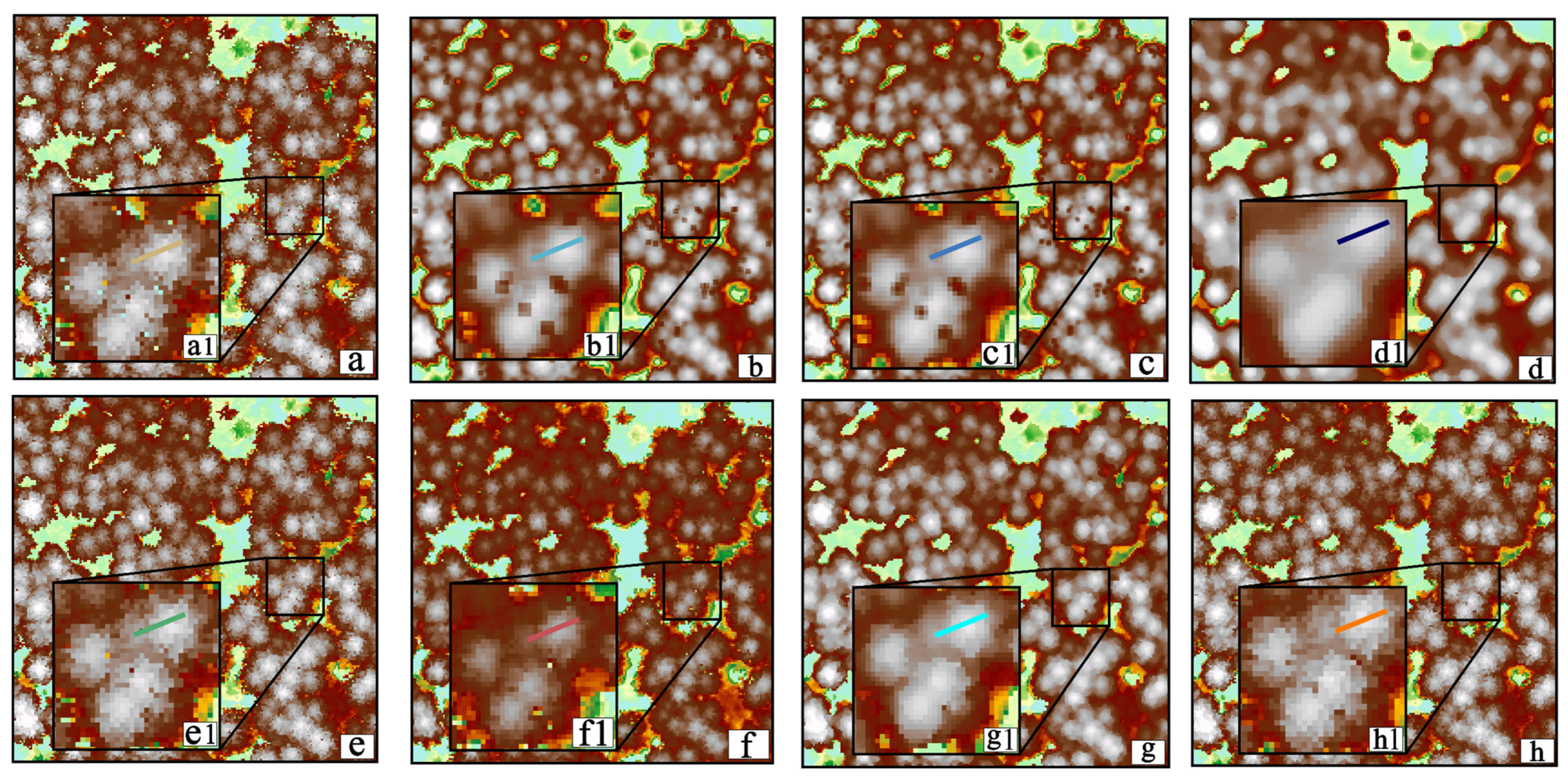
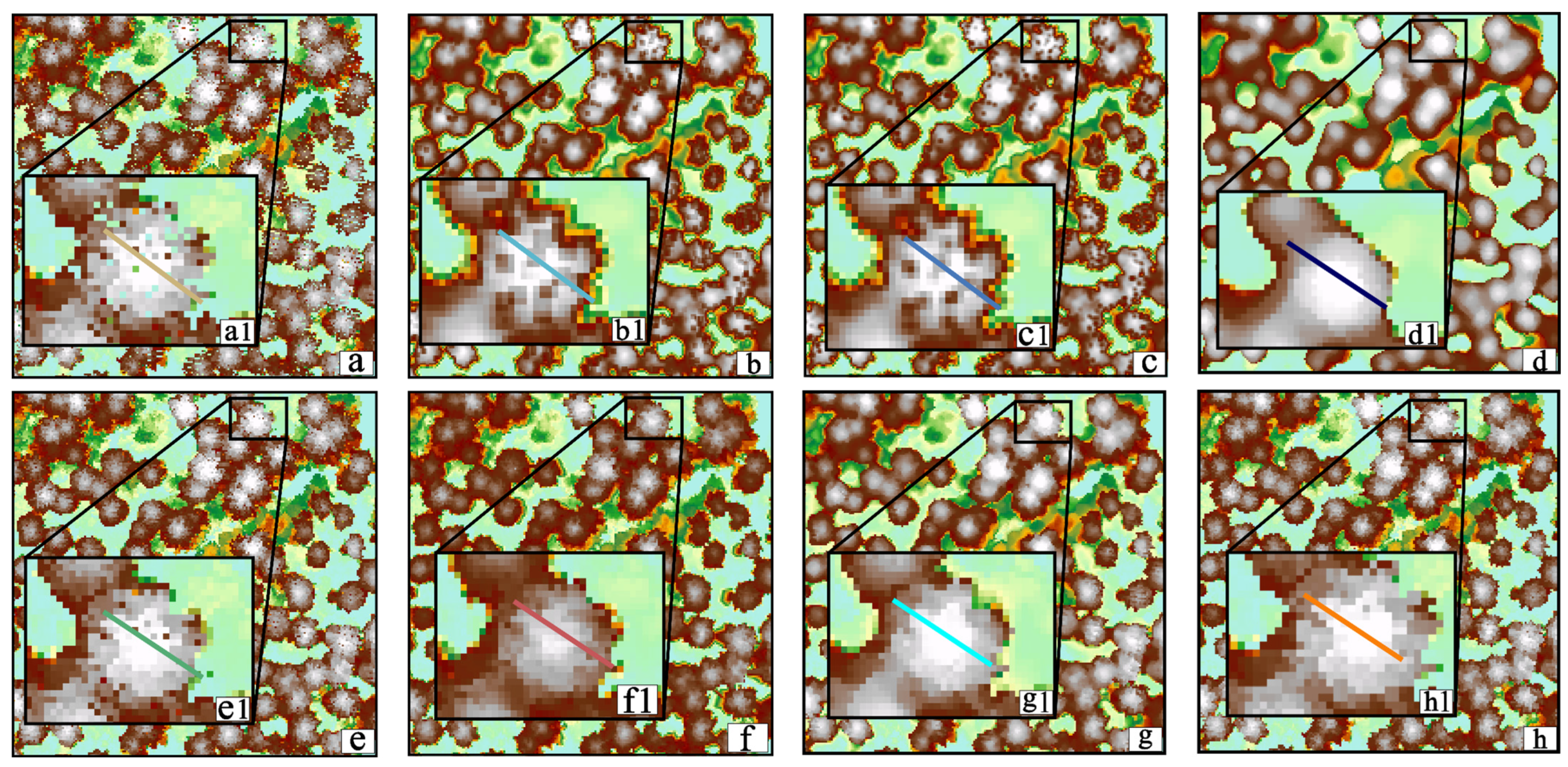
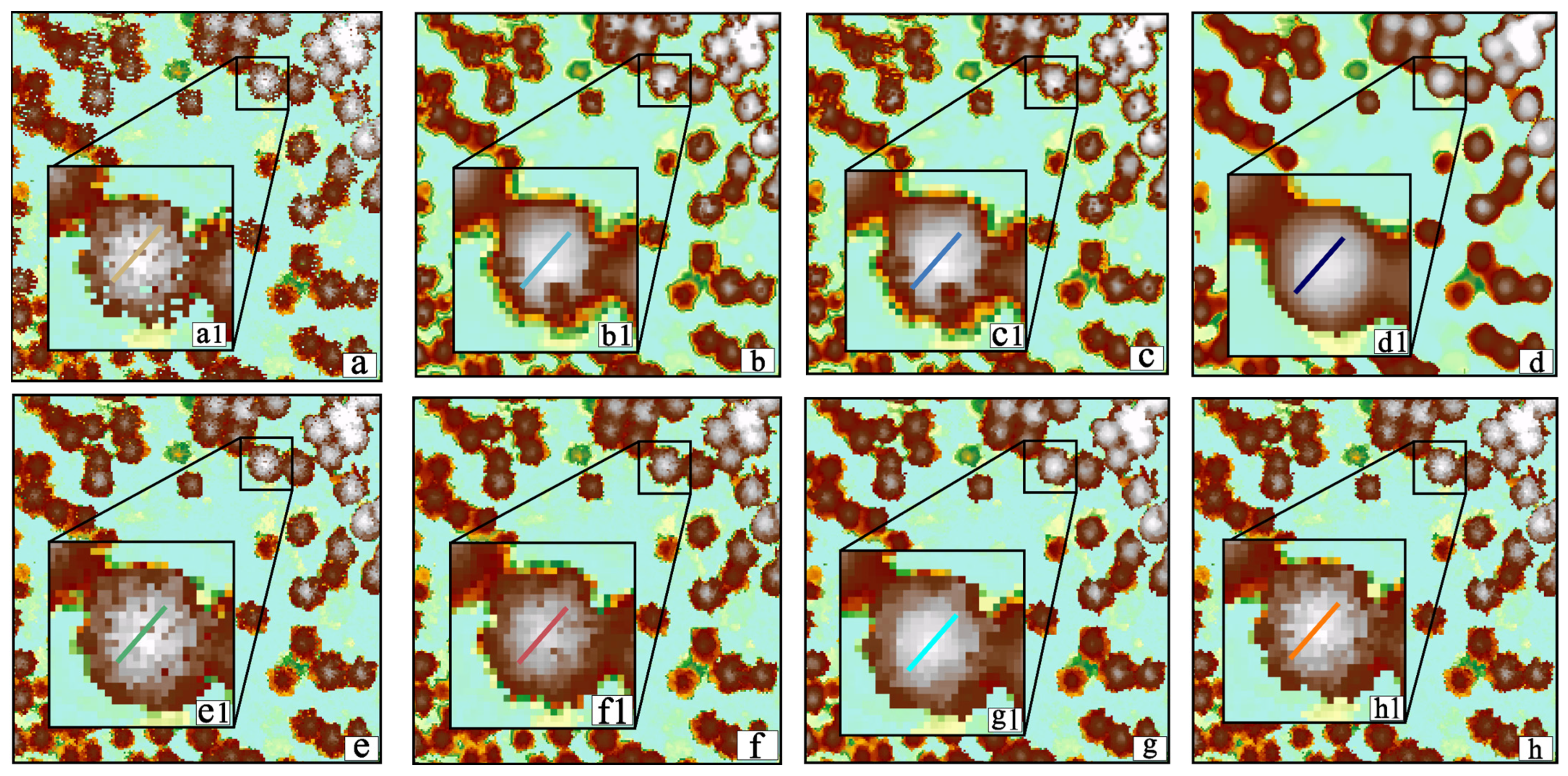
3.3. Comparison and Analysis
3.3.1. Visual Evaluation
3.3.2. Quantitative Evaluation
3.4. Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, D.H.; Ko, C.U.; Kim, D.G.; Kang, J.T.; Park, J.M.; Cho, H.J. Automated Segmentation of Individual Tree Structures Using Deep Learning over LiDAR Point Cloud Data. Forests 2023, 14, 1159. [Google Scholar] [CrossRef]
- Whelan, A.W.; Cannon, J.B.; Bigelow, S.W.; Rutledge, B.T.; Meador, A.J.S. Improving generalized models of forest structure in complex forest types using area-and voxel-based approaches from lidar. Remote Sens. Environ. 2023, 284, 113362. [Google Scholar] [CrossRef]
- Oehmcke, S.; Li, L.; Trepekli, K.; Revenga, J.C.; Nord-Larsen, T.; Gieseke, F.; Igel, C. Deep point cloud regression for above-ground forest biomass estimation from airborne LiDAR. Remote Sens. Environ. 2024, 302, 113968. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, J.; Skidmore, A.K.; Premier, J.; Heurich, M. A voxel matching method for effective leaf area index estimation in temperate deciduous forests from leaf-on and leaf-off airborne LiDAR data. Remote Sens. Environ. 2020, 240, 111696. [Google Scholar] [CrossRef]
- Schneider, F.D.; Kükenbrink, D.; Schaepman, M.E.; Schimel, D.S.; Morsdorf, F. Quantifying 3D structure and occlusion in dense tropical and temperate forests using close-range LiDAR. Agric. For. Meteorol. 2019, 268, 249–257. [Google Scholar] [CrossRef]
- Liu, Q.; Fu, L.; Wang, G.; Li, S.; Li, Z.; Chen, E.; Hu, K. Improving estimation of forest canopy cover by introducing loss ratio of laser pulses using airborne LiDAR. IEEE Trans. Geosci. Remote Sens. 2019, 58, 567–585. [Google Scholar] [CrossRef]
