Preharvest Durum Wheat Yield, Protein Content, and Protein Yield Estimation Using Unmanned Aerial Vehicle Imagery and Pléiades Satellite Data in Field Breeding Experiments
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Site and Experimental Design of the Study
2.2. Data Acquisition
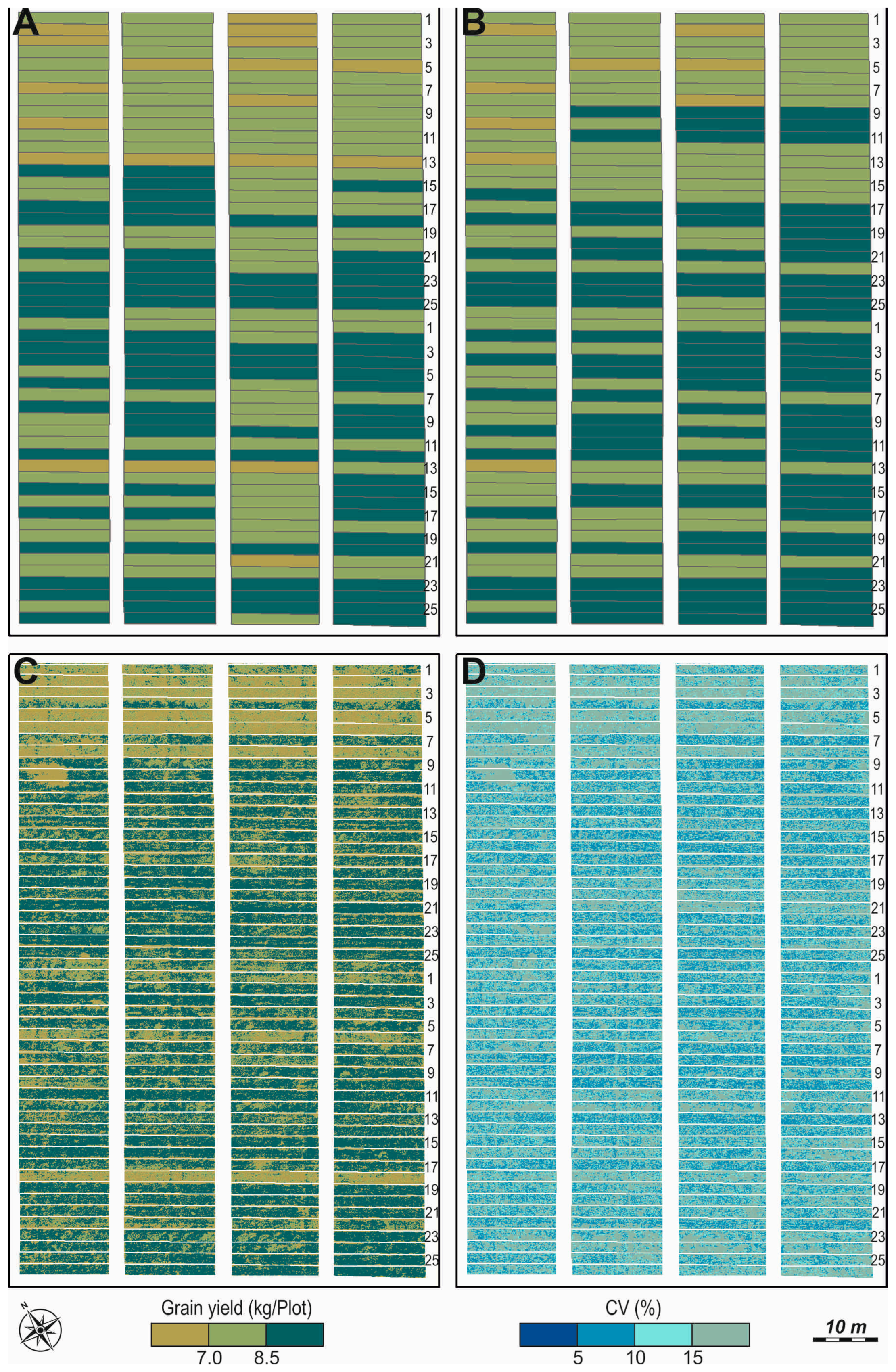
2.2.1. Grain Yield (GY)
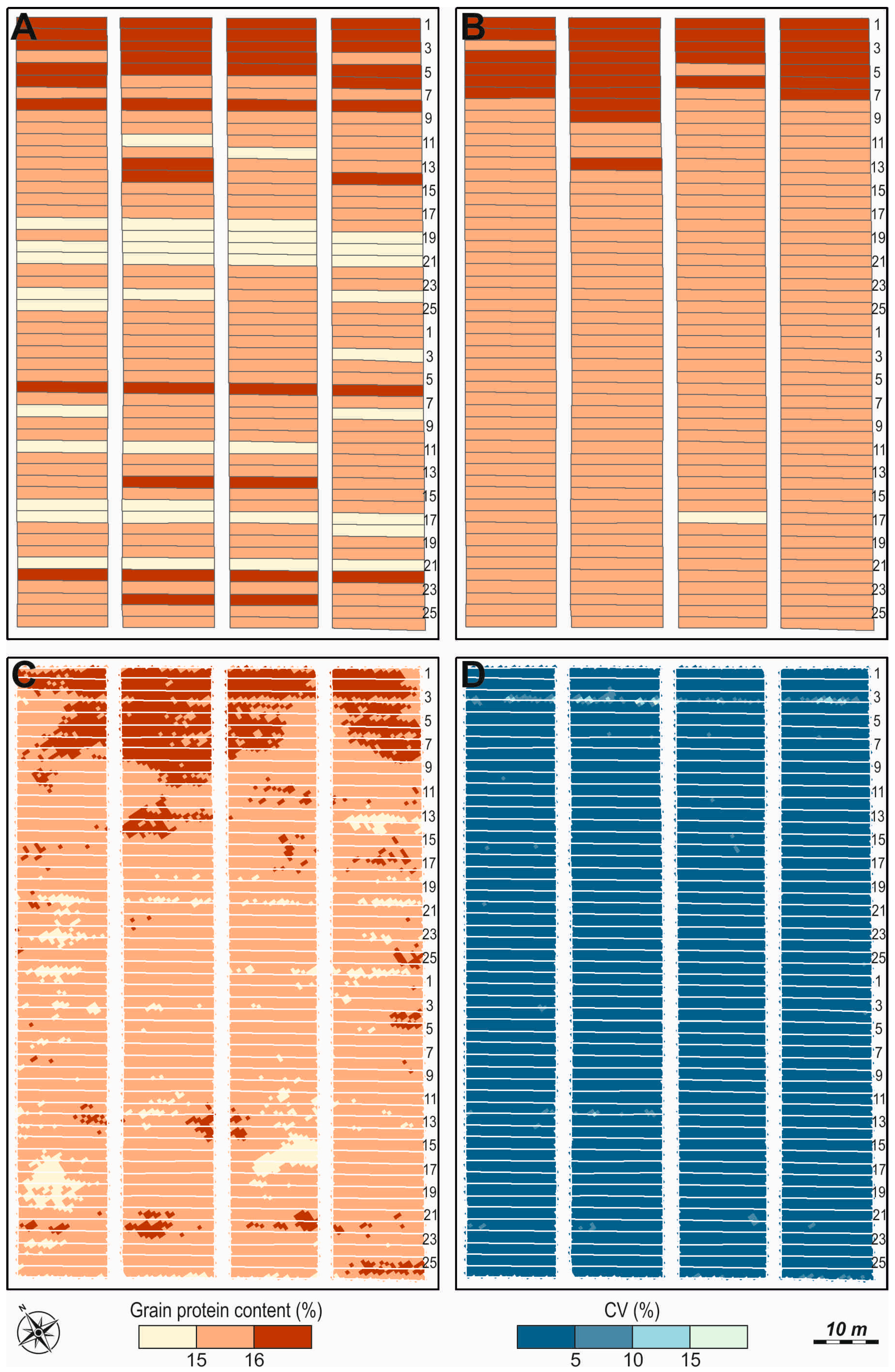
2.2.2. Grain Protein Content (GPC)
2.2.3. Protein Yield (PY)
2.2.4. Multispectral Image Acquisition and Pre-Processing
2.3. Modeling Approach
| Index | Equation 1 | Reference | Selected Studies Utilizing the Index for Wheat Parameters’ Evaluation |
|---|---|---|---|
| Simple ratio (SR) 2 | [60] | GY [61,62] | |
| Reciprocal ratio vegetation index (repRVI) 2 | [60] | GY [34] | |
| Normalized-difference vegetation index (NDVI) 2 | [63] | GY [36,64,65,66]; GPC [66,67]; and biomass and LAI [45] | |
| Green normalized-difference vegetation index (GNDVI) 2 | [68] | GY [25,28,69] and PY [25] | |
| Structure-insensitive pigment index (SIPI) | [70] | GPC [67] | |
| Normalized-difference red-edge index (reNDVI) | [71] | GPC [72]; assessing the grain-filling process: [73]; and GY, PY, and biomass [26] | |
| Normalized-green–red-difference index (NGRDI) | [74] | GY [69] | |
| Modified normalized-difference blue index (mNDblue) | [75] | GY [34] | |
| MERIS terrestrial chlorophyll index (MTCI) | [76] | GPC [77] GY [34] | |
| Normalized green–blue-difference index (NGBDI) | [78] | GY [79] | |
| Triangular greenness index (TGI) | [80] | GY [69] | |
| Triangular vegetation index (TVI) | [81] | GY and GPC [82] | |
| Optimized soil-adjusted vegetation index (OSAVI) 2 | [83] | GPC [84] | |
| Two-band enhanced vegetation index (EVI2) 2 | [85] | GY [86] | |
| Visible atmospherically resistant index (VARI) | [87] | Vegetation fraction [88] | |
| Three-band vegetation index (3BSI-Tian) 2 | [30,89] | GY [30] | |
| Red-edge chlorophyll index (CIred-edge) | [90] | GY [86] LAI [88] | |
| Green chlorophyll index (CIgreen) | [90] | GPC [91] | |
| Plant senescence reflectance index (PSRI) 2 | [92] | Assess the grain-filling process [73]; GPC [67]; | |
