Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
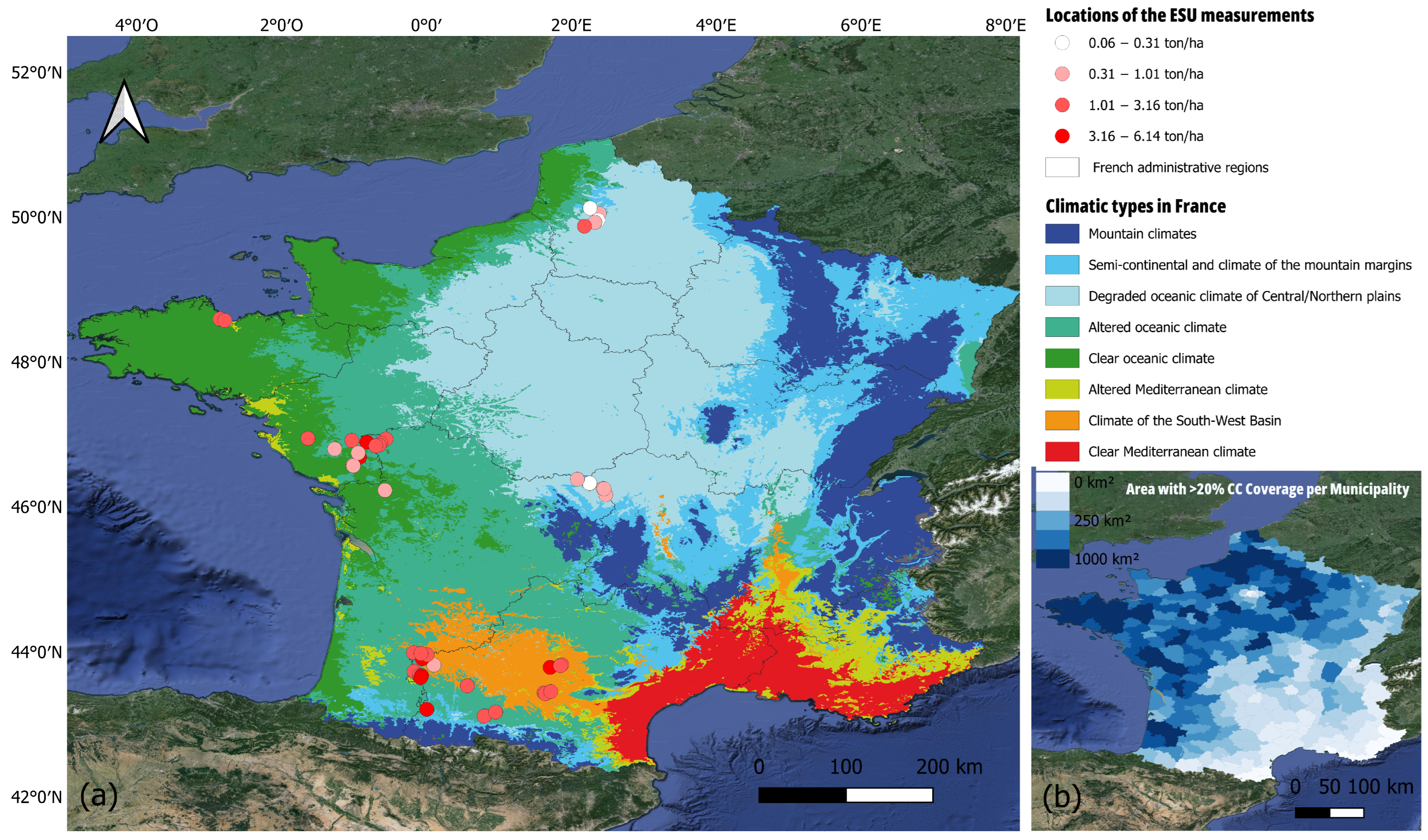
2.1. Study Area
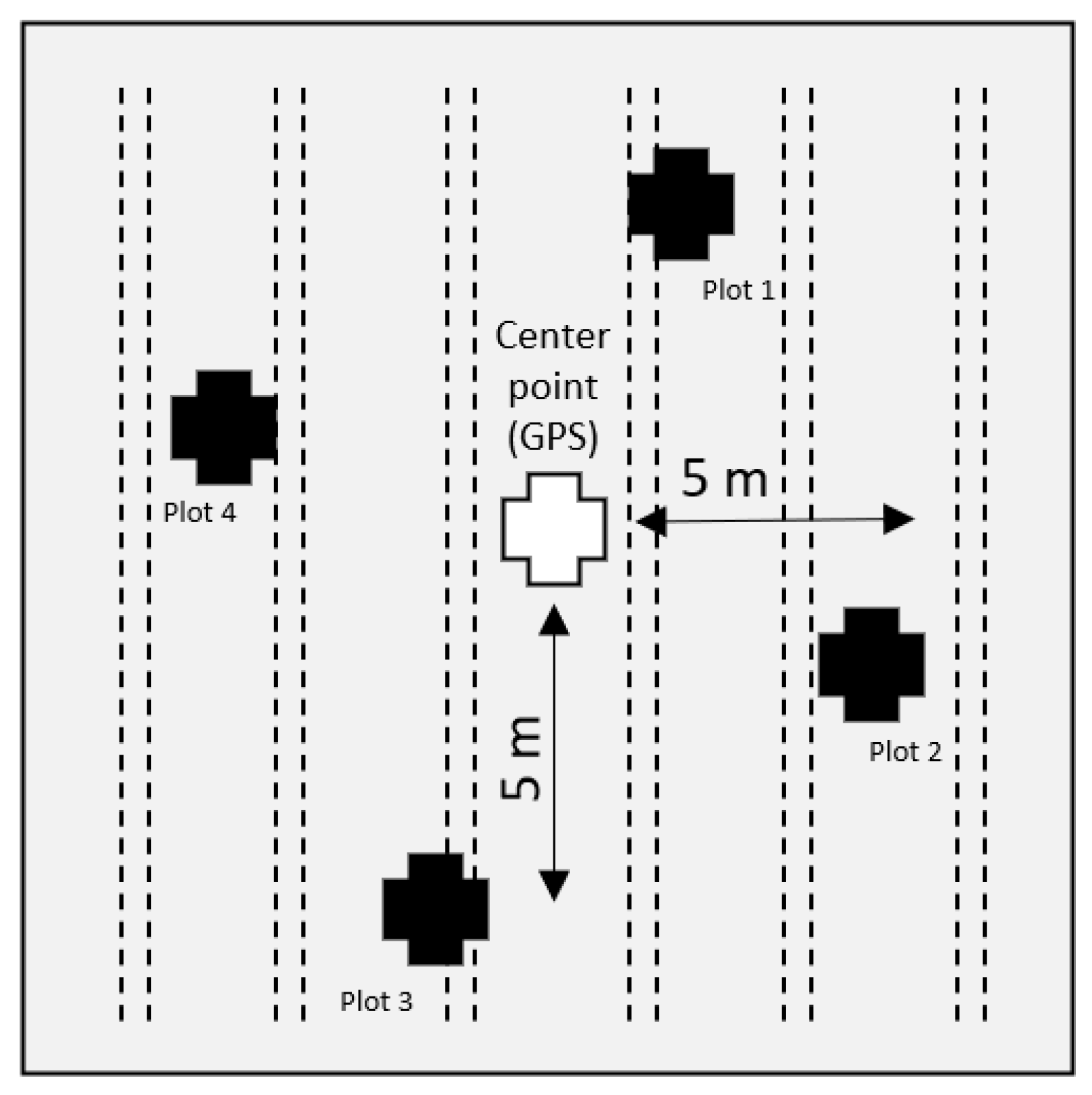
2.2. Field Data Sampling Procedures and Cover Crop Characteristcs
2.3. Satellite Data
2.4. Methods
2.4.1. Empirical Relationships with Spectral Bands and Vegetation Indices Using the Last Available Image before the Sampling
2.4.2. Multivariate Analyses Using Machine Learning Algorithms
2.5. Validation
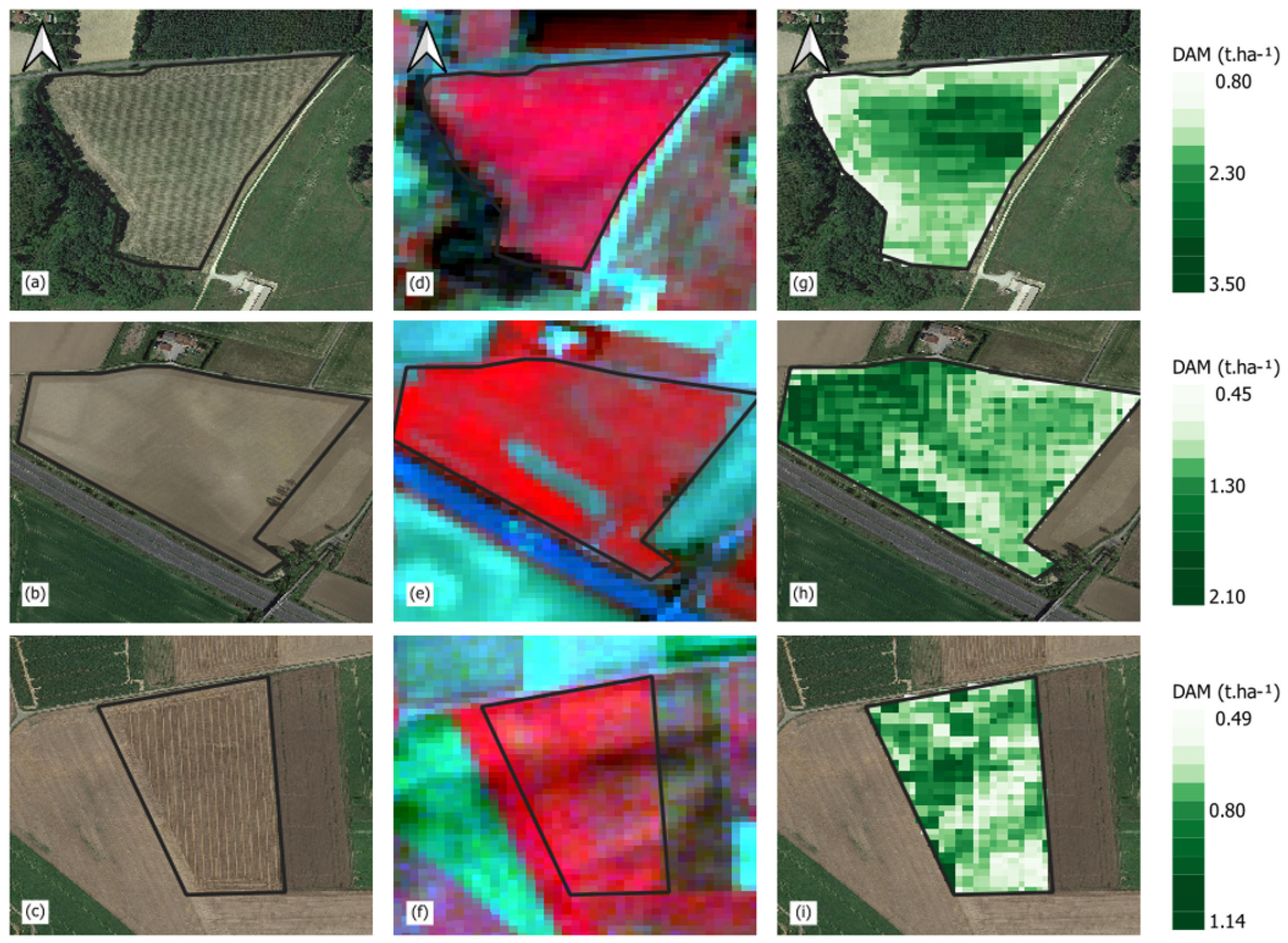
2.6. Model Spatialization of the Final Dry Biomass on the Experimental Fields across France
3. Results
3.1. Cover Crop Characteristics
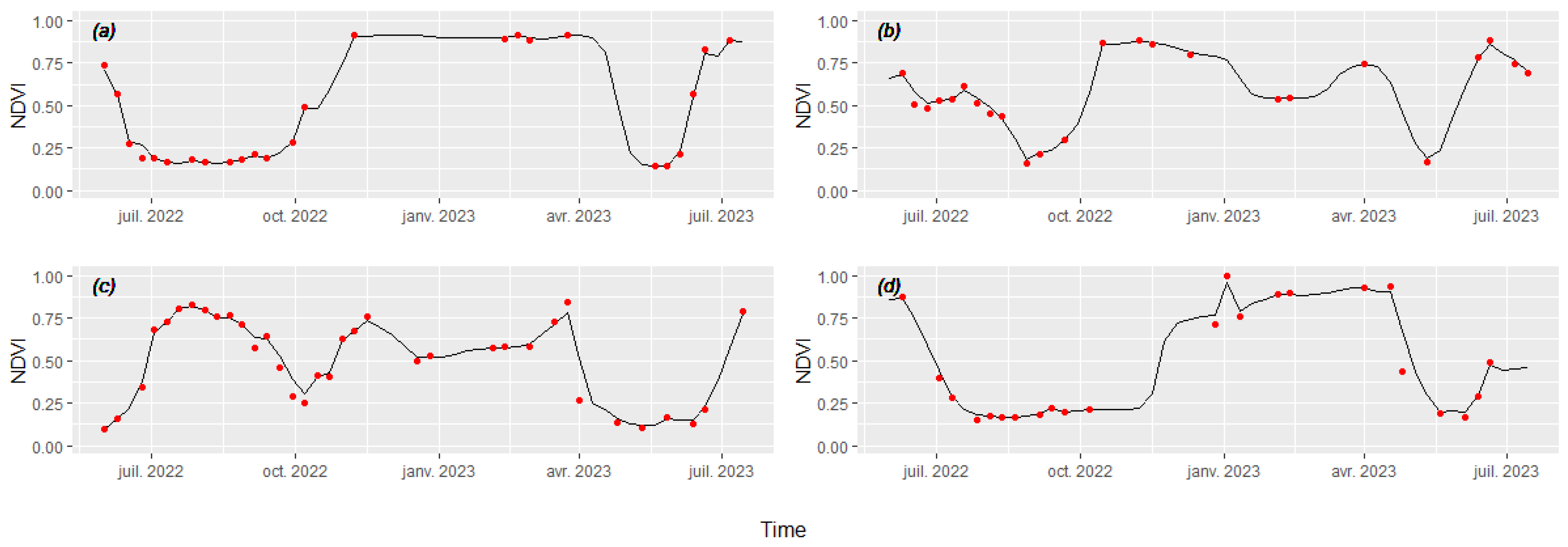
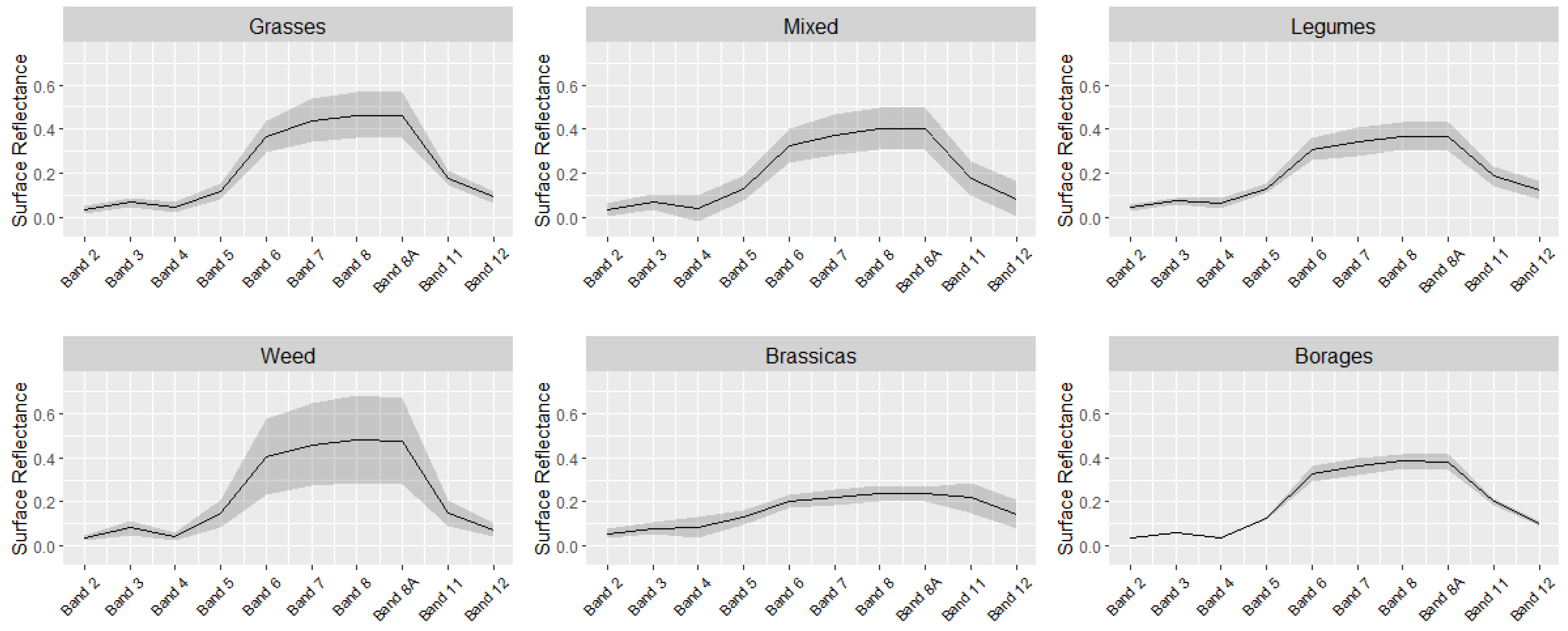
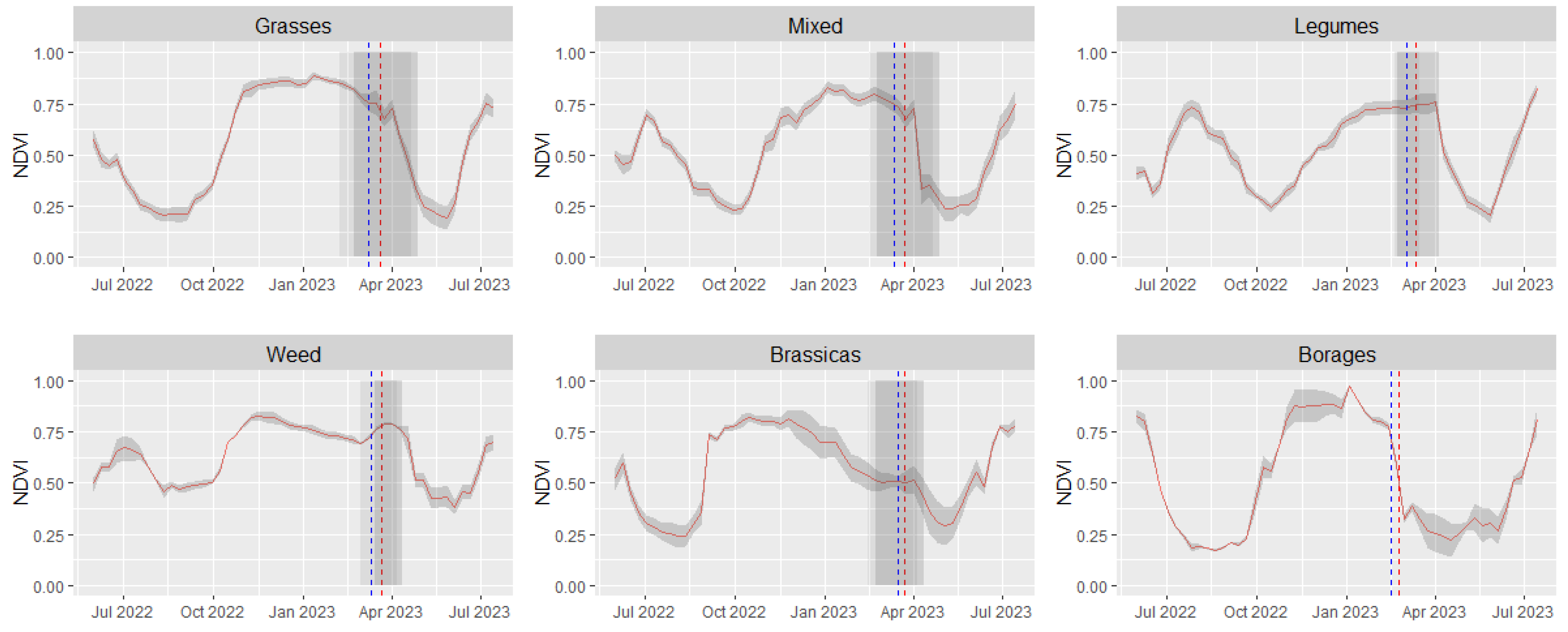
3.2. Cover Crop Satellite-Based Phenology and Spectral Behavior
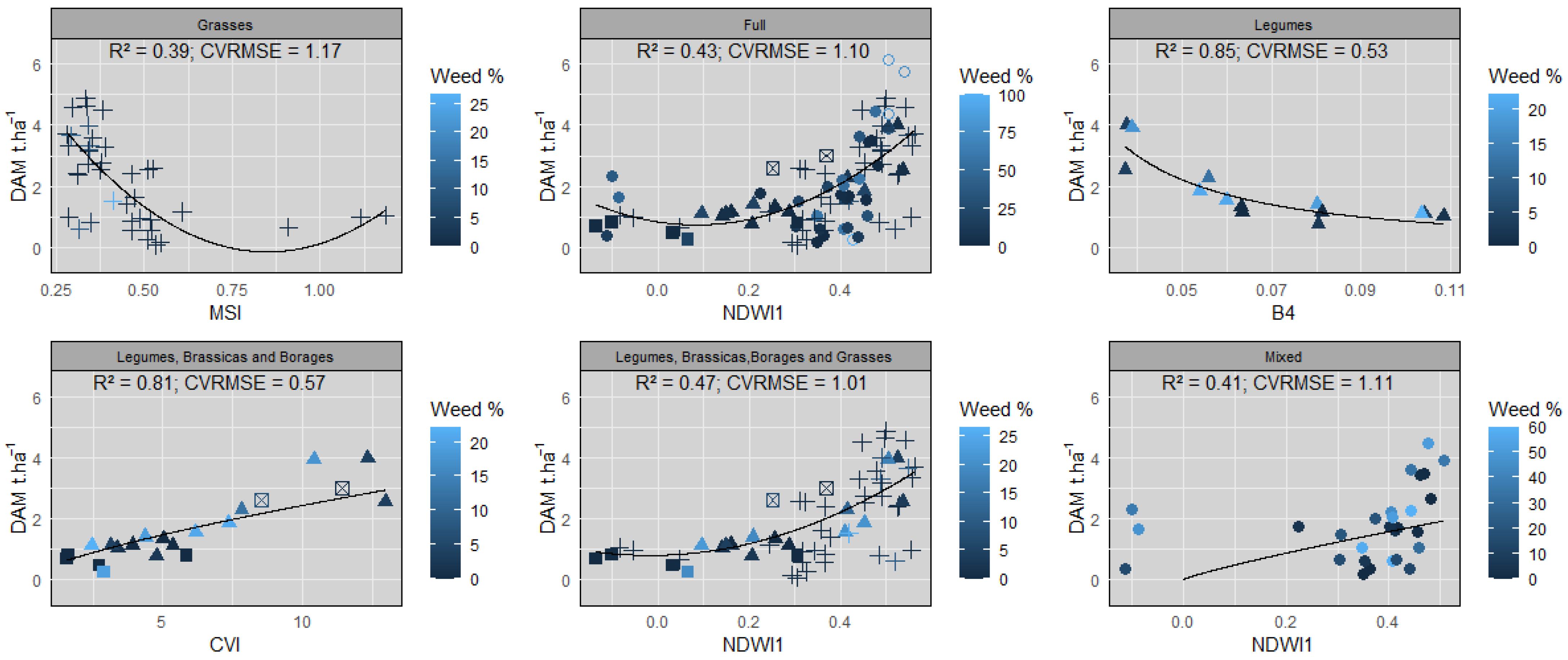
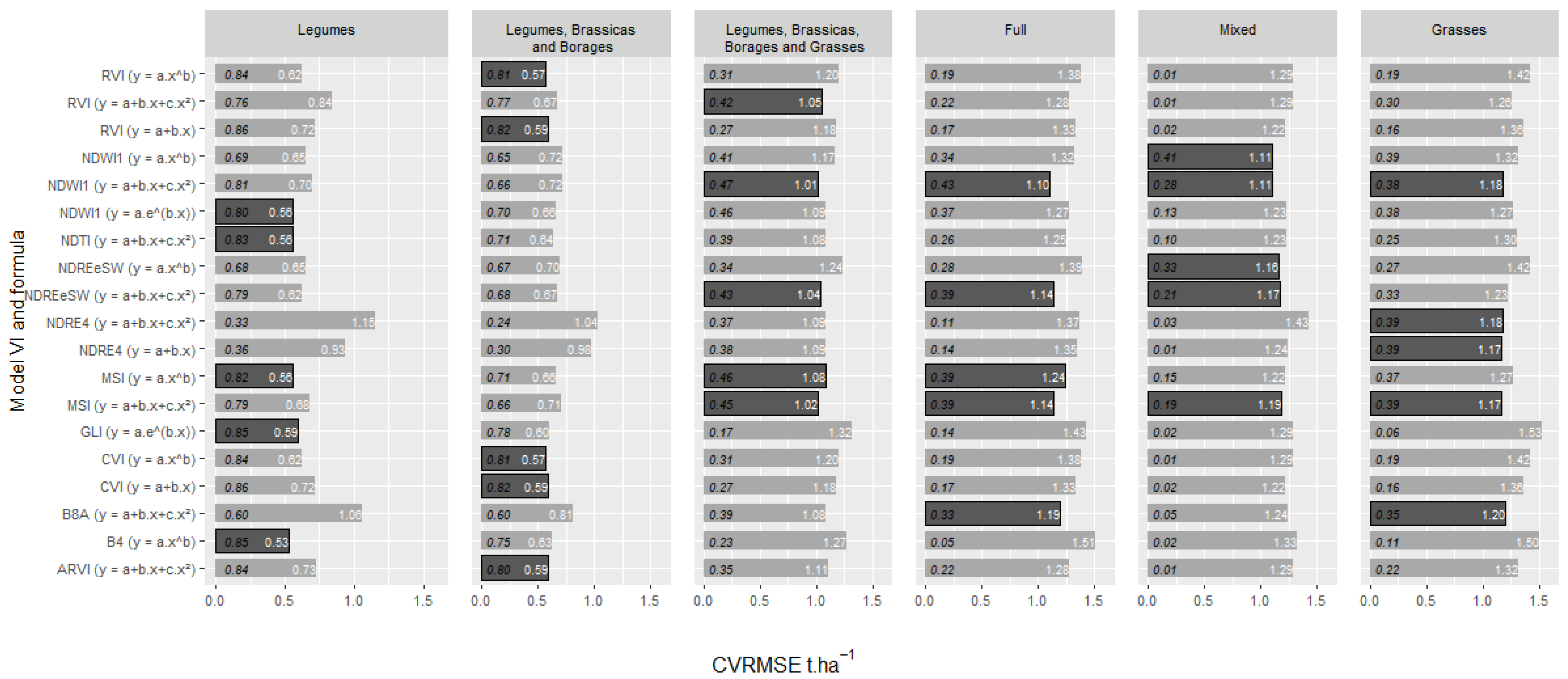
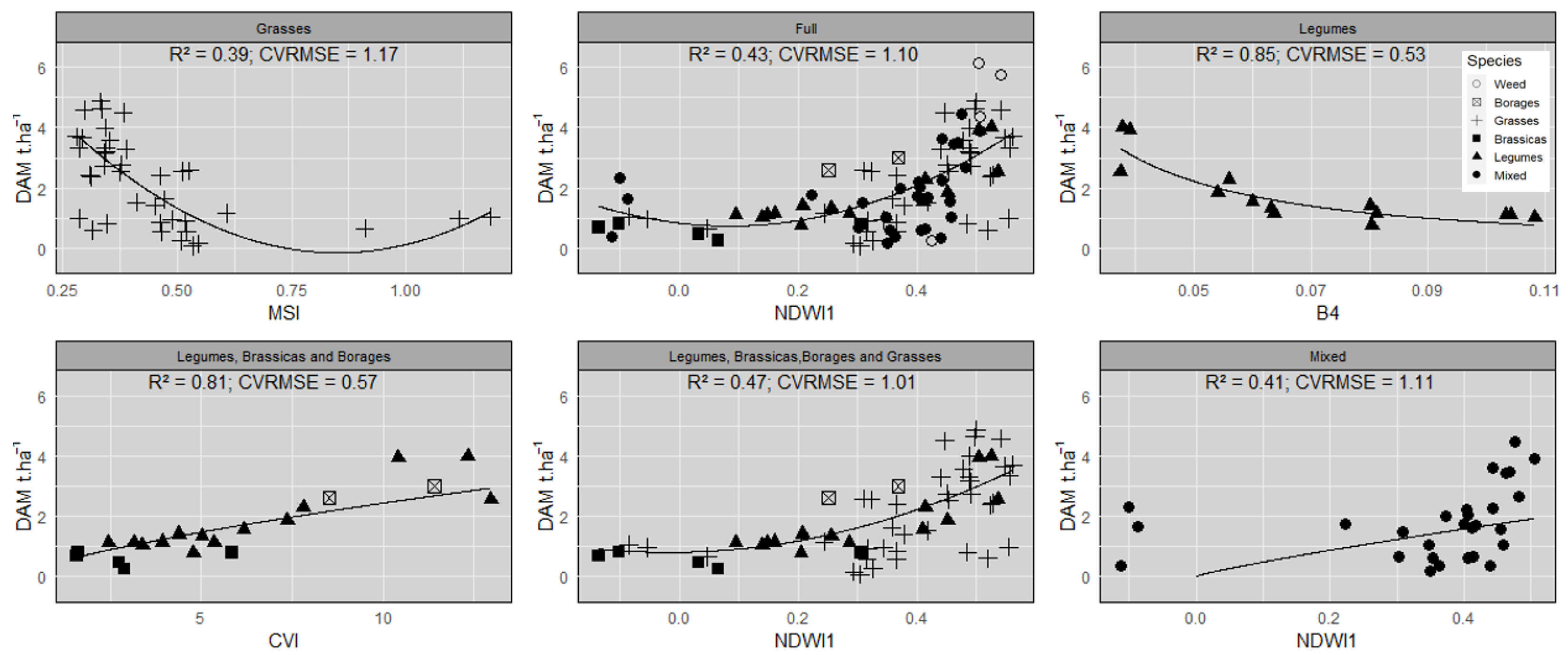
3.3. Assessment of the Empirical Relationships between VI/SB Values and DAM
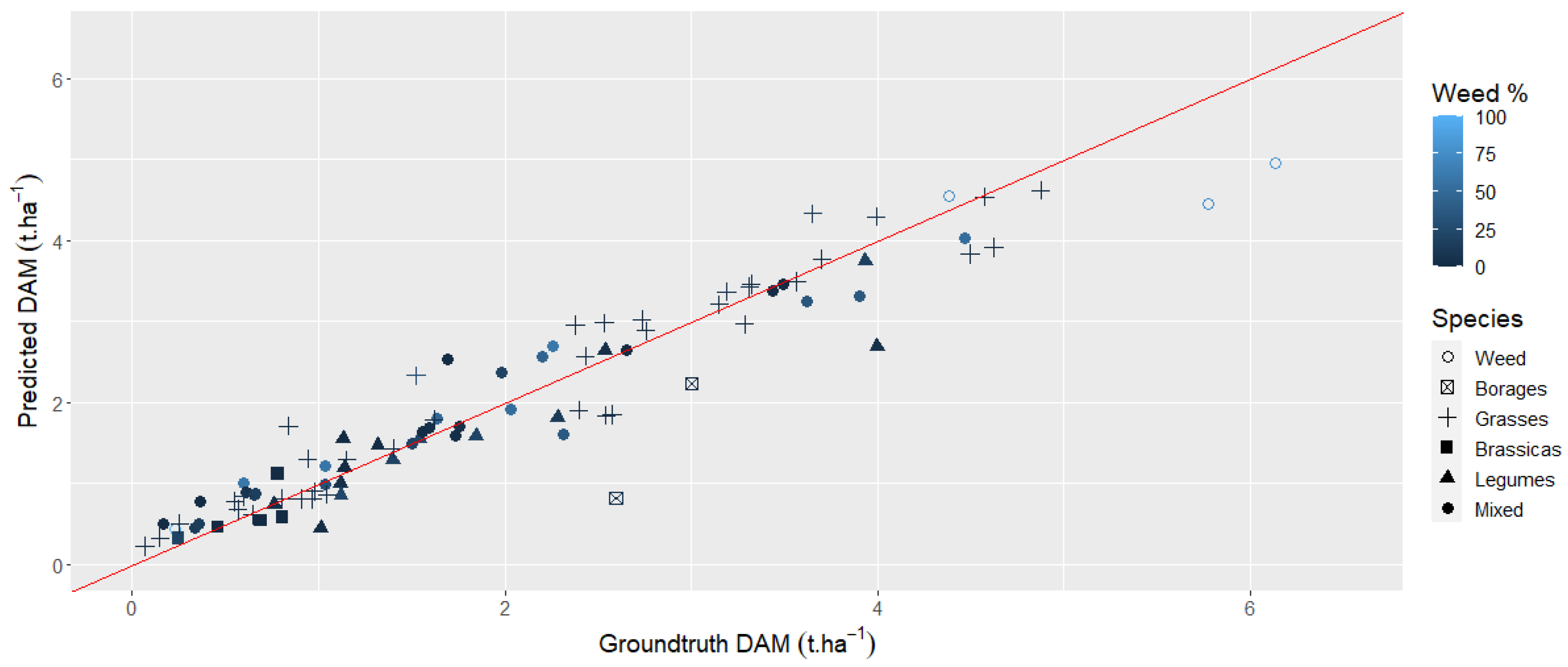
3.4. Multivariate Analyses Considering the Last Available Images with all the Spectral Bands and VIs Using Machine Learning Algorithms
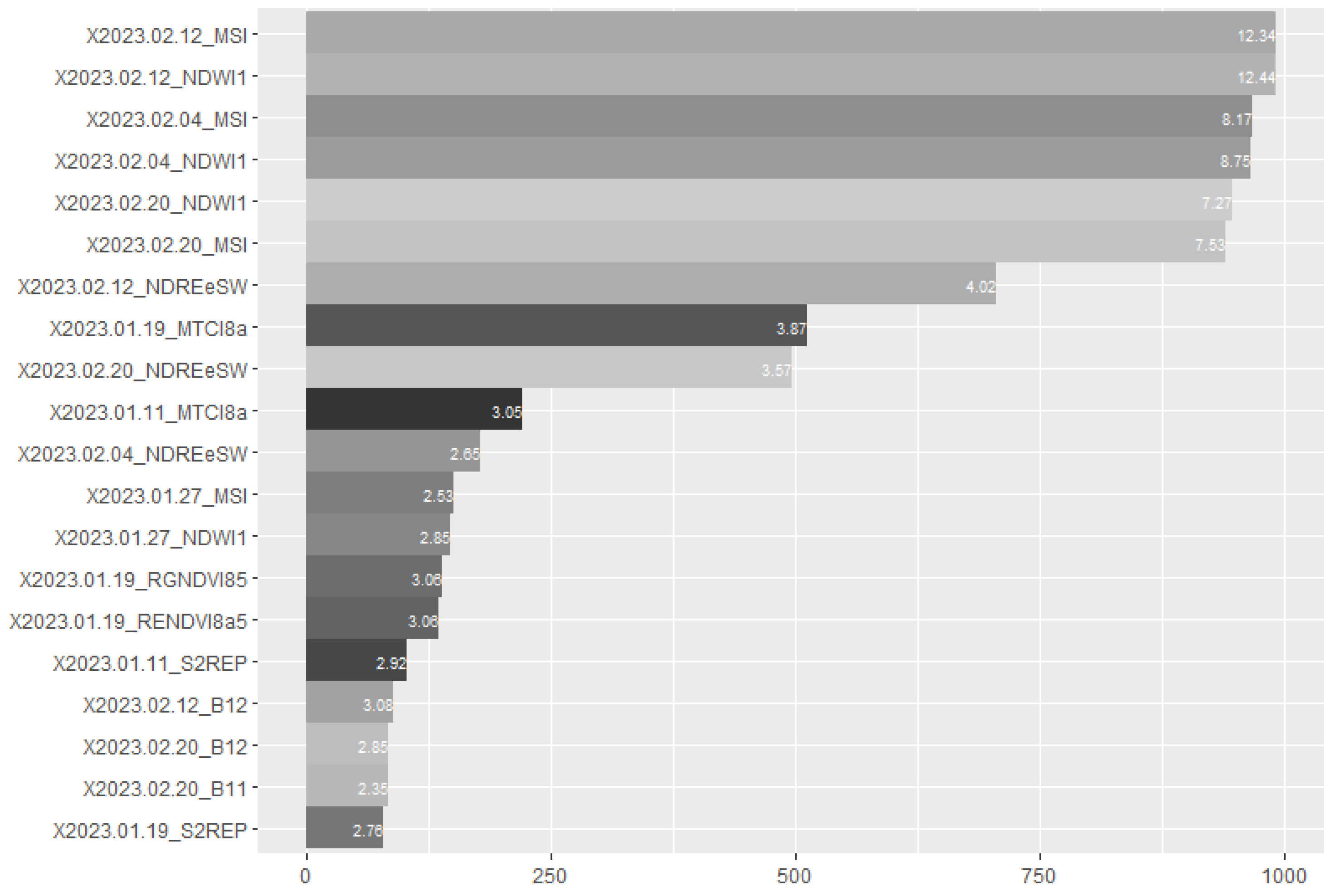
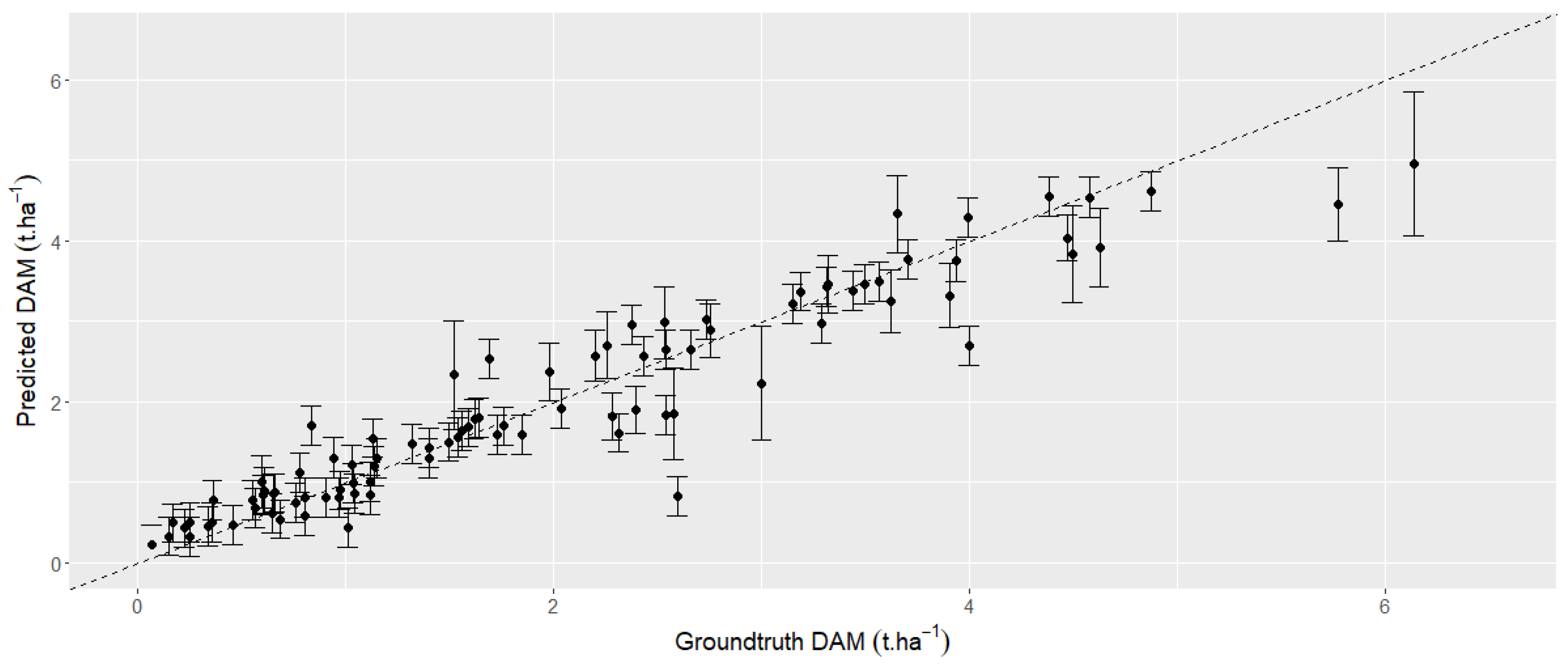
3.5. Multivariate Analyses Using the Dense Time Series with All the Spectral Bands and VIs Using Machine Learning Algorithms
4. Discussion
4.1. Key Contributions of the Study
4.2. Potential Applications of the Proposed Method
4.3. Limitations and Prospects for Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Vegetation Index | Formula | Reference |
|---|---|---|
| Normalized Difference Vegetation Index (NDVI) | (B8 − B4)/(B8 + B4) | Rouse et al. [93] |
| Enhanced Vegetation Index (EVI) | 2.5 × (B8 − B4)/ ((B8 + 6 × B4 − 7.5 × B2) + 1) | Liu and Huete [94] |
| Chlorophyll Vegetation Index (CVI) | (B8/B3) (B4/B3) | Vincini et al. [95] |
| Enhanced Vegetation Index 2 (EVI2) | 2.5 × (B8 − B4)/ ((B8 + 2.4 × B4) + 1) | Sentinel Hub |
| Moisture stress index (MSI) | (B6 − B12)/ (B6 + B12) | Rock, B.N. et al. [96] |
| Normalized Difference of Red-edge and SWIR2 (NDReSw) | B11/B8A | Radoux et al. [97] |
| Normalized Difference Water Index 1 (NDWI1) | (B8A − B11)/ (B8A + B11) | Gao et al. [98] |
| Sentinel 2 red-edge position (S2REP) | 705 + 35 × ((((B7 + B4)/2) − B5)/ (B6 − B5)) | Frampton [99] |
| Soil-Adjusted Vegetation Index (SAVI) | (1 + L) × ((B8−B4)/(B8 + B4 + L) [L = 1] | Huete [100] |
| MERIS Terrestrial Chlorophyll Index (MTCI8a) | (B8A − B5)/ (B5 − B4) | Dash and Curran [101] |
| Red-edge normalized difference vegetation index (RENDVI8a5) | (B8A − B5)/(B8A + B5) | Gitelson et al. [102] |
| Normalized Difference Index (RENDVI85) | (B8 − B5)/(B8 + B5) | Delegido et al. [56] |
| Ratio Vegetation Index (RVI) | B8/B4 | Birth et al. [103] |
| NDTI (Normalized Difference Tillage Index) | (B11 − B12)/(B11 + B12) | Van Deventer et al. [104] |