- Cai, S.; Zhang, W.; Jin, S.; Shao, J.; Li, L.; Yu, S.; Yan, G. Improving the estimation of canopy cover from UAV-LiDAR data using a pit-free CHM-based method. Int. J. Digit. Earth 2021, 14, 1477–1492. [Google Scholar] [CrossRef]
- Tian, Y.; Huang, H.; Zhou, G.; Zhang, Q.; Tao, J.; Zhang, Y.; Lin, J. Aboveground mangrove biomass estimation in Beibu Gulf using machine learning and UAV remote sensing. Sci. Total Environ. 2021, 781, 146816. [Google Scholar] [CrossRef]
- Zhao, K.; Suarez, J.C.; Garcia, M.; Hu, T.; Wang, C.; Londo, A. Utility of multitemporal lidar for forest and carbon monitoring: Tree growth, biomass dynamics, and carbon flux. Remote Sens. Environ. 2018, 204, 883–897. [Google Scholar] [CrossRef]
- Qi, Z.; Li, S.; Pang, Y.; Zheng, G.; Kong, D.; Li, Z. Assessing spatiotemporal variations of forest carbon density using bi-temporal discrete aerial laser scanning data in Chinese boreal forests. For. Ecosyst. 2023, 10, 100135. [Google Scholar] [CrossRef]
- Hao, J.; Li, X.; Wu, H.; Yang, K.; Zeng, Y.; Wang, Y.; Pan, Y. Extraction and analysis of tree canopy height information in high-voltage transmission-line corridors by using integrated optical remote sensing and LiDAR. Geod. Geodyn. 2023, 14, 292–303. [Google Scholar] [CrossRef]
- Erfanifard, Y.; Stereńczak, K.; Kraszewski, B.; Kamińska, A. Development of a robust canopy height model derived from ALS point clouds for predicting individual crown attributes at the species level. Int. J. Remote Sens. 2018, 39, 9206–9227. [Google Scholar] [CrossRef]
- Xiao, C.; Qin, R.; Huang, X. Treetop detection using convolutional neural networks trained through automatically generated pseudo labels. Int. J. Remote Sens. 2020, 41, 3010–3030. [Google Scholar] [CrossRef]
- Mu, Y.; Zhou, G.; Wang, H. Canopy lidar point cloud data k-means clustering watershed segmentation method. ISPRS Annals of the Photogrammetry. Remote Sens. Spat. Inf. Sci. 2020, 6, 67–73. [Google Scholar] [CrossRef]