| Enhanced vegetation index (EVI) 2 | [93] | GY [53,94,95]; GPC [67,84]; And biomass and LAI [45] | |
| Soil-adjusted vegetation index (SAVI) 2 | [96] | GY [65,94,95] and biomass and LAI [45] | |
| Difference vegetation index (DVI) 2 | [74] | GPC [84] |
3. Results
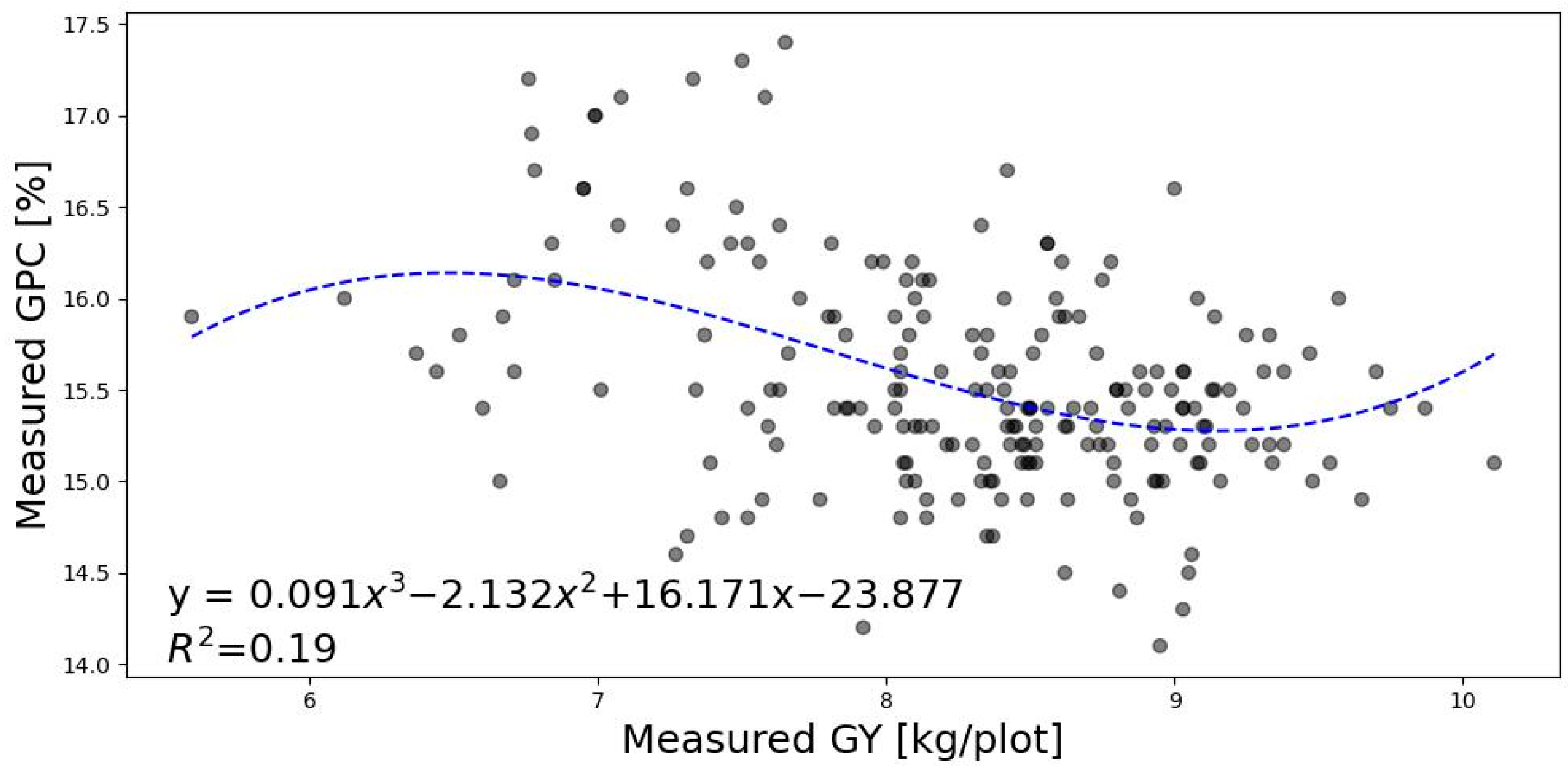
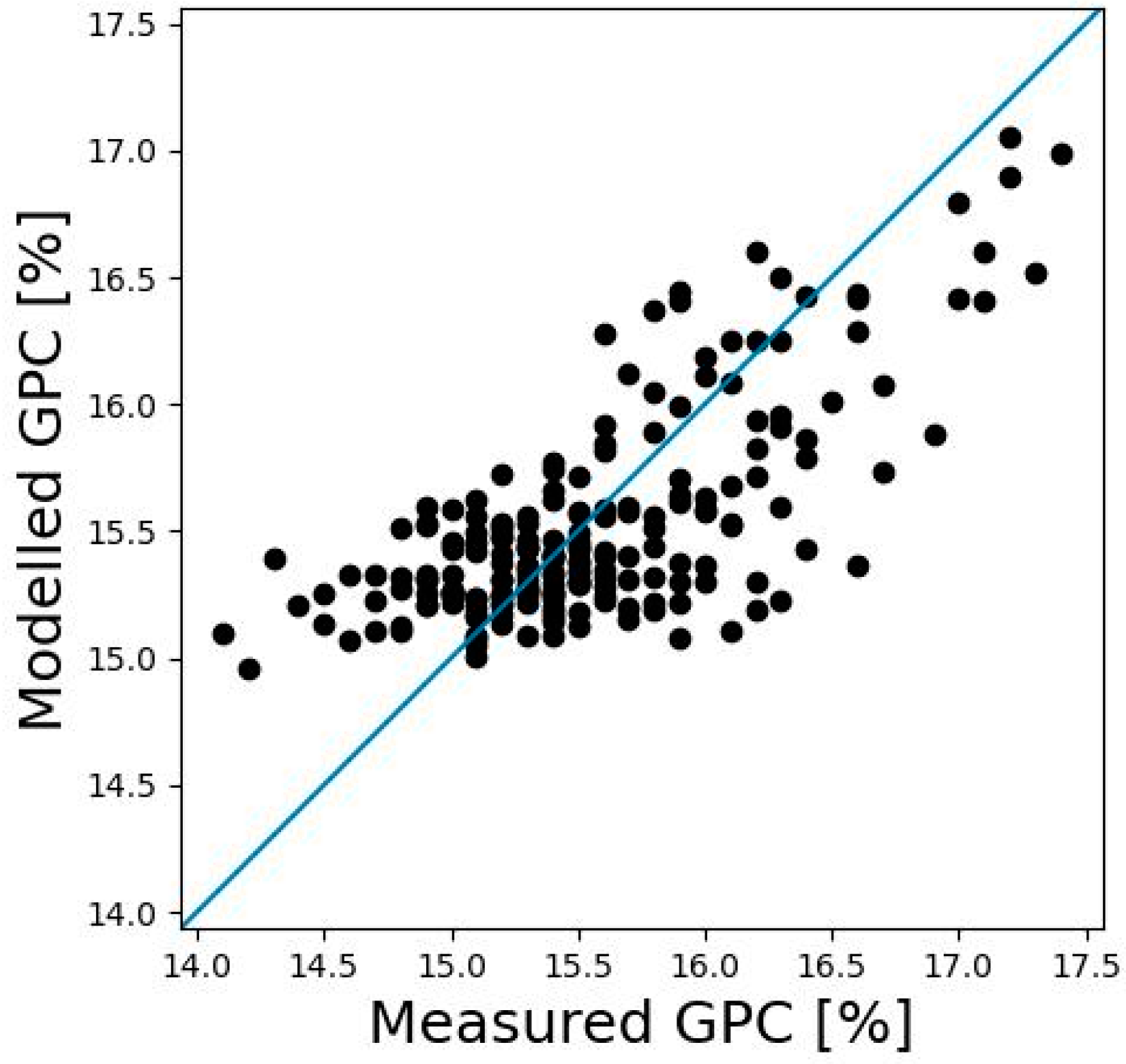
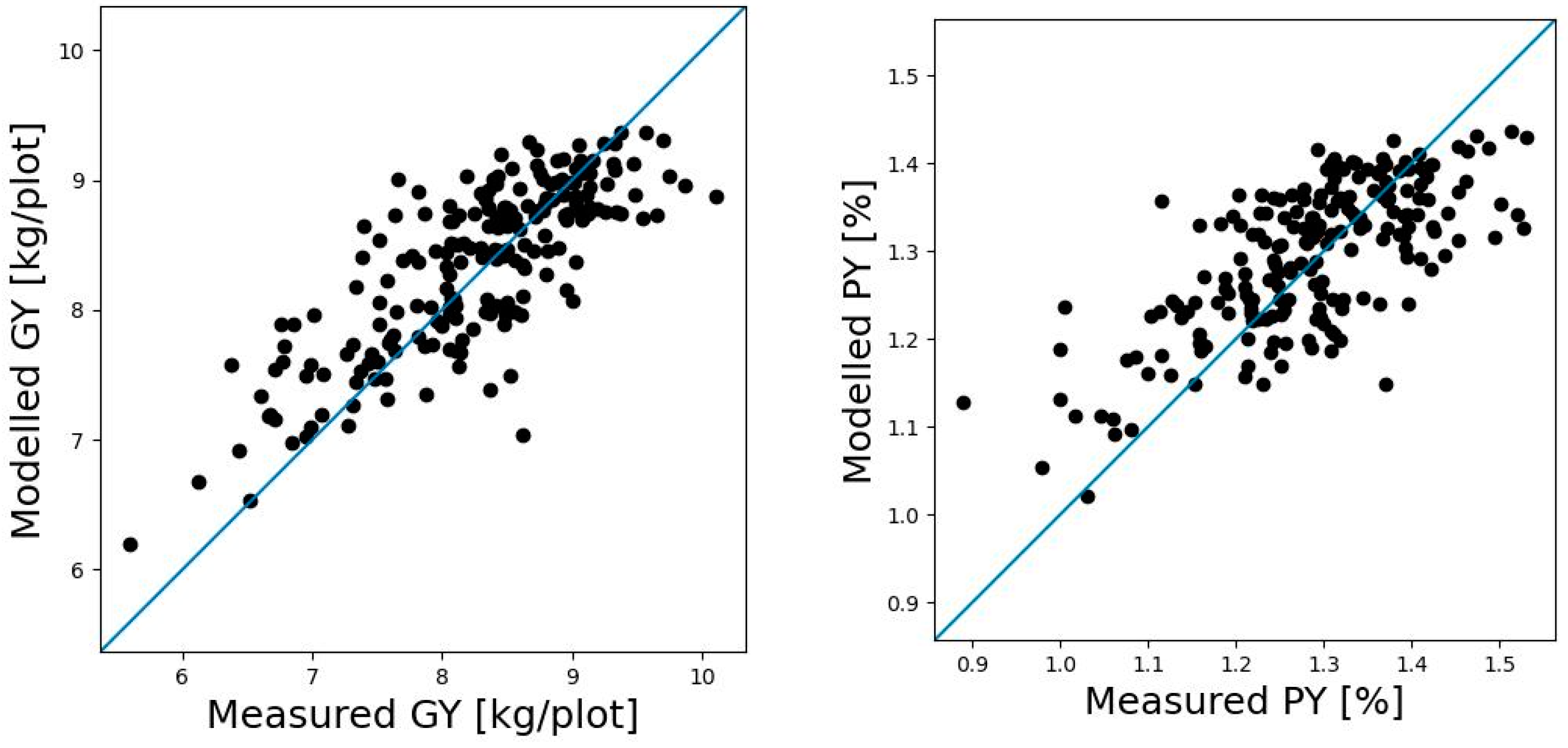
3.1. Relationship between GY, GCP, and PY
3.2. Models per Date
3.2.1. Models with Pléiades Data per Date
3.2.2. Models with P4M Data per Date
3.3. Model for the Growing Period of April–June 2022
3.3.1. Models with Pléiades Data across Multiple Dates
3.3.2. Models with P4M Data across Multiple Dates
4. Discussion
4.1. Predicting GY with Pléiades and P4M Data
4.2. Predicting GPC with Pléiades and P4M Data
4.3. Predicting PY with Pléiades and P4M Data
4.4. Limitations, Challenges, and Future Opportunities
- (1)
- Few applications of satellite imagery for phenotyping have been reported so far [38,39,40,108]. These studies’ findings support the use of high-resolution satellite imagery for phenotyping applications. Our results using Pléiades data are not conclusive but suggest that some potential for the prediction of GPC exists. High variability among genotypes may have contributed to the observed relations with the satellite data. Training models with a larger number of spectral bands may improve the accuracy of the predictions [31]. A red-edge band may be particularly useful since it would permit the calculation of VIs that were shown as good predictors in this study.
- (2)
- The selection of the machine learning method may be of key importance in modeling GY and other crop parameters, as previous studies have emphasized [31]. The reason is that the best-performing method may differ across traits and data sets. In this study, one machine learning method was tested, GPR. This method was selected based on the good results achieved in previous studies [30] and its wide application to the prediction of vegetation parameters.
- (3)