| NDWI (Normalized Difference Water Index) | (B3−B8)/(B3 + B8) | McFeeters [105] |
| Soil-Adjusted and Atmospherically Resistant Vegetation Index (SARVI) | (1 + L) × (B8 − (B4 − (B4 − B2)))/(B8 + (B4 − (B4 − B2)) + L) [L = 1] | Kaufman et al. [106] |
| Green Leaf Area Index (GLI) | (2 × B3 − B4 − B2)/(2 × B3 + B4 + B2) | Louhaichi et al. [97] |
| Normalized Difference Red-Edge Index (NDRE1) | (B8 − B5)/(B8 + B5) | Gitelson et al. [102] |
| Normalized Difference Red-Edge Index (NDRE2) | (B8 − B6)/(B8 + B6) | Gitelson et al. [102] |
| Normalized Difference Red-Edge Index (NDRE3) | (B8 − B7)/(B8 + B7) | Gitelson et al. [102] |
| Normalized Difference Red-Edge Index (NDRE4) | (B6 − B5)/(B6 + B5) | Gitelson et al. [102] |
| Sentinel-2 LAI Green Index (SeLI) | (B8A − B5)/(B8A + B5) | Pasqualotto et al. [107] |
| Atmospherically Resistant Vegetation Index (ARVI) | (B8 − (B4 − × (B4 − B2)))/ (B8 + (B4 − × (B4 − B2))) [ = 1] | Kaufman et al. [106] |

References
- Ceschia, E.; Béziat, P.; Dejoux, J.F.; Aubinet, M.; Bernhofer, C.; Bodson, B.; Buchmann, N.; Carrara, A.; Cellier, P.; Di Tommasi, P.; et al. Management effects on net ecosystem carbon and GHG budgets at European crop sites. Agric. Ecosyst. Environ. 2010, 139, 363–383. [Google Scholar] [CrossRef]
- Rosegrant, M.W.; Koo, J.; Cenacchi, N.; Ringler, C.; Robertson, R.D.; Fisher, M.; Cox, C.M.; Garrett, K.; Perez, N.D.; Sabbagh, P. Food Security in a World of Natural Resource Scarcity: The Role of Agricultural Technologies; Intl Food Policy Res Inst: Washington DC, USA, 2014. [Google Scholar]
- Chabbi, A.; Lehmann, J.; Ciais, P.; Loescher, H.W.; Cotrufo, M.F.; Don, A.; SanClements, M.; Schipper, L.; Six, J.; Smith, P.; et al. Aligning agriculture and climate policy. Nat. Clim. Chang. 2017, 7, 307–309. [Google Scholar] [CrossRef]
- Varvel, G.E. Soil organic carbon changes in diversified rotations of the western Corn Belt. Soil Sci. Soc. Am. J. 2006, 70, 426–433. [Google Scholar] [CrossRef]
- Poeplau, C.; Don, A. Carbon sequestration in agricultural soils via cultivation of cover crops–A meta-analysis. Agric. Ecosyst. Environ. 2015, 200, 33–41. [Google Scholar] [CrossRef]
- Lugato, E.; Cescatti, A.; Jones, A.; Ceccherini, G.; Duveiller, G. Maximising climate mitigation potential by carbon and radiative agricultural land management with cover crops. Environ. Res. Lett. 2020, 15, 094075. [Google Scholar] [CrossRef]
- Vereecken, H.; Schnepf, A.; Hopmans, J.W.; Javaux, M.; Or, D.; Roose, T.; Vanderborght, J.; Young, M.; Amelung, W.; Aitkenhead, M.; et al. Modeling soil processes: Review, key challenges, and new perspectives. Vadose Zone J. 2016, 15, vzj2015.09.0131. [Google Scholar] [CrossRef]
- Steinbeiss, S.; Gleixner, G.; Antonietti, M. Effect of biochar amendment on soil carbon balance and soil microbial activity. Soil Biol. Biochem. 2009, 41, 1301–1310. [Google Scholar] [CrossRef]