- Estornell, J.; Hadas, E.; Martí, J.; López-Cortés, I. Tree extraction and estimation of walnut structure parameters using airborne LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2021, 96, 102273. [Google Scholar] [CrossRef]
- Liu, H.; Dong, P. A new method for generating canopy height models from discrete-return LiDAR point clouds. Remote Sens. Lett. 2014, 5, 575–582. [Google Scholar] [CrossRef]
- Chang, K.T.; Lin, C.; Lin, Y.C.; Liu, J.K. Accuracy Assessment of Crown Delineation Methods for the Individual Trees Using LIDAR Data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 585–588. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y.A. Generating pit-free canopy height models from airborne lidar. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Duan, Z.; Zeng, Y.; Zhao, D.; Wu, B.; Zhao, Y.; Zhu, J. Method of removing pits of canopy height model from airborne laser radar. Trans. Chin. Soc. Agric. Eng. 2014, 30, 209–217. [Google Scholar]
- Zhao, D.; Pang, Y.; Li, Z.; Sun, G. Filling invalid values in a lidar-derived canopy height model with morphological crown control. Int. J. Remote Sens. 2013, 34, 4636–4654. [Google Scholar] [CrossRef]
- Kucharczyk, M.; Hugenholtz, C.H.; Zou, X. UAV–LiDAR accuracy in vegetated terrain. J. Unmanned Veh. Syst. 2018, 6, 212–234. [Google Scholar] [CrossRef]
- Axelsson, P. DEM generation from laser scanner data using adaptive TIN models. Int. Arch. Photogramm. Remote Sens. 2000, 33, 110–117. [Google Scholar]
- LaRue, E.A.; Fahey, R.; Fuson, T.L.; Foster, J.R.; Matthes, J.H.; Krause, K.; Hardiman, B.S. Evaluating the sensitivity of forest structural diversity characterization to LiDAR point density. Ecosphere 2022, 13, e4209. [Google Scholar] [CrossRef]