- Our study utilized data from 52 genotypes within a single growing season. We posit that enhancing the training set with additional genotypes could enhance the predictive capacity of the developed models [31], and incorporating data from multiple growing seasons would bolster the robustness of these models. Conducting experimental studies in the form of CVT under natural conditions could facilitate these improvements. Accurately predicting GY and GPC is integral to the monitoring of such trials.
- (4)
- Assessing the accuracy of measured data and retrieved products is a significant challenge in phenotyping. This crucial information is essential for optimizing the potential benefits of applied remote sensing technology. Quantitative measurements form the foundation of both proximal and remote sensing approaches, and it is imperative to provide associated uncertainty estimates [109]. Ensuring the traceability of measurements and derived products to international standards is essential to facilitate the generation of actionable information. While our study delved into the uncertainties of the retrieval algorithm, additional efforts are required to gauge the impact of uncertainties throughout the entire value chain, encompassing remote sensing observations and the propagation of uncertainty as a product uncertainty budget.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poutanen, K.S.; Kårlund, A.O.; Gómez-Gallego, C.; Johansson, D.P.; Scheers, N.M.; Marklinder, I.M.; Eriksen, A.K.; Silventoinen, P.C.; Nordlund, E.; Sozer, N.; et al. Grains—A Major Source of Sustainable Protein for Health. Nutr. Rev. 2022, 80, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, H.; Rosado, P.; Roser, M. Environmental Impacts of Food Production. Our World Data. 2022. Available online: https://ourworldindata.org/environmental-impacts-of-food (accessed on 11 September 2023).
- Shewry, P. Increasing the Health Benefits of Wheat. FEBS J. 2009, 276, 71. [Google Scholar] [CrossRef]
- FAOSTAT—Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database. 2016. Available online: http://faostat.fao.org (accessed on 11 September 2023).