- Su, Y.Z.; Wang, F.; Suo, D.R.; Zhang, Z.H.; Du, M.W. Long-term effect of fertilizer and manure application on soil-carbon sequestration and soil fertility under the wheat–wheat–maize cropping system in northwest China. Nutr. Cycl. Agroecosyst. 2006, 75, 285–295. [Google Scholar] [CrossRef]
- Karhu, K.; Mattila, T.; Bergström, I.; Regina, K. Biochar addition to agricultural soil increased CH4 uptake and water holding capacity–Results from a short-term pilot field study. Agric. Ecosyst. Environ. 2011, 140, 309–313. [Google Scholar] [CrossRef]
- Wall, D.H.; Nielsen, U.N.; Six, J. Soil biodiversity and human health. Nature 2015, 528, 69–76. [Google Scholar] [CrossRef]
- Rejesus, R.M.; Aglasan, S.; Knight, L.G.; Cavigelli, M.A.; Dell, C.J.; Lane, E.D.; Hollinger, D.Y. Economic dimensions of soil health practices that sequester carbon: Promising research directions. J. Soil Water Conserv. 2021, 76, 55A–60A. [Google Scholar] [CrossRef]
- Kaye, J.P.; Quemada, M. Using cover crops to mitigate and adapt to climate change. A review. Agron. Sustain. Dev. 2017, 37, 1–17. [Google Scholar] [CrossRef]
- Carrer, D.; Pique, G.; Ferlicoq, M.; Ceamanos, X.; Ceschia, E. What is the potential of cropland albedo management in the fight against global warming? A case study based on the use of cover crops. Environ. Res. Lett. 2018, 13, 044030. [Google Scholar] [CrossRef]
- Pique, G.; Carrer, D.; Lugato, E.; Fieuzal, R.; Garisoain, R.; Ceschia, E. About the Assessment of Cover Crop Albedo Potential Cooling Effect: Risk of the Darkening Feedback Loop Effects. Remote. Sens. 2023, 15, 3231. [Google Scholar] [CrossRef]
- Ceschia, E.; Mary, B.; Ferlicoq, M.; Pique, G.; Carrer, D.; Dejoux, J.; Dedieu, G. Potentiel d’atténuation des changements climatiques par les couverts intermédiaires. Innov. Agron 2017, 62, 43–58. [Google Scholar]
- Soil Science Society of America. Glossary of soil science terms 2008. In ASA-CSSA-SSSA; Soil Science Society of America: Madison, WI, USA, 2008. [Google Scholar]
- Abdalla, M.; Hastings, A.; Cheng, K.; Yue, Q.; Chadwick, D.; Espenberg, M.; Truu, J.; Rees, R.M.; Smith, P. A critical review of the impacts of cover crops on nitrogen leaching, net greenhouse gas balance and crop productivity. Glob. Chang. Biol. 2019, 25, 2530–2543. [Google Scholar] [CrossRef]
- Thapa, R.; Mirsky, S.B.; Tully, K.L. Cover crops reduce nitrate leaching in agroecosystems: A global meta-analysis. J. Environ. Qual. 2018, 47, 1400–1411. [Google Scholar] [CrossRef]
- De Baets, S.; Poesen, J.; Meersmans, J.; Serlet, L. Cover crops and their erosion-reducing effects during concentrated flow erosion. Catena 2011, 85, 237–244. [Google Scholar] [CrossRef]
- Gao, F.; Anderson, M.C.; Hively, W.D. Detecting cover crop end-of-season using VENμS and sentinel-2 Satellite imagery. Remote. Sens. 2020, 12, 3524. [Google Scholar] [CrossRef]