- Swanson, C.; Merrick, T.; Abelev, A.; Liang, R.; Vermillion, M.; Buma, W.; Li, R. Effects of point density on interpretability of lidar-derived forest structure metrics in two temperate forests. bioRxiv 2024. [Google Scholar] [CrossRef]
- Shamsoddini, A.; Turner, R.; Trinder, J.C. Improving lidar-based forest structure mapping with crown-level pit removal. J. Spat. Sci. 2013, 58, 29–51. [Google Scholar] [CrossRef]
- Mielcarek, M.; Stereńczak, K.; Khosravipour, A. Testing and evaluating different LiDAR-derived canopy height model generation methods for tree height estimation. Int. J. Appl. Earth Obs. Geoinf. 2018, 71, 132–143. [Google Scholar] [CrossRef]
- Barnes, C.; Balzter, H.; Barrett, K.; Eddy, J.; Milner, S.; Suárez, J.C. Individual tree crown delineation from airborne laser scanning for diseased larch forest stands. Remote Sens. 2017, 9, 231. [Google Scholar] [CrossRef]
- Ben-Arie, J.R.; Hay, G.J.; Powers, R.P.; Castilla, G.; St-Onge, B. Development of a pit filling algorithm for LiDAR canopy height models. Comput. Geosci. 2009, 35, 1940–1949. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M. Generating spike-free digital surface models using LiDAR raw point clouds: A new approach for forestry applications. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 104–114. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Li, Y.; Yue, T.; Wang, X. Robust and parameter-free algorithm for constructing pit-free canopy height models. ISPRS Int. J. Geo-Inf. 2017, 6, 219. [Google Scholar] [CrossRef]
- Hao, Y.; Zhen, Z.; Li, F.; Zhao, Y. A graph-based progressive morphological filtering (GPMF) method for generating canopy height models using ALS data. Int. J. Appl. Earth Obs. Geoinf. 2019, 79, 84–96. [Google Scholar] [CrossRef]
- Rousseeuw, P.J.; Hubert, M. Robust statistics for outlier detection. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 73–79. [Google Scholar] [CrossRef]