- Ren, J.; Sun, D.; Chen, L.; You, F.M.; Wang, J.; Peng, Y.; Nevo, E.; Sun, D.; Luo, M.-C.; Peng, J. Genetic Diversity Revealed by Single Nucleotide Polymorphism Markers in a Worldwide Germplasm Collection of Durum Wheat. Int. J. Mol. Sci. 2013, 14, 7061–7088. [Google Scholar] [CrossRef] [PubMed]
- Dechev, D.; Bozhanova, V.; Yanev, S.; Delchev, G.; Panayotova, G.; Saldzhiev, I.; Nedyalkova, S.; Hadzhiivanova, B.; Taneva, K. Achievements and Problems of Durum Wheat Breeding and Technologies. Field Crops Stud. 2011, 6, 201–216. [Google Scholar]
- Ceglar, A.; Toreti, A.; Zampieri, M.; Royo, C. Global Loss of Climatically Suitable Areas for Durum Wheat Growth in the Future. Environ. Res. Lett. 2021, 16, 104049. [Google Scholar] [CrossRef]
- Rharrabti, Y.; Villegas, D.; Royo, C.; Martos-Núñez, V.; García del Moral, L.F. Durum Wheat Quality in Mediterranean Environments: II. Influence of Climatic Variables and Relationships between Quality Parameters. Field Crops Res. 2003, 80, 133–140. [Google Scholar] [CrossRef]
- Ben Mariem, S.; González-Torralba, J.; Collar, C.; Aranjuelo, I.; Morales, F. Durum Wheat Grain Yield and Quality under Low and High Nitrogen Conditions: Insights into Natural Variation in Low- and High-Yielding Genotypes. Plants 2020, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Dragov, R.G. Combining Ability for Quantitative Traits Related to Productivity in Durum Wheat. Vavilov J. Genet. Breed. 2022, 26, 515–523. [Google Scholar] [CrossRef]
- Sharma, A.; Garg, S.; Sheikh, I.; Vyas, P.; Dhaliwal, H.S. Effect of Wheat Grain Protein Composition on End-Use Quality. J. Food Sci. Technol. 2020, 57, 2771–2785. [Google Scholar] [CrossRef]
- Goutam, U.; Kukreja, S.; Tiwari, R.; Chaudhury, A.; Gupta, R.K.; Yadav, R. Biotechnological Approaches for Grain Quality Improvement in Wheat: Present Status and Future Possibilities. Aust. J. Crop Sci. 2013, 7, 469–483. [Google Scholar]
- Ruiz, M.; Vázquez, J.F.; Carrillo, J.M. Genetic Bases of Grain Quality. In Durum Wheat Breeding; CRC Press, Food Products Press: New York, NY, USA, 2005; ISBN 978-0-429-18029-3. [Google Scholar]
- Olmos, S.; Distelfeld, A.; Chicaiza, O.; Schlatter, A.R.; Fahima, T.; Echenique, V.; Dubcovsky, J. Precise Mapping of a Locus Affecting Grain Protein Content in Durum Wheat. Theor. Appl. Genet. 2003, 107, 1243–1251. [Google Scholar] [CrossRef]
- Roselló, M.; Royo, C.; Álvaro, F.; Villegas, D.; Nazco, R.; Soriano, J.M. Pasta-Making Quality QTLome From Mediterranean Durum Wheat Landraces. Front. Plant Sci. 2018, 9, 1512. [Google Scholar] [CrossRef]
- Blanco, A.; Simeone, R.; Gadaleta, A. Detection of QTLs for Grain Protein Content in Durum Wheat. Theor. Appl. Genet. 2006, 112, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Leiser, W.L.; Kazman, E.; Longin, C.F.H. Genetic Control of Protein Content and Sedimentation Volume in European Winter Wheat Cultivars. Theor. Appl. Genet. 2016, 129, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Rapp, M.; Lein, V.; Lacoudre, F.; Lafferty, J.; Müller, E.; Vida, G.; Bozhanova, V.; Ibraliu, A.; Thorwarth, P.; Piepho, H.P.; et al. Simultaneous Improvement of Grain Yield and Protein Content in Durum Wheat by Different Phenotypic Indices and Genomic Selection. Theor. Appl. Genet. 2018, 131, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.-M.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The Molecular-Genetics of Nitrogen Assimilation into Amino Acids in Higher Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Serret, M.D.; Pie, J.B.; Shah, S.S.; Li, Z. Relative Contribution of Nitrogen Absorption, Remobilization, and Partitioning to the Ear During Grain Filling in Chinese Winter Wheat. Front. Plant Sci. 2018, 9, 1351. [Google Scholar] [CrossRef]
- Lammerts van Bueren, E.T.; Struik, P.C. Diverse Concepts of Breeding for Nitrogen Use Efficiency. A Review. Agron. Sustain. Dev. 2017, 37, 50. [Google Scholar] [CrossRef]
- Xynias, I.N.; Mylonas, I.; Korpetis, E.G.; Ninou, E.; Tsaballa, A.; Avdikos, I.D.; Mavromatis, A.G. Durum Wheat Breeding in the Mediterranean Region: Current Status and Future Prospects. Agronomy 2020, 10, 432. [Google Scholar] [CrossRef]
- Monaghan, J.M.; Snape, J.W.; Chojecki, A.J.S.; Kettlewell, P.S. The Use of Grain Protein Deviation for Identifying Wheat Cultivars with High Grain Protein Concentration and Yield. Euphytica 2001, 122, 309–317. [Google Scholar] [CrossRef]
- Koekemoer, F.P.; Labuschagne, M.T.; Van Deventer, C.S. A Selection Strategy for Combining High Grain Yield and High Protein Content in South African Wheat Cultivars. Cereal Res. Commun. 1999, 27, 107–114. [Google Scholar] [CrossRef]
- Xue, L.-H.; Cao, W.-X.; Yang, L.-Z. Predicting Grain Yield and Protein Content in Winter Wheat at Different N Supply Levels Using Canopy Reflectance Spectra. Pedosphere 2007, 17, 646–653. [Google Scholar] [CrossRef]
- Dalla Marta, A.; Grifoni, D.; Mancini, M.; Orlando, F.; Guasconi, F.; Orlandini, S. Durum Wheat In-Field Monitoring and Early-Yield Prediction: Assessment of Potential Use of High Resolution Satellite Imagery in a Hilly Area of Tuscany, Central Italy. J. Agric. Sci. 2015, 153, 68–77. [Google Scholar] [CrossRef]
- Machwitz, M.; Pieruschka, R.; Berger, K.; Schlerf, M.; Aasen, H.; Fahrner, S.; Jiménez-Berni, J.; Baret, F.; Rascher, U. Bridging the Gap Between Remote Sensing and Plant Phenotyping—Challenges and Opportunities for the Next Generation of Sustainable Agriculture. Front. Plant Sci. 2021, 12, 749374. [Google Scholar] [CrossRef] [PubMed]
- Kyratzis, A.C.; Skarlatos, D.P.; Menexes, G.C.; Vamvakousis, V.F.; Katsiotis, A. Assessment of Vegetation Indices Derived by UAV Imagery for Durum Wheat Phenotyping under a Water Limited and Heat Stressed Mediterranean Environment. Front. Plant Sci. 2017, 8, 1114. [Google Scholar] [CrossRef]
- Gracia-Romero, A.; Kefauver, S.C.; Fernandez-Gallego, J.A.; Vergara-Díaz, O.; Nieto-Taladriz, M.T.; Araus, J.L. UAV and Ground Image-Based Phenotyping: A Proof of Concept with Durum Wheat. Remote Sens. 2019, 11, 1244. [Google Scholar] [CrossRef]