- Borrelli, P.; Panagos, P. An indicator to reflect the mitigating effect of Common Agricultural Policy on soil erosion. Land Use Policy 2020, 92, 104467. [Google Scholar] [CrossRef]
- Kathage, J.; Smit, B.; Janssens, B.; Haagsma, W.; Adrados, J.L. How much is policy driving the adoption of cover crops? Evidence from four EU regions. Land Use Policy 2022, 116, 106016. [Google Scholar] [CrossRef] [PubMed]
- de L’agriculture et de L’alimentation, M. GRAPH’AGRI. Available online: https://agreste.agriculture.gouv.fr/agreste-web/ (accessed on 1 October 2023).
- Ols, C.; Hervé, J.C.; Bontemps, J.D. Recent growth trends of conifers across Western Europe are controlled by thermal and water constraints and favored by forest heterogeneity. Sci. Total. Environ. 2020, 742, 140453. [Google Scholar] [CrossRef]
- Fendrich, A.N.; Matthews, F.; Van Eynde, E.; Carozzi, M.; Li, Z.; d’Andrimont, R.; Lugato, E.; Martin, P.; Ciais, P.; Panagos, P. From regional to parcel scale: A high-resolution map of cover crops across Europe combining satellite data with statistical surveys. Sci. Total. Environ. 2023, 873, 162300. [Google Scholar] [CrossRef] [PubMed]
- Bockstaller, C.; Sirami, C.; Sheeren, D.; Keichinger, O.; Arnaud, L.; Favreau, A.; Angevin, F.; Laurent, D.; Marchand, G.; De Laroche, E.; et al. Apports de la télédétection au calcul d’indicateurs agri-environnementaux au service de la PAC, des agriculteurs et porteurs d’enjeu. Innov. Agron. 2021, 83, 43–59. [Google Scholar]
- TUCKER, C.J. A critical review of remote sensing and other methods for non-destructive estimation of standing crop biomass. Grass Forage Sci. 1980, 35, 177–182. [Google Scholar] [CrossRef]
- Zheng, D.; Rademacher, J.; Chen, J.; Crow, T.; Bresee, M.; Le Moine, J.; Ryu, S.R. Estimating aboveground biomass using Landsat 7 ETM+ data across a managed landscape in northern Wisconsin, USA. Remote. Sens. Environ. 2004, 93, 402–411. [Google Scholar] [CrossRef]
- Hively, W.; Lang, M.; McCarty, G.; Keppler, J.; Sadeghi, A.; McConnell, L. Using satellite remote sensing to estimate winter cover crop nutrient uptake efficiency. J. Soil Water Conserv. 2009, 64, 303–313. [Google Scholar] [CrossRef]
- Günlü, A.; Ercanli, I.; Başkent, E.; Çakır, G. Estimating aboveground biomass using Landsat TM imagery: A case study of Anatolian Crimean pine forests in Turkey. Ann. For. Res. 2014, 289–298. [Google Scholar]
- Kross, A.; McNairn, H.; Lapen, D.; Sunohara, M.; Champagne, C. Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 235–248. [Google Scholar] [CrossRef]
- Punalekar, S.M.; Verhoef, A.; Quaife, T.; Humphries, D.; Bermingham, L.; Reynolds, C. Application of Sentinel-2A data for pasture biomass monitoring using a physically based radiative transfer model. Remote. Sens. Environ. 2018, 218, 207–220. [Google Scholar] [CrossRef]