- Zhang, W.; Cai, S.; Liang, X.; Shao, J.; Hu, R.; Yu, S.; Yan, G. Cloth simulation-based construction of pit-free canopy height models from airborne LiDAR data. For. Ecosyst. 2020, 7, 1. [Google Scholar] [CrossRef]
- Song, Q.; Xiang, M.; Hovis, C.; Zhou, Q.; Lu, M.; Tang, H.; Wu, W. Object-based feature selection for crop classification using multi-temporal high-resolution imagery. Int. J. Remote Sens. 2019, 40, 2053–2068. [Google Scholar] [CrossRef]
- Liu, L.; Lim, S.; Shen, X.; Yebra, M. A multiscale morphological algorithm for improvements to canopy height models. Comput. Geosci. 2019, 130, 20–31. [Google Scholar] [CrossRef]
- Yin, D.; Wang, L. Individual mangrove tree measurement using UAV-based LiDAR data: Possibilities and challenges. Remote Sens. Environ. 2019, 223, 34–49. [Google Scholar] [CrossRef]
- Qiao, L.; Gao, D.; Zhao, R.; Tang, W.; An, L.; Li, M.; Sun, H. Improving estimation of LAI dynamic by fusion of morphological and vegetation indices based on UAV imagery. Comput Electron Agr. 2022, 192, 106603. [Google Scholar] [CrossRef]
- Holmgren, J.; Lindberg, E.; Olofsson, K.; Persson, H.J. Tree crown segmentation in three dimensions using density models derived from airborne laser scanning. Int. J. Remote Sens. 2022, 43, 299–329. [Google Scholar] [CrossRef]
- Rizaev, I.G.; Pogorelov, A.V.; Krivova, M.A. A technique to increase the efficiency of artefacts identification in lidar-based canopy height models. Int. J. Remote Sens. 2016, 37, 1658–1670. [Google Scholar] [CrossRef]
- Marshall, J.A.; Roering, J.J.; Gavin, D.G.; Granger, D.E. Late Quaternary Climatic Controls on Erosion Rates and Geomorphic Processes in Western Oregon, USA. Geol. Soc. Am. Bull. 2017, 129, 715–731. [Google Scholar] [CrossRef]
- Cloud Compare. Available online: http://www.cloudcompare.org/main.html (accessed on 25 March 2022).