- Ganeva, D.; Roumenina, E.; Dimitrov, P.; Gikov, A.; Jelev, G.; Dragov, R.; Bozhanova, V.; Taneva, K. Phenotypic Traits Estimation and Preliminary Yield Assessment in Different Phenophases of Wheat Breeding Experiment Based on UAV Multispectral Images. Remote Sens. 2022, 14, 1019. [Google Scholar] [CrossRef]
- Vatter, T.; Gracia-Romero, A.; Kefauver, S.C.; Nieto-Taladriz, M.T.; Aparicio, N.; Araus, J.L. Preharvest Phenotypic Prediction of Grain Quality and Yield of Durum Wheat Using Multispectral Imaging. Plant J. 2022, 109, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Feng, L.; Chen, S.; Zhang, C.; Zhang, Y.; He, Y. A Comprehensive Review on Recent Applications of Unmanned Aerial Vehicle Remote Sensing with Various Sensors for High-Throughput Plant Phenotyping. Comput. Electron. Agric. 2021, 182, 106033. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Tao, X.; Chen, X.; Li, X. Rapid Prediction of Winter Wheat Yield and Nitrogen Use Efficiency Using Consumer-Grade Unmanned Aerial Vehicles Multispectral Imagery. Front. Plant Sci. 2022, 13, 1032170. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field High-Throughput Phenotyping: The New Crop Breeding Frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A Rapid Monitoring of NDVI across the Wheat Growth Cycle for Grain Yield Prediction Using a Multi-Spectral UAV Platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Zhang, C.; Marzougui, A.; Sankaran, S. High-Resolution Satellite Imagery Applications in Crop Phenotyping: An Overview. Comput. Electron. Agric. 2020, 175, 105584. [Google Scholar] [CrossRef]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A Direct Comparison of Remote Sensing Approaches for High-Throughput Phenotyping in Plant Breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, S.; Quirós, J.J.; Miklas, P.N. Unmanned Aerial System and Satellite-Based High Resolution Imagery for High-Throughput Phenotyping in Dry Bean. Comput. Electron. Agric. 2019, 165, 104965. [Google Scholar] [CrossRef]
- Meng, X.; Shen, H.; Li, H.; Zhang, L.; Fu, R. Review of the Pansharpening Methods for Remote Sensing Images Based on the Idea of Meta-Analysis: Practical Discussion and Challenges. Inf. Fusion 2019, 46, 102–113. [Google Scholar] [CrossRef]
- Gleyzes, A.; Perret, L.; Cazala-Houcade, E. Pleiades System Is Fully Operational in Orbit. In Proceedings of the EARSeL Symposium, Matera, Italy, 3–6 June 2013; Volume 33, pp. 445–460. [Google Scholar]
- Coeurdevey, L.; Fernandez, K. Pleiades Imagery—User Guide, V2.0; Airbus Defence and Space Intelligence: Paris, France, 2012. [Google Scholar]
- Latry, C.; Fourest, S.; Thiebaut, C. Restoration Technique for Pléades-HR Panchromatic Images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, XXXIX-B1, 555–560. [Google Scholar] [CrossRef]
- Kokhan, S.; Vostokov, A. Using Vegetative Indices to Quantify Agricultural Crop Characteristics. J. Ecol. Eng. 2020, 21, 120–127. [Google Scholar] [CrossRef]
- Bolton, D.K.; Friedl, M.A. Forecasting Crop Yield Using Remotely Sensed Vegetation Indices and Crop Phenology Metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Herrmann, I.; Bdolach, E.; Montekyo, Y.; Rachmilevitch, S.; Townsend, P.A.; Karnieli, A. Assessment of Maize Yield and Phenology by Drone-Mounted Superspectral Camera. Precis. Agric. 2020, 21, 51–76. [Google Scholar] [CrossRef]
- Dimov, D.; Uhl, J.H.; Löw, F.; Seboka, G.N. Sugarcane Yield Estimation through Remote Sensing Time Series and Phenology Metrics. Smart Agric. Technol. 2022, 2, 100046. [Google Scholar] [CrossRef]
- Nazir, A.; Ullah, S.; Saqib, Z.A.; Abbas, A.; Ali, A.; Iqbal, M.S.; Hussain, K.; Shakir, M.; Shah, M.; Butt, M.U. Estimation and Forecasting of Rice Yield Using Phenology-Based Algorithm and Linear Regression Model on Sentinel-II Satellite Data. Agriculture 2021, 11, 1026. [Google Scholar] [CrossRef]
- Qader, S.H.; Dash, J.; Atkinson, P.M. Forecasting Wheat and Barley Crop Production in Arid and Semi-Arid Regions Using Remotely Sensed Primary Productivity and Crop Phenology: A Case Study in Iraq. Sci. Total Environ. 2018, 613–614, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Evans, F.H.; Shen, J. Long-Term Hindcasts of Wheat Yield in Fields Using Remotely Sensed Phenology, Climate Data and Machine Learning. Remote Sens. 2021, 13, 2435. [Google Scholar] [CrossRef]
- Richetti, J.; Judge, J.; Boote, K.J.; Johann, J.A.; Uribe-Opazo, M.A.; Becker, W.R.; Paludo, A.; de Silva, L.C.A. Using Phenology-Based Enhanced Vegetation Index and Machine Learning for Soybean Yield Estimation in Paraná State, Brazil. J. Appl. Remote Sens. 2018, 12, 026029. [Google Scholar] [CrossRef]
- Rodrigues, F.A.; Blasch, G.; Defourny, P.; Ortiz-Monasterio, J.I.; Schulthess, U.; Zarco-Tejada, P.J.; Taylor, J.A.; Gérard, B. Multi-Temporal and Spectral Analysis of High-Resolution Hyperspectral Airborne Imagery for Precision Agriculture: Assessment of Wheat Grain Yield and Grain Protein Content. Remote Sens. 2018, 10, 930. [Google Scholar] [CrossRef]
- IUSS Working Group. WRB World Reference Base for Soil Resources 2014. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. Update 2015; FAO—Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- ISO 20483:2013; Determination of the Nitrogen Content and Calculation of the Crude Protein Content—Kjeldahl Method. Bulgarian Institute for Standardization Cereals and Pulses: Sofia, Bulgaria, 2013.
- Cubero-Castan, M.; Schneider-Zapp, K.; Bellomo, M.; Shi, D.; Rehak, M.; Strecha, C. Assessment of The Radiometric Accuracy in a Target Less Work Flow Using Pix4D Software. In Proceedings of the 2018 9th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 23–26 September 2018; pp. 1–4. [Google Scholar]