- Cate, R.; Artley, J.; Phinney, D. Quantitative Estimation of Plant Characteristics Using Spectral Measurement: A Survey of the Literature; Lockheed Engineering And Management Services Company, Inc.: Bethesda, MD, USA, 1980. [Google Scholar]
- Stenberg, P.; Rautiainen, M.; Manninen, T.; Voipio, P.; Smolander, H. Reduced simple ratio better than NDVI for estimating LAI in Finnish pine and spruce stands. Silva Fenn. 2004, 38, 3–14. [Google Scholar] [CrossRef]
- Chen, P.F.; Nicolas, T.; Wang, J.H.; Philippe, V.; Huang, W.J.; Li, B.G. New index for crop canopy fresh biomass estimation. Spectrosc. Spectr. Anal. 2010, 30, 512–517. [Google Scholar]
- Hosseini, M.; McNairn, H.; Mitchell, S.; Robertson, L.D.; Davidson, A.; Homayouni, S. Synthetic aperture radar and optical satellite data for estimating the biomass of corn. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101933. [Google Scholar] [CrossRef]
- Prabhakara, K.; Hively, W.D.; McCarty, G.W. Evaluating the relationship between biomass, percent groundcover and remote sensing indices across six winter cover crop fields in Maryland, United States. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 88–102. [Google Scholar] [CrossRef]
- Thieme, A.; Hively, W.D.; Gao, F.; Jennewein, J.; Mirsky, S.; Soroka, A.; Keppler, J.; Bradley, D.; Skakun, S.; McCarty, G.W. Remote sensing evaluation of winter cover crop springtime performance and the impact of delayed termination. Agron. J. 2023, 115, 442–458. [Google Scholar] [CrossRef]
- Fan, X.; Vrieling, A.; Muller, B.; Nelson, A. Winter cover crops in Dutch maize fields: Variability in quality and its drivers assessed from multi-temporal Sentinel-2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2020, 91, 102139. [Google Scholar] [CrossRef]
- Xia, Y.; Guan, K.; Copenhaver, K.; Wander, M. Estimating cover crop biomass nitrogen credits with Sentinel-2 imagery and sites covariates. Agron. J. 2021, 113, 1084–1101. [Google Scholar] [CrossRef]
- Jennewein, J.S.; Lamb, B.T.; Hively, W.D.; Thieme, A.; Thapa, R.; Goldsmith, A.; Mirsky, S.B. Integration of Satellite-Based Optical and Synthetic Aperture Radar Imagery to Estimate Winter Cover Crop Performance in Cereal Grasses. Remote. Sens. 2022, 14, 2077. [Google Scholar] [CrossRef]
- Goffart, D.; Curnel, Y.; Planchon, V.; Goffart, J.P.; Defourny, P. Field-scale assessment of Belgian winter cover crops biomass based on Sentinel-2 data. Eur. J. Agron. 2021, 126, 126278. [Google Scholar] [CrossRef]
- Holzhauser, K.; Räbiger, T.; Rose, T.; Kage, H.; Kühling, I. Estimation of Biomass and N Uptake in Different Winter Cover Crops from UAV-Based Multispectral Canopy Reflectance Data. Remote. Sens. 2022, 14, 4525. [Google Scholar] [CrossRef]
- Kharel, T.P.; Bhandari, A.B.; Mubvumba, P.; Tyler, H.L.; Fletcher, R.S.; Reddy, K.N. Mixed-Species Cover Crop Biomass Estimation Using Planet Imagery. Sensors 2023, 23, 1541. [Google Scholar] [CrossRef]