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Impact of tree-oriented growth order in marker-controlled region growing for individual tree crown delineation using airborne laser scanner (ALS) data. Remote Sens. 2014, 6, 555–579. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Stehman, S.V.; Zhang, L. Agent-based region growing for individual tree crown delineation from airborne laser scanning (ALS) data. Int. J. Remote Sens. 2015, 36, 1965–1993. [Google Scholar] [CrossRef]
- Granholm, A.H.; Lindgren, N.; Olofsson, K.; Nyström, M.; Allard, A.; Olsson, H. Estimating vertical canopy cover using dense image-based point cloud data in four vegetation types in southern Sweden. Int. J. Remote Sens. 2017, 38, 1820–1838. [Google Scholar] [CrossRef]
- Oh, S.; Jung, J.; Shao, G.; Shao, G.; Gallion, J.; Fei, S. High-resolution canopy height model generation and validation using USGS 3DEP LiDAR data in Indiana, USA. Remote Sens. 2022, 14, 935. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, K.; McDermid, G.J.; Hay, G.J. The influence of sampling density on geographically weighted regression: A case study using forest canopy height and optical data. Int. J. Remote Sens. 2012, 33, 2909–2924. [Google Scholar] [CrossRef]
- Zhou, G.; Song, B.; Liang, P.; Xu, J.; Yue, T. Voids filling of DEM with multiattention generative adversarial network model. Remote Sens. 2022, 14, 1206. [Google Scholar] [CrossRef]
- Quan, Y.; Li, M.; Hao, Y.; Wang, B. Comparison and evaluation of different pit-filling methods for generating high resolution canopy height model using UAV laser scanning data. Remote Sens. 2021, 13, 2239. [Google Scholar] [CrossRef]
- Bonnet, S.; Lisein, J.; Lejeune, P. Comparison of UAS photogrammetric products for tree detection and characterization of coniferous stands. Int. J. Remote Sens. 2017, 38, 5310–5337. [Google Scholar] [CrossRef]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.M.; Dalponte, M.; Pfeifer, N. A benchmark of lidar-based single tree detection methods using heterogeneous forest data from the alpine space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef]








| Test Area | Number of Trees | Tree Height(m) | ) | |
|---|---|---|---|---|
| Average of Height | Std of Height | |||
| 1 | 191 | 43.39 | 5.28 | 16.02 |
| 2 | 104 | 36.32 | 4.53 | 13.18 |
| 3 | 75 | 35.92 | 6.69 | 11.64 |
| Methods | Evaluation Parameters | |||
|---|---|---|---|---|
| R2 | RMSE | MBE | ||
| Test Area 1 | Raw CHM | 0.79 | 1.30 | 1.20 |
| Mean filter | 0.83 | 1.17 | 0.62 | |
| Gaussian filter | 0.87 | 1.05 | 0.88 | |
| Median filter | 0.92 | 0.67 | 0.48 | |
| Pit free | 0.94 | 0.70 | 0.28 | |
| Spike free | 0.98 | 0.37 | 0.16 | |
| GPMF | 0.98 | 0.30 | 0.13 | |