- Rivera, J.P.; Verrelst, J.; Delegido, J.; Veroustraete, F.; Moreno, J. On the Semi-Automatic Retrieval of Biophysical Parameters Based on Spectral Index Optimization. Remote Sens. 2014, 6, 4927–4951. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI Estimation Using Parametric, Non-Parametric and Physical Retrieval Methods—A Comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Richter, K.; Hank, T.B.; Mauser, W.; Atzberger, C. Derivation of Biophysical Variables from Earth Observation Data: Validation and Statistical Measures. J. Appl. Remote Sens. 2012, 6, 063557. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Y.; Yao, X.; Zhu, Y.; Cao, W. Predicting Grain Yield and Protein Content in Wheat by Fusing Multi-Sensor and Multi-Temporal Remote-Sensing Images. Field Crops Res. 2014, 164, 178–188. [Google Scholar] [CrossRef]
- Serrano, L.; Filella, I.; Peñuelas, J. Remote Sensing of Biomass and Yield of Winter Wheat under Different Nitrogen Supplies. Crop Sci. 2000, 40, 723–731. [Google Scholar] [CrossRef]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring Vegetation Systems in the Great Plains with ERTS. In Proceedings of the Goddard Space Flight Center 3D ERTS-1 Symposium, Washington, DC, USA, 10–14 December 1973; NASA Special Publication: Washington, DC, USA; Volume 1. [Google Scholar]
- Walsh, O.S.; Marshall, J.M.; Nambi, E.; Jackson, C.A.; Ansah, E.O.; Lamichhane, R.; McClintick-Chess, J.; Bautista, F. Wheat Yield and Protein Estimation with Handheld and Unmanned Aerial Vehicle-Mounted Sensors. Agronomy 2023, 13, 207. [Google Scholar] [CrossRef]
- Nagy, A.; Szabó, A.; Adeniyi, O.D.; Tamás, J. Wheat Yield Forecasting for the Tisza River Catchment Using Landsat 8 NDVI and SAVI Time Series and Reported Crop Statistics. Agronomy 2021, 11, 652. [Google Scholar] [CrossRef]
- Stoy, P.C.; Khan, A.M.; Wipf, A.; Silverman, N.; Powell, S.L. The Spatial Variability of NDVI within a Wheat Field: Information Content and Implications for Yield and Grain Protein Monitoring. PLoS ONE 2022, 17, e0265243. [Google Scholar] [CrossRef]
- Tan, C.; Zhou, X.; Zhang, P.; Wang, Z.; Wang, D.; Guo, W.; Yun, F. Predicting Grain Protein Content of Field-Grown Winter Wheat with Satellite Images and Partial Least Square Algorithm. PLoS ONE 2020, 15, e0228500. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Segarra, J.; Araus, J.L.; Kefauver, S.C. Farming and Earth Observation: Sentinel-2 Data to Estimate within-Field Wheat Grain Yield. Int. J. Appl. Earth Obs. Geoinf. 2022, 107, 102697. [Google Scholar] [CrossRef]
- Penuelas, J.; Baret, F.; Filella, I. Semi-Empirical Indices to Assess Carotenoids/Chlorophyll a Ratio from Leaf Spectral Reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Gitelson, A.; Merzlyak, M.N. Quantitative Estimation of Chlorophyll-a Using Reflectance Spectra: Experiments with Autumn Chestnut and Maple Leaves. J. Photochem. Photobiol. B 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Veverka, D.; Chatterjee, A.; Carlson, M. Comparisons of Sensors to Predict Spring Wheat Grain Yield and Protein Content. Agron. J. 2021, 113, 2091–2101. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Sun, Y.; Su, Y.; Li, Y.; Yi, Y.; Liu, Y. Research on Dynamic Monitoring of Grain Filling Process of Winter Wheat from Time-Series Planet Imageries. Agronomy 2022, 12, 2451. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Jay, S.; Maupas, F.; Bendoula, R.; Gorretta, N. Retrieving LAI, Chlorophyll and Nitrogen Contents in Sugar Beet Crops from Multi-Angular Optical Remote Sensing: Comparison of Vegetation Indices and PROSAIL Inversion for Field Phenotyping. Field Crops Res. 2017, 210, 33–46. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS Terrestrial Chlorophyll Index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Zhao, H.; Song, X.; Yang, G.; Li, Z.; Zhang, D.; Feng, H. Monitoring of Nitrogen and Grain Protein Content in Winter Wheat Based on Sentinel-2A Data. Remote Sens. 2019, 11, 1724. [Google Scholar] [CrossRef]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.T.; Mcmurtrey, J.E.; Walthall, C.L. Evaluation of Digital Photography from Model Aircraft for Remote Sensing of Crop Biomass and Nitrogen Status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Du, M.; Noguchi, N. Monitoring of Wheat Growth Status and Mapping of Wheat Yield’s within-Field Spatial Variations Using Color Images Acquired from UAV-Camera System. Remote Sens. 2017, 9, 289. [Google Scholar] [CrossRef]
- Hunt, E.R., Jr.; Daughtry, C.S.T.; Eitel, J.U.H.; Long, D.S. Remote Sensing Leaf Chlorophyll Content Using a Visible Band Index. Agron. J. 2011, 103, 1090–1099. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing Prediction Power and Stability of Broadband and Hyperspectral Vegetation Indices for Estimation of Green Leaf Area Index and Canopy Chlorophyll Density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Colecchia, S.A.; De Vita, P.; Rinaldi, M. Effects of Tillage Systems in Durum Wheat under Rainfed Mediterranean Conditions. Cereal Res. Commun. 2015, 43, 704–716. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of Soil-Adjusted Vegetation Indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Xu, X.; Teng, C.; Zhao, Y.; Du, Y.; Zhao, C.; Yang, G.; Jin, X.; Song, X.; Gu, X.; Casa, R.; et al. Prediction of Wheat Grain Protein by Coupling Multisource Remote Sensing Imagery and ECMWF Data. Remote Sens. 2020, 12, 1349. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a Two-Band Enhanced Vegetation Index without a Blue Band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Zhou, X.; Kono, Y.; Win, A.; Matsui, T.; Tanaka, T.S.T. Predicting Within-Field Variability in Grain Yield and Protein Content of Winter Wheat Using UAV-Based Multispectral Imagery and Machine Learning Approaches. Plant Prod. Sci. 2021, 24, 137–151. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Roumenina, E.; Jelev, G.; Dimitrov, P.; Filchev, L.; Kamenova, I.; Gikov, A.; Banov, M.; Krasteva, V.; Kercheva, M.; Kolchakov, V. Qualitative evaluation and within-field mapping of winter wheat crop condition using multispectral remote sensing data. Bulg. J. Agric. Sci. 2020, 26, 1129–1142. [Google Scholar]