- Kümmerer, R.; Noack, P.O.; Bauer, B. Using High-Resolution UAV Imaging to Measure Canopy Height of Diverse Cover Crops and Predict Biomass. Remote. Sens. 2023, 15, 1520. [Google Scholar] [CrossRef]
- Roth, R.T.; Chen, K.; Scott, J.R.; Jung, J.; Yang, Y.; Camberato, J.J.; Armstrong, S.D. Prediction of Cereal Rye Cover Crop Biomass and Nutrient Accumulation Using Multi-Temporal Unmanned Aerial Vehicle Based Visible-Spectrum Vegetation Indices. Remote. Sens. 2023, 15, 580. [Google Scholar] [CrossRef]
- Gao, F.; Jennewein, J.; Hively, W.D.; Soroka, A.; Thieme, A.; Bradley, D.; Keppler, J.; Mirsky, S.; Akumaga, U. Near real-time detection of winter cover crop termination using harmonized Landsat and Sentinel-2 (HLS) to support ecosystem assessment. Sci. Remote. Sens. 2023, 7, 100073. [Google Scholar] [CrossRef]
- Thieme, A.; Yadav, S.; Oddo, P.C.; Fitz, J.M.; McCartney, S.; King, L.; Keppler, J.; McCarty, G.W.; Hively, W.D. Using NASA Earth observations and Google Earth Engine to map winter cover crop conservation performance in the Chesapeake Bay watershed. Remote. Sens. Environ. 2020, 248, 111943. [Google Scholar] [CrossRef]
- Najem, S.; Baghdadi, N.; Bazzi, H.; Lalande, N.; Bouchet, L. Detection and Mapping of Cover Crops using Sentinel-1 SAR Remote Sensing data. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2023, 17, 1446–1461. [Google Scholar] [CrossRef]
- Wang, S.; Guan, K.; Zhang, C.; Jiang, C.; Zhou, Q.; Li, K.; Qin, Z.; Ainsworth, E.A.; He, J.; Wu, J.; et al. Airborne hyperspectral imaging of cover crops through radiative transfer process-guided machine learning. Remote. Sens. Environ. 2023, 285, 113386. [Google Scholar] [CrossRef]
- Basche, A.D.; Archontoulis, S.V.; Kaspar, T.C.; Jaynes, D.B.; Parkin, T.B.; Miguez, F.E. Simulating long-term impacts of cover crops and climate change on crop production and environmental outcomes in the Midwestern United States. Agric. Ecosyst. Environ. 2016, 218, 95–106. [Google Scholar] [CrossRef]
- Joly, D.; Brossard, T.; Cardot, H.; Cavailhes, J.; Hilal, M.; Wavresky, P. Les types de climats en France, une construction spatiale. Cybergeo Eur. J. Geogr. 2010. [Google Scholar] [CrossRef]
- Main-Knorn, M.; Pflug, B.; Louis, J.; Debaecker, V.; Müller-Wilm, U.; Gascon, F. Sen2Cor for sentinel-2. In Proceedings of the Image and Signal Processing for Remote Sensing XXIII. SPIE, Warsaw, Poland, 11–13 September 2017; Volume 10427, pp. 37–48. [Google Scholar]
- Jaramaz, D.; Perovic, V.; Belanovic, S.; Saljnikov, E.; Cakmak, D.; Mrvic, V.; Zivotic, L. The ESA Sentinel-2 mission vegetation variables for remote sensing of plant monitoring. In Proceedings of the Conference Proceedings 2nd International Scientific Conference, Belgrade, Serbia, 14–17 May 2013; pp. 950–961. [Google Scholar]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of sentinel-2 red-edge bands for empirical estimation of green LAI and chlorophyll content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef] [PubMed]