| Our method | 0.99 | 0.29 | 0.06 | |
| Test Area 2 | Raw CHM | 0.72 | 1.48 | 1.40 |
| Mean filter | 0.78 | 1.31 | 1.00 | |
| Gaussian filter | 0.76 | 1.36 | 0.77 | |
| Median filter | 0.89 | 1.12 | 0.53 | |
| Pit free | 0.86 | 1.05 | 0.60 | |
| Spike free | 0.98 | 0.41 | 0.16 | |
| GPMF | 0.98 | 0.33 | 0.13 | |
| Our method | 0.98 | 0.31 | 0.12 | |
| Test Area 3 | Raw CHM | 0.80 | 1.17 | 1.54 |
| Mean filter | 0.87 | 1.08 | 0.94 | |
| Gaussian filter | 0.89 | 1.00 | 0.79 | |
| Median filter | 0.92 | 0.88 | 0.56 | |
| Pit free | 0.93 | 0.77 | 0.44 | |
| Spike free | 0.98 | 0.47 | 0.14 | |
| GPMF | 0.99 | 0.30 | 0.18 | |
| Our method | 0.98 | 0.36 | 0.12 | |
| Methods | Indicator | Test Area 1 | Test Area 2 | Test Area 3 |
|---|---|---|---|---|
| Raw CHM | P | 0.69 | 0.71 | 0.61 |
| R | 0.75 | 0.75 | 0.64 | |
| F1 score | 0.72 | 0.73 | 0.62 | |
| Mean filter | P | 0.79 | 0.79 | 0.80 |
| R | 0.76 | 0.75 | 0.70 | |
| F1 score | 0.77 | 0.77 | 0.75 | |
| Gaussian filter | P | 0.79 | 0.78 | 0.83 |
| R | 0.78 | 0.77 | 0.75 | |
| F1 score | 0.78 | 0.77 | 0.79 | |
| Median filter | P | 0.77 | 0.73 | 0.76 |
| R | 0.86 | 0.88 | 0.89 | |
| F1 score | 0.81 | 0.80 | 0.82 | |
| Pit free | P | 0.85 | 0.86 | 0.82 |
| R | 0.85 | 0.87 | 0.89 | |
| F1 score | 0.85 | 0.86 | 0.86 | |
| Spike free | P | 0.86 | 0.82 | 0.82 |
| R | 0.85 | 0.88 | 0.84 | |
| F1 score | 0.85 | 0.85 | 0.83 | |
| GPMF | P | 0.86 | 0.82 | 0.87 |
| R | 0.85 | 0.89 | 0.92 | |
| F1 score | 0.85 | 0.85 | 0.89 | |
| Our method | P | 0.83 | 0.89 | 0.89 |
| R | 0.84 | 0.86 | 0.93 | |
| F1 score | 0.83 | 0.88 | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, G.; Li, H.; Huang, J.; Gao, E.; Song, T.; Han, X.; Zhu, S.; Liu, J. Weighted Differential Gradient Method for Filling Pits in Light Detection and Ranging (LiDAR) Canopy Height Model. Remote Sens. 2024, 16, 1304. https://doi.org/10.3390/rs16071304
Zhou G, Li H, Huang J, Gao E, Song T, Han X, Zhu S, Liu J. Weighted Differential Gradient Method for Filling Pits in Light Detection and Ranging (LiDAR) Canopy Height Model. Remote Sensing. 2024; 16(7):1304. https://doi.org/10.3390/rs16071304
Chicago/Turabian StyleZhou, Guoqing, Haowen Li, Jing Huang, Ertao Gao, Tianyi Song, Xiaoting Han, Shuaiguang Zhu, and Jun Liu. 2024. "Weighted Differential Gradient Method for Filling Pits in Light Detection and Ranging (LiDAR) Canopy Height Model" Remote Sensing 16, no. 7: 1304. https://doi.org/10.3390/rs16071304
APA StyleZhou, G., Li, H., Huang, J., Gao, E., Song, T., Han, X., Zhu, S., & Liu, J. (2024). Weighted Differential Gradient Method for Filling Pits in Light Detection and Ranging (LiDAR) Canopy Height Model. Remote Sensing, 16(7), 1304. https://doi.org/10.3390/rs16071304