- Tian, Y.-C.; Gu, K.-J.; Chu, X.; Yao, X.; Cao, W.-X.; Zhu, Y. Comparison of Different Hyperspectral Vegetation Indices for Canopy Leaf Nitrogen Concentration Estimation in Rice. Plant Soil 2014, 376, 193–209. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Wolters, S.; Söderström, M.; Piikki, K.; Börjesson, T.; Pettersson, C.-G. Predicting Grain Protein Concentration in Winter Wheat (Triticum aestivum L.) Based on Unpiloted Aerial Vehicle Multispectral Optical Remote Sensing. Acta Agric. Scand. Sect. B Soil Plant Sci. 2022, 72, 788–802. [Google Scholar] [CrossRef]
- Ren, S.; Chen, X.; An, S. Assessing Plant Senescence Reflectance Index-Retrieved Vegetation Phenology and Its Spatiotemporal Response to Climate Change in the Inner Mongolian Grassland. Int. J. Biometeorol. 2017, 61, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Guo, C.; Tang, Y.; Lu, J.; Zhu, Y.; Cao, W.; Cheng, T.; Zhang, L.; Tian, Y. Predicting Wheat Productivity: Integrating Time Series of Vegetation Indices into Crop Modeling via Sequential Assimilation. Agric. For. Meteorol. 2019, 272–273, 69–80. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, L.; Zhou, X.; Zhu, Y.; Cao, W.; Qiu, X.; Cheng, T.; Tian, Y. Integrating Remote Sensing Information with Crop Model to Monitor Wheat Growth and Yield Based on Simulation Zone Partitioning. Precis. Agric. 2018, 19, 55–78. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Rasmussen, C.; Williams, C. Gaussian Processes for Machine Learning; The MIT Press: New York, NY, USA, 2006. [Google Scholar]
- Berger, K.; Verrelst, J.; Féret, J.-B.; Wang, Z.; Wocher, M.; Strathmann, M.; Danner, M.; Mauser, W.; Hank, T. Crop Nitrogen Monitoring: Recent Progress and Principal Developments in the Context of Imaging Spectroscopy Missions. Remote Sens. Environ. 2020, 242, 111758. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Moreno, J.; Camps-Valls, G. Gaussian Processes Uncertainty Estimates in Experimental Sentinel-2 LAI and Leaf Chlorophyll Content Retrieval. ISPRS J. Photogramm. Remote Sens. 2013, 86, 157–167. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Gitelson, A.; Delegido, J.; Moreno, J.; Camps-Valls, G. Spectral Band Selection for Vegetation Properties Retrieval Using Gaussian Processes Regression. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 554–567. [Google Scholar] [CrossRef]
- Galvão, L.S.; Breunig, F.M.; dos Santos, J.R.; de Moura, Y.M. View-Illumination Effects on Hyperspectral Vegetation Indices in the Amazonian Tropical Forest. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 291–300. [Google Scholar] [CrossRef]
- Zhu, G.; Ju, W.; Chen, J.M.; Liu, Y. A Novel Moisture Adjusted Vegetation Index (MAVI) to Reduce Background Reflectance and Topographical Effects on LAI Retrieval. PLoS ONE 2014, 9, e102560. [Google Scholar] [CrossRef]
- Bastos, L.M.; Froes de Borja Reis, A.; Sharda, A.; Wright, Y.; Ciampitti, I.A. Current Status and Future Opportunities for Grain Protein Prediction Using On- and Off-Combine Sensors: A Synthesis-Analysis of the Literature. Remote Sens. 2021, 13, 5027. [Google Scholar] [CrossRef]
- Shewry, P.R.; Mitchell, R.A.C.; Tosi, P.; Wan, Y.; Underwood, C.; Lovegrove, A.; Freeman, J.; Toole, G.A.; Mills, E.N.C.; Ward, J.L. An Integrated Study of Grain Development of Wheat (cv. Hereward). J. Cereal Sci. 2012, 56, 21–30. [Google Scholar] [CrossRef]
- Aranguren, M.; Castellón, A.; Aizpurua, A. Wheat Grain Protein Content under Mediterranean Conditions Measured with Chlorophyll Meter. Plants 2021, 10, 374. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bellido, R.J.; Shepherd, C.E.; Barraclough, P.B. Predicting Post-Anthesis N Requirements of Bread Wheat with a Minolta SPAD Meter. Eur. J. Agron. 2004, 20, 313–320. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, T.; Foulkes, M.J.; Orford, S.; Singh, A.M.; Wingen, L.U.; Karnam, V.; Nair, L.S.; Mandal, P.K.; Griffiths, S.; et al. Nitrogen Uptake and Remobilization from Pre- and Post-Anthesis Stages Contribute towards Grain Yield and Grain Protein Concentration in Wheat Grown in Limited Nitrogen Conditions. CABI Agric. Biosci. 2023, 4, 12. [Google Scholar] [CrossRef]
- Sankaran, S.; Zhang, C.; Hurst, J.P.; Marzougui, A.; Veeranampalayam-Sivakumar, A.N.; Li, J.; Schnable, J.; Shi, Y. Investigating the Potential of Satellite Imagery for High-Throughput Field Phenotyping Applications. In Proceedings of the Autonomous Air and Ground Sensing Systems for Agricultural Optimization and Phenotyping V, SPIE Defence + Commercial Sensing, Online, 27 April–9 May 2020; Volume 11414, p. 1141402. [Google Scholar]
- Mittaz, J.; Merchant, C.J.; Woolliams, E.R. Applying Principles of Metrology to Historical Earth Observations from Satellites. Metrologia 2019, 56, 032002. [Google Scholar] [CrossRef]



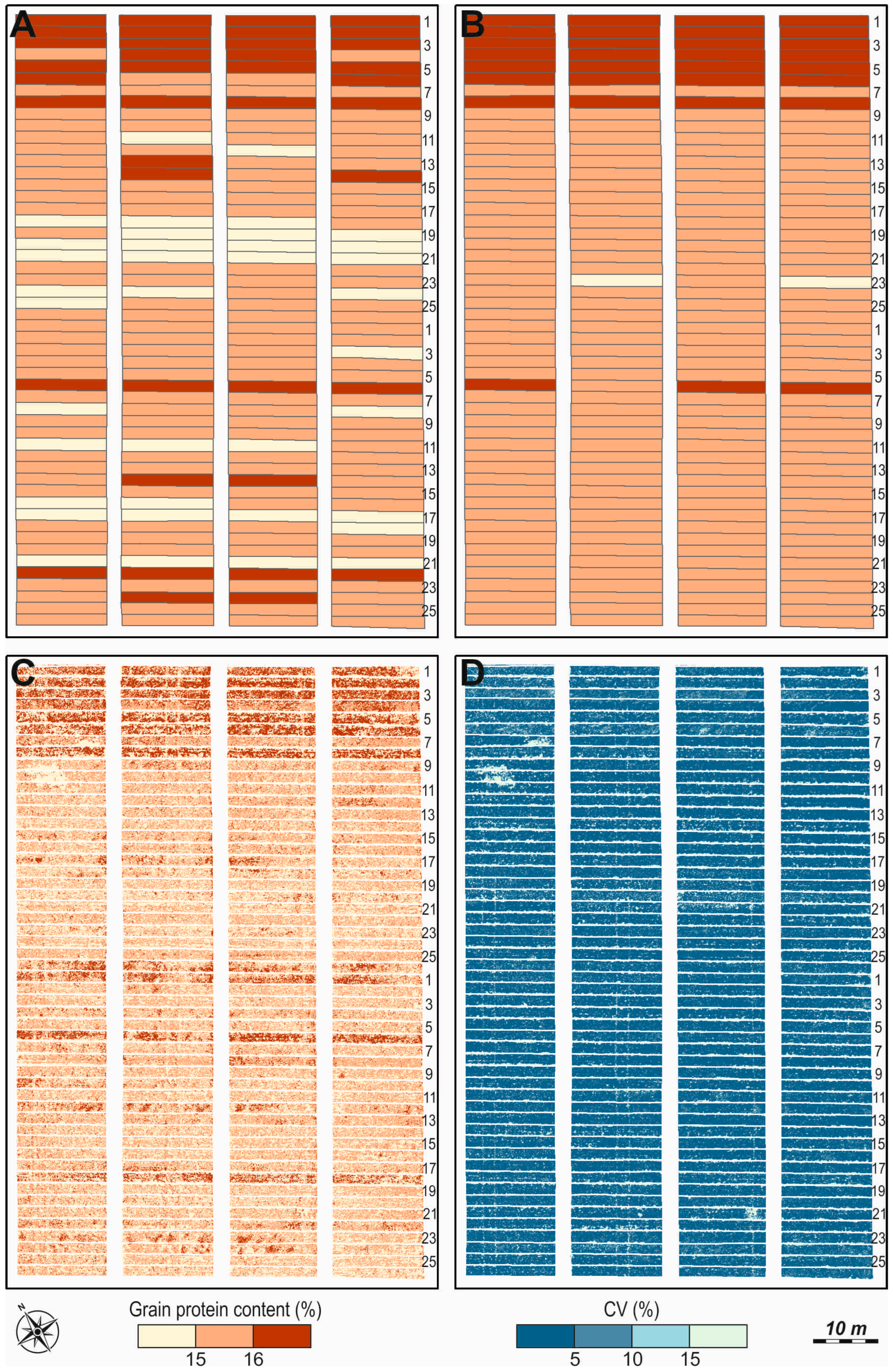
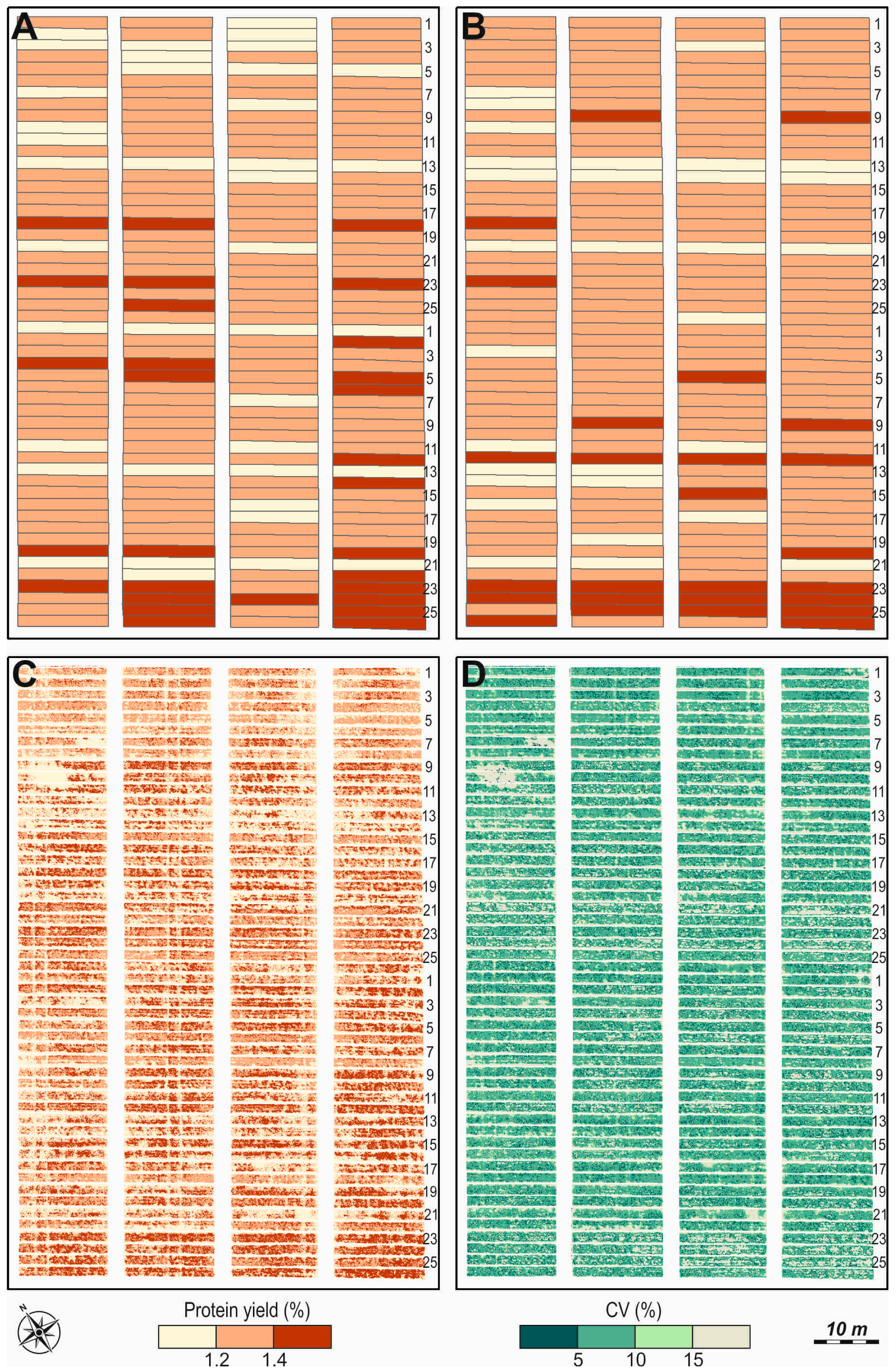

| Months | Mean Daily Air Temperature, °C | Monthly Amount of Precipitation, mm | ||
|---|---|---|---|---|
| 2021–2022 | 1928–2022 | 2021–2022 | 1928–2022 | |
| October | 11.3 | 12.7 | 150.5 | 38.6 |
| November | 7.9 | 7 | 14.2 | 47.3 |
| December | 3.9 | 1.4 | 108.8 | 54.0 |
| January | 1.8 | −0.2 | 21.4 | 44.3 |
| February | 4.2 | 1.7 | 40.1 | 37.7 |
| March | 4.2 | 5.7 | 22.4 | 37.0 |
| April | 12.2 | 11.8 | 36.0 | 45.3 |
| May | 17.3 | 16.9 | 29.4 | 64.1 |
| June | 22.0 | 20.7 | 80.5 | 65.4 |
| July | 25.1 | 23.1 | 7.7 | 54.1 |
| Sum | 109.9 | 100.8 | 511 | 487.8 |
| % | 109 | 100 | 104.7 | 100 |
| Number of Measurements | Min. | Max. | Mean | Std. Dev. | CV % | |
|---|---|---|---|---|---|---|
| GY (kg/plot) | 208 | 5.59 | 10.11 | 8.27 | 0.79 | 9.60 |
| GPC (%) | 208 | 14.10 | 17.40 | 15.55 | 0.60 | 3.87 |
| PY (%) | 208 | 0.89 | 1.53 | 1.28 | 0.11 | 8.86 |
| Date | Growth Stage | BBCH Code | P4M | Pléiades |
|---|---|---|---|---|
| 5 April 2022 | Beginning of stem elongation | BBCH 30–31 | ✓ | |
| 9 April 2022 | Mid-stem elongation | BBCH 34 | ✓ | |
| 28 April 2022 | Mid-boot | BBCH 43 | ✓ | |
| 5 May 2022 | 20% of inflorescence emerged | BBCH 52 | ✓ | |
| 19 May 2022 | Beginning of flowering | BBCH 61 | ✓ | |
| 26 May 2022 | Watery ripe | BBCH 71 | ✓ | |
| 31 May 2022 | Watery ripe | BBCH 71 | ✓ | |
| 15 June 2022 | Medium to late milk | BBCH 75–77 | ✓ | |
| 19 June 2022 | Medium to late milk | BBCH 75–77 | ✓ |
| Band Name | P4M | Pléiades 1A and 1B | ||