- Frantz, D.; Haß, E.; Uhl, A.; Stoffels, J.; Hill, J. Improvement of the Fmask algorithm for Sentinel-2 images: Separating clouds from bright surfaces based on parallax effects. Remote. Sens. Environ. 2018, 215, 471–481. [Google Scholar] [CrossRef]
- Zupanc, A. Improving Cloud Detection with Machine Learning. 2020. Available online: https://medium.com/sentinel-hub/improving-cloud-detection-with-machine-learning-c09dc5d7cf13 (accessed on 5 February 2023).
- Rufin, P.; Rabe, A.; Nill, L.; Hostert, P. GEE timeseries explorer for qgis–instant access to petabytes of earth observation data. Int. Arch. Photogramm. Remote. Sens. Spat. Inf. Sci. 2021, 46, 155–158. [Google Scholar] [CrossRef]
- Schwieder, M.; Leitão, P.J.; da Cunha Bustamante, M.M.; Ferreira, L.G.; Rabe, A.; Hostert, P. Mapping Brazilian savanna vegetation gradients with Landsat time series. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 361–370. [Google Scholar] [CrossRef]
- do Nascimento Bendini, H.; Fonseca, L.M.G.; Schwieder, M.; Körting, T.S.; Rufin, P.; Sanches, I.D.A.; Leitão, P.J.; Hostert, P. Detailed agricultural land classification in the Brazilian cerrado based on phenological information from dense satellite image time series. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101872. [Google Scholar] [CrossRef]
- Swoish, M.; Filho, J.F.D.C.L.; Reiter, M.S.; Campbell, J.B.; Thomason, W.E. Comparing satellites and vegetation indices for cover crop biomass estimation. Comput. Electron. Agric. 2022, 196, 106900. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; Version 2023.03.1; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support vector machine. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Flynn, K.C.; Baath, G.; Lee, T.O.; Gowda, P.; Northup, B. Hyperspectral reflectance and machine learning to monitor legume biomass and nitrogen accumulation. Comput. Electron. Agric. 2023, 211, 107991. [Google Scholar] [CrossRef]
- Li, H.; Song, X.P.; Hansen, M.C.; Becker-Reshef, I.; Adusei, B.; Pickering, J.; Wang, L.; Wang, L.; Lin, Z.; Zalles, V.; et al. Development of a 10-m resolution maize and soybean map over China: Matching satellite-based crop classification with sample-based area estimation. Remote. Sens. Environ. 2023, 294, 113623. [Google Scholar] [CrossRef]
- Blickensdörfer, L.; Schwieder, M.; Pflugmacher, D.; Nendel, C.; Erasmi, S.; Hostert, P. Mapping of crop types and crop sequences with combined time series of Sentinel-1, Sentinel-2 and Landsat 8 data for Germany. Remote. Sens. Environ. 2022, 269, 112831. [Google Scholar] [CrossRef]
- Dimitriadou, E.; Hornik, K.; Leisch, F.; Meyer, D.; Weingessel, A.; Leisch, M.F. The e1071 Package. In Misc Functions of Department of Statistics (e1071), TU Wien; 2006; pp. 297–304. Available online: https://rdocumentation.org/packages/e1071/versions/1.7-14 (accessed on 1 March 2023).