|---|---|---|---|---|
| Central Wavelength (nm) | Band Width (nm) | Central Wavelength (nm) | Band Width (nm) | |
| Panchromatic | – | – | 650 | 390 |
| Blue (B) | 450 | 32 | 490 | 120 |
| Green (G) | 560 | 32 | 560 | 120 |
| Red I | 650 | 32 | 650 | 120 |
| Red-edge (RE) | 730 | 32 | – | – |
| Near-infrared (NIR) | 840 | 52 | 840 | 200 |
| Date/Growth Stage | Parameter | Independent Features of the Best Model | Cross-Validation | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | nRMSE | rRMSE | R2 | RMSE | nRMSE | rRMSE | |||
| 9 April 2022/mid-stem-elongation | GY | TVI and VARI | 0.91 | 0.66 | 6.49 | 8.35 | 0.18 | 0.72 | 24.13 | 9.04 |
| GPC | NDVI, repRVI, VARI, and 3BSI-Tian | 0.99 | 0.45 | 2.57 | 3.07 | 0.51 | 0.41 | 13.77 | 2.65 | |
| PY | TVI and VARI | 0.90 | 0.1 | 6.72 | 8.43 | 0.07 | 0.11 | 25.70 | 8.86 | |
| 28 April 2022/mid-boot | GY | TVI and EVI | 0.91 | 0.64 | 6.37 | 8.20 | 0.30 | 0.73 | 24.47 | 9.17 |
| GPC | EVI2, VARI, and SAVI | 0.98 | 0.47 | 2.70 | 3.22 | 0.49 | 0.45 | 14.91 | 2.87 | |
| PY | TVI and TGI | 0.91 | 0.10 | 6.57 | 8.25 | 0.17 | 0.11 | 25.82 | 8.90 | |
| 5 May 2022/20% of inflorescence emerged | GY | SR, CIgreen, and TGI | 0.91 | 0.65 | 6.45 | 8.30 | 0.29 | 0.69 | 23.14 | 8.67 |
| GPC | TVI, VARI, EVI, and DVI | 0.99 | 0.47 | 2.69 | 3.20 | 0.58 | 0.40 | 13.22 | 2.54 | |
| PY | repRVI, TGI, and NDVI | 0.90 | 0.10 | 6.60 | 8.29 | 0.16 | 0.11 | 24.78 | 8.54 | |
| 19 May 2022/beginning of flowering | GY | All bands except blue, green, and SIPI | 0.89 | 0.71 | 6.99 | 8.99 | 0.12 | 0.70 | 23.37 | 8.76 |
| GPC | CIgreen, SR, DVI, EVI2, NIR, and SAVI | 0.98 | 0.48 | 2.79 | 3.32 | 0.46 | 0.50 | 16.63 | 3.20 | |
| PY | NDVI, SR, repSVI, TGI, OSAVI, 3BSI-Tian, GNDVI, SAVI, and EVI2 | 0.90 | 0.10 | 6.65 | 8.35 | 0.11 | 0.10 | 24.30 | 8.37 | |
| 31 May 2022/watery ripe | GY | SR | 0.90 | 0.68 | 6.76 | 8.69 | 0.15 | 0.72 | 23.97 | 8.98 |
| GPC | CIgreen, SR, and GNDVI | 0.98 | 0.52 | 3.00 | 3.58 | 0.34 | 0.49 | 16.35 | 3.14 | |
| PY | All bands except blue, green, red, and NGBDI | 0.90 | 0.10 | 6.65 | 8.35 | 0.16 | 0.11 | 24.77 | 8.54 | |
| 19 June 2022/medium to late milk | GY | GNDVI and DVI | 0.88 | 0.72 | 7.44 | 9.58 | 0.21 | 0.83 | 27.64 | 10.36 |
| GPC | GNDVI and DVI | 0.98 | 0.56 | 3.24 | 3.86 | 0.02 | 0.79 | 26.44 | 5.09 | |
| PY | DVI, GNDVI, and NGBDI | 0.87 | 0.12 | 7.64 | 9.60 | 0.08 | 0.14 | 32.24 | 11.11 | |
| Date/ Growth Stage | Parameter | Independent Features of the Best Model | Cross-Validation | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | nRMSE | rRMSE | R2 | RMSE | nRMSE | rRMSE | |||
| 5 April 2022/ beginning of stem elongation | GY | mNDblue, NIR, and TVI | 0.92 | 0.61 | 6.04 | 7.77 | 0.38 | 0.69 | 22.85 | 8.57 |
| GPC | PSRI and 3BSI-Tian | 0.98 | 0.53 | 3.04 | 3.62 | 0.38 | 0.46 | 15.42 | 2.97 | |
| PY | mNDblue and TVI | 0.92 | 0.09 | 6.61 | 8.30 | 0.41 | 0.09 | 21.86 | 7.53 | |
| 26 May 2022/ watery ripe | GY | CIred-edge, SR, CIgreen, and EVI | 0.94 | 0.54 | 5.31 | 6.83 | 0.55 | 0.58 | 19.27 | 7.22 |
| GPC | All bands | 0.98 | 0.50 | 2.85 | 3.39 | 0.29 | 0.50 | 16.56 | 3.19 | |
| PY | SR, CIred-edge, and reNDVI | 0.95 | 0.07 | 4.64 | 5.83 | 0.62 | 0.08 | 18.53 | 6.38 | |
| 15 June 2022/ medium to late milk | GY | 3BSI-Tian, CIred-edge, NGBDI, TGI, NGRDI, repRVI, PSRI, VARI, NDVI, OSAVI, reNDVI, SAVI, EVI, and TVI | 0.94 | 0.52 | 5.19 | 6.68 | 0.67 | 0.46 | 10.26 | 5.61 |
| GPC | repRVI, mNDblue, GNDVI, Red, and NGBDI | 0.98 | 0.46 | 2.66 | 3.17 | 0.52 | 0.42 | 12.63 | 2.68 | |
| PY | VARI, NIR, repRVI, NGBDI, CIred-edge, and NGRDI | 0.94 | 0.08 | 5.35 | 6.73 | 0.52 | 0.08 | 12.41 | 6.21 | |
| Parameter | Independent Features of the Best Model | Cross-Validation | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | nRMSE | rRMSE | R2 | RMSE | nRMSE | rRMSE | ||
| GY | GNDVI, PSRI, DVI, EVI2, and SAVI | 0.91 | 0.66 | 6.40 | 8.23 | 0.25 | 0.68 | 22.52 | 8.44 |
| GPC | GNDVI, repRVI, SR, SAVI, OSAVI, NDVI, EVI2, 3BSI-Tian, and DVI | 0.99 | 0.45 | 2.58 | 3.07 | 0.58 | 0.40 | 13.13 | 2.53 |
| PY | DVI, GNDVI, and PSRI | 0.90 | 0.10 | 6.73 | 8.45 | 0.13 | 0.10 | 24.14 | 8.32 |
| Parameter | Independent Features of the Best Model | Cross-Validation | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | nRMSE | rRMSE | R2 | RMSE | nRMSE | rRMSE | ||
| GY | repRVI, GNDVI, MTCI, PSRI, SR, and EVI | 0.94 | 0.54 | 5.30 | 6.52 | 0.57 | 0.59 | 19.77 | 7.41 |
| GPC | EVI, OSAVI, 3BSI-Tian, GNDVI, and EVI2 | 0.99 | 0.42 | 2.43 | 2.89 | 0.47 | 0.43 | 14.43 | 2.78 |
| PY | repRVI and MTCI | 0.94 | 0.08 | 5.25 | 6.60 | 0.51 | 0.09 | 20.30 | 6.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganeva, D.; Roumenina, E.; Dimitrov, P.; Gikov, A.; Bozhanova, V.; Dragov, R.; Jelev, G.; Taneva, K. Preharvest Durum Wheat Yield, Protein Content, and Protein Yield Estimation Using Unmanned Aerial Vehicle Imagery and Pléiades Satellite Data in Field Breeding Experiments. Remote Sens. 2024, 16, 559. https://doi.org/10.3390/rs16030559
Ganeva D, Roumenina E, Dimitrov P, Gikov A, Bozhanova V, Dragov R, Jelev G, Taneva K. Preharvest Durum Wheat Yield, Protein Content, and Protein Yield Estimation Using Unmanned Aerial Vehicle Imagery and Pléiades Satellite Data in Field Breeding Experiments. Remote Sensing. 2024; 16(3):559. https://doi.org/10.3390/rs16030559
Chicago/Turabian StyleGaneva, Dessislava, Eugenia Roumenina, Petar Dimitrov, Alexander Gikov, Violeta Bozhanova, Rangel Dragov, Georgi Jelev, and Krasimira Taneva. 2024. "Preharvest Durum Wheat Yield, Protein Content, and Protein Yield Estimation Using Unmanned Aerial Vehicle Imagery and Pléiades Satellite Data in Field Breeding Experiments" Remote Sensing 16, no. 3: 559. https://doi.org/10.3390/rs16030559
APA StyleGaneva, D., Roumenina, E., Dimitrov, P., Gikov, A., Bozhanova, V., Dragov, R., Jelev, G., & Taneva, K. (2024). Preharvest Durum Wheat Yield, Protein Content, and Protein Yield Estimation Using Unmanned Aerial Vehicle Imagery and Pléiades Satellite Data in Field Breeding Experiments. Remote Sensing, 16(3), 559. https://doi.org/10.3390/rs16030559