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Efron, B. The Jackknife, the Bootstrap and Other Resampling Plans; Department of Statistics Stanford Iniversity: Stanford, CA, USA, 1982. [Google Scholar]
- Efron, B. Estimation and accuracy after model selection. J. Am. Stat. Assoc. 2014, 109, 991–1007. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wijmer, T.; Al Bitar, A.; Arnaud, L.; Fieuzal, R.; Ceschia, E. AgriCarbon-EO: V1. 0.1: Large Scale and High Resolution Simulation of Carbon Fluxes by Assimilation of Sentinel-2 and Landsat-8 Reflectances using a Bayesian approach. EGUsphere 2023, 2023, 1–41. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote. Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Blackmore, S.; Godwin, R.J.; Fountas, S. The analysis of spatial and temporal trends in yield map data over six years. Biosyst. Eng. 2003, 84, 455–466. [Google Scholar] [CrossRef]
- Grieve, B.D.; Duckett, T.; Collison, M.; Boyd, L.; West, J.; Yin, H.; Arvin, F.; Pearson, S. The challenges posed by global broadacre crops in delivering smart agri-robotic solutions: A fundamental rethink is required. Glob. Food Secur. 2019, 23, 116–124. [Google Scholar] [CrossRef]
- Nowak, B.; Marliac, G.; Michaud, A. Estimation of winter soil cover by vegetation before spring-sown crops for mainland France using multispectral satellite imagery. Environ. Res. Lett. 2021, 16, 064024. [Google Scholar] [CrossRef]
- Breunig, F.M.; Galvão, L.S.; Dalagnol, R.; Dauve, C.E.; Parraga, A.; Santi, A.L.; Flora, D.P.D.; Chen, S. Delineation of management zones in agricultural fields using cover–crop biomass estimates from PlanetScope data. Int. J. Appl. Earth Obs. Geoinf. 2020, 85, 102004. [Google Scholar] [CrossRef]
- Constantin, J.; Minette, S.; Vericel, G.; Jordan-Meille, L.; Justes, E. MERCI: A simple method and decision-support tool to estimate availability of nitrogen from a wide range of cover crops to the next cash crop. Plant Soil 2023, 1–19. [Google Scholar] [CrossRef]
- Paustian, K.; Collier, S.; Baldock, J.; Burgess, R.; Creque, J.; DeLonge, M.; Dungait, J.; Ellert, B.; Frank, S.; Goddard, T.; et al. Quantifying carbon for agricultural soil management: From the current status toward a global soil information system. Carbon Manag. 2019, 10, 567–587. [Google Scholar] [CrossRef]
- Smith, P.; Lanigan, G.; Kutsch, W.L.; Buchmann, N.; Eugster, W.; Aubinet, M.; Ceschia, E.; Béziat, P.; Yeluripati, J.B.; Osborne, B.; et al. Measurements necessary for assessing the net ecosystem carbon budget of croplands. Agric. Ecosyst. Environ. 2010, 139, 302–315. [Google Scholar] [CrossRef]
- Soenen, B. Méthode LBC Grandes Cultures (version 1.1) LABEL BAS-CARBONE Méthode Grandes Cultures. Available online: https://www.ecologie.gouv.fr/sites/default/files/M%C3%A9thode%20LBC%20Grandes%20cultures.pdf (accessed on 1 December 2023).
- Lamichhane, J.R.; Alletto, L. Ecosystem services of cover crops: A research roadmap. Trends Plant Sci. 2022. [Google Scholar] [CrossRef] [PubMed]
- Allies, A.; Roumiguie, A.; Dejoux, J.F.; Fieuzal, R.; Jacquin, A.; Veloso, A.; Champolivier, L.; Baup, F. Evaluation of multiorbital SAR and multisensor optical data for empirical estimation of rapeseed biophysical parameters. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2021, 14, 7268–7283. [Google Scholar] [CrossRef]
- Baup, F.; Ameline, M.; Fieuzal, R.; Frappart, F.; Corgne, S.; Berthoumieu, J.F. Temporal evolution of corn mass production based on agro-meteorological modelling controlled by satellite optical and SAR images. Remote. Sens. 2019, 11, 1978. [Google Scholar] [CrossRef]
- Betbeder, J.; Fieuzal, R.; Baup, F. Assimilation of LAI and dry biomass data from optical and SAR images into an agro-meteorological model to estimate soybean yield. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2016, 9, 2540–2553. [Google Scholar] [CrossRef]
- Fieuzal, R.; Baup, F. Estimation of leaf area index and crop height of sunflowers using multi-temporal optical and SAR satellite data. Int. J. Remote. Sens. 2016, 37, 2780–2809. [Google Scholar] [CrossRef]
- Fieuzal, R.; Baup, F.; Marais-Sicre, C. Monitoring wheat and rapeseed by using synchronous optical and radar satellite data—From temporal signatures to crop parameters estimation. Adv. Remote Sens. 2013, 2, 33222. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Liu, H.Q.; Huete, A. A feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote. Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Rock, B.N.; Vogelmann, J.E.; Williams, D. Field and airborne spectral characterization of suspected damage in red spruce (Picea rubens) from vermont. In Proceedings of the Machine Processing of Remotely Sensed Data Symposium, West Lafayette, IN, USA, 25–27 June 1985. [Google Scholar]
- Radoux, J.; Chomé, G.; Jacques, D.C.; Waldner, F.; Bellemans, N.; Matton, N.; Lamarche, C.; d’Andrimont, R.; Defourny, P. Sentinel-2’s potential for sub-pixel landscape feature detection. Remote. Sens. 2016, 8, 488. [Google Scholar] [CrossRef]
- Gao, B.C. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote. Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote. Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote. Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 540–5413. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-a using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Birth, G.S.; McVey, G.R. Measuring the color of growing turf with a reflectance spectrophotometer 1. Agron. J. 1968, 60, 640–643. [Google Scholar] [CrossRef]
- Van Deventer, A.; Ward, A.; Gowda, P.; Lyon, J. Using thematic mapper data to identify contrasting soil plains and tillage practices. Photogramm. Eng. Remote. Sens. 1997, 63, 87–93. [Google Scholar]
- McFeeters, S.K. The use of the Normalized Difference Water Index (NDWI) in the delineation of open water features. Int. J. Remote. Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote. Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Pasqualotto, N.; Delegido, J.; Van Wittenberghe, S.; Rinaldi, M.; Moreno, J. Multi-crop green LAI estimation with a new simple Sentinel-2 LAI Index (SeLI). Sensors 2019, 19, 904. [Google Scholar] [CrossRef]

| Parameter | Maximum | Minimum | Mean | Median | SD |
|---|---|---|---|---|---|
| Species (number site−1) | 5.00 | 1.00 | 1.90 | 2.00 | 1.00 |
| Grasses (percentage of total biomass−1) | 100.00 | 0.00 | 50.28 | 54.33 | 42.33 |
| Legumes (percentage of total biomass−1) | 100.00 | 0.00 | 26.35 | 13.19 | 32.04 |
| Brassicas (percentage of total biomass−1) | 100.00 | 0.00 | 6.31 | 0.00 | 21.80 |
| Borages (percentage of total biomass−1) | 97.82 | 0.00 | 3.48 | 0.00 | 15.31 |
| Flaxes (percentage of total biomass−1) | 34.04 | 0.00 | 0.45 | 0.00 | 3.63 |
| Weeds (percentage of total biomass−1) | 100.00 | 0.00 | 13.13 | 1.34 | 21.83 |
| Group of Species | Observations | Dry Biomass (t·ha−1) | Water Content (%) | Weed (%) |
|---|---|---|---|---|
| Grasses | 39 | 2.40 ± 1.42 | 80.36 ± 4.88 | 2.13 |
| Legumes | 14 | 1.36 ± 1.04 | 86.24 ± 2.65 | 9.25 |
| Brassicas | 5 | 0.69 ± 0.24 | 82.39 ± 1.28 | 3.94 |
| Borages | 2 | 2.80 ± 0.29 | 85.72 ± 1.68 | 3.65 |
| Mixed | 28 | 1.67 ± 1.18 | 83.53 ± 3.64 | 22.60 |
| Weeds | 4 | 5.08 ± 2.71 | 82.70 ± 5.31 | 83.70 |
| Machine Learning Model | Tuned Parameters | r2 | MAE | CVRMSE |
|---|---|---|---|---|
| Random Forest (RF) | maxnodes: 20; ntree: 100 | 0.55 | 0.73 | 0.98 |
| Support Vector Machine (SVM) | kernel: radial; cost: 1; gamma: 0.1 | 0.53 | 0.71 | 0.97 |
| eXtreme Gradient Boosting (XGBoost) | nfold: 5; nrounds: 10; eta: 0.3; max depth: 10; subsample: 0.7 | 0.46 | 0.81 | 1.08 |
| Multivariate Linear Regression | - | 0.24 | 1.12 | 1.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
do Nascimento Bendini, H.; Fieuzal, R.; Carrere, P.; Clenet, H.; Galvani, A.; Allies, A.; Ceschia, É. Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning. Remote Sens. 2024, 16, 834. https://doi.org/10.3390/rs16050834
do Nascimento Bendini H, Fieuzal R, Carrere P, Clenet H, Galvani A, Allies A, Ceschia É. Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning. Remote Sensing. 2024; 16(5):834. https://doi.org/10.3390/rs16050834
Chicago/Turabian Styledo Nascimento Bendini, Hugo, Rémy Fieuzal, Pierre Carrere, Harold Clenet, Aurelie Galvani, Aubin Allies, and Éric Ceschia. 2024. "Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning" Remote Sensing 16, no. 5: 834. https://doi.org/10.3390/rs16050834
APA Styledo Nascimento Bendini, H., Fieuzal, R., Carrere, P., Clenet, H., Galvani, A., Allies, A., & Ceschia, É. (2024). Estimating Winter Cover Crop Biomass in France Using Optical Sentinel-2 Dense Image Time Series and Machine Learning. Remote Sensing, 16(5), 834. https://doi.org/10.3390/rs16050834






