Tree Crown Segmentation and Diameter at Breast Height Prediction Based on BlendMask in Unmanned Aerial Vehicle Imagery
Abstract
1. Introduction
1.1. Research Significance and Background
1.2. Research Landscape
1.2.1. Research Status of Crown Width Extraction
- Lidar-based Crown Width Extraction [16]: Lidar technology allows for the highly precise collection of three-dimensional information about ground and canopy surfaces. It is widely used in crown width extraction. Various algorithms, including altitude-threshold-based [17], topological-relation-based [18], and morphological-operation-based [19] approaches, analyze laser point cloud data to extract tree crown information.
- Image-processing-based Crown Width Extraction [20]: This method uses remote sensing images to extract crown width. By analyzing color, texture, and shape attributes within remote sensing images, the automatic extraction of crown width is achieved.
- Machine-learning-based Crown Width Extraction [21]: Recent advancements in machine learning algorithms have led to increased exploration of these methods for crown width extraction. Researchers create training sample sets and utilize supervised learning algorithms such as vector machines [22] and random forests [23] to enable the automatic detection and segmentation of crowns.
1.2.2. Research Status of Deep Learning in Forestry Segmentation
1.2.3. Research Status of DBH Prediction of Trees
1.3. Primary Research Focus
- Utilizing the orthophoto map of Beijing Jingyue Ecological Forest Farm as experimental data to use the BlendMask network for segmenting individual crowns and detecting the count of Pinus tabulaeformis trees.
- Assessing the prediction results of the model using relevant accuracy evaluation metrics.
- Fitting an optimal relationship model between the DBH and crown width of trees using a Bayesian neural network, leveraging DBH measurements of sample trees collected in the field and the calculated crown mask area obtained from segmentation.
2. Research Area and Data Acquisition
2.1. Field Investigation and Data Acquisition
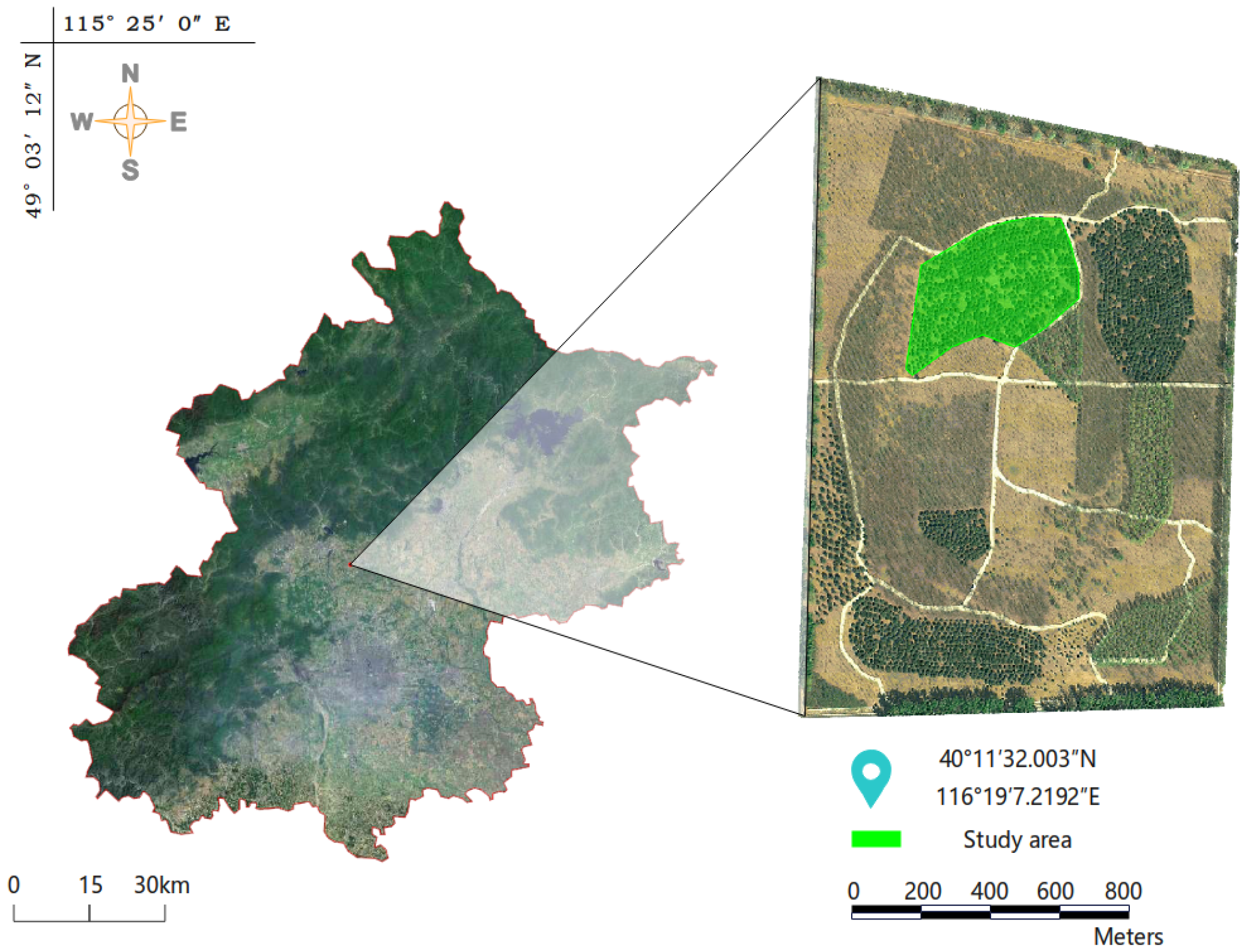
2.1.1. Research Area
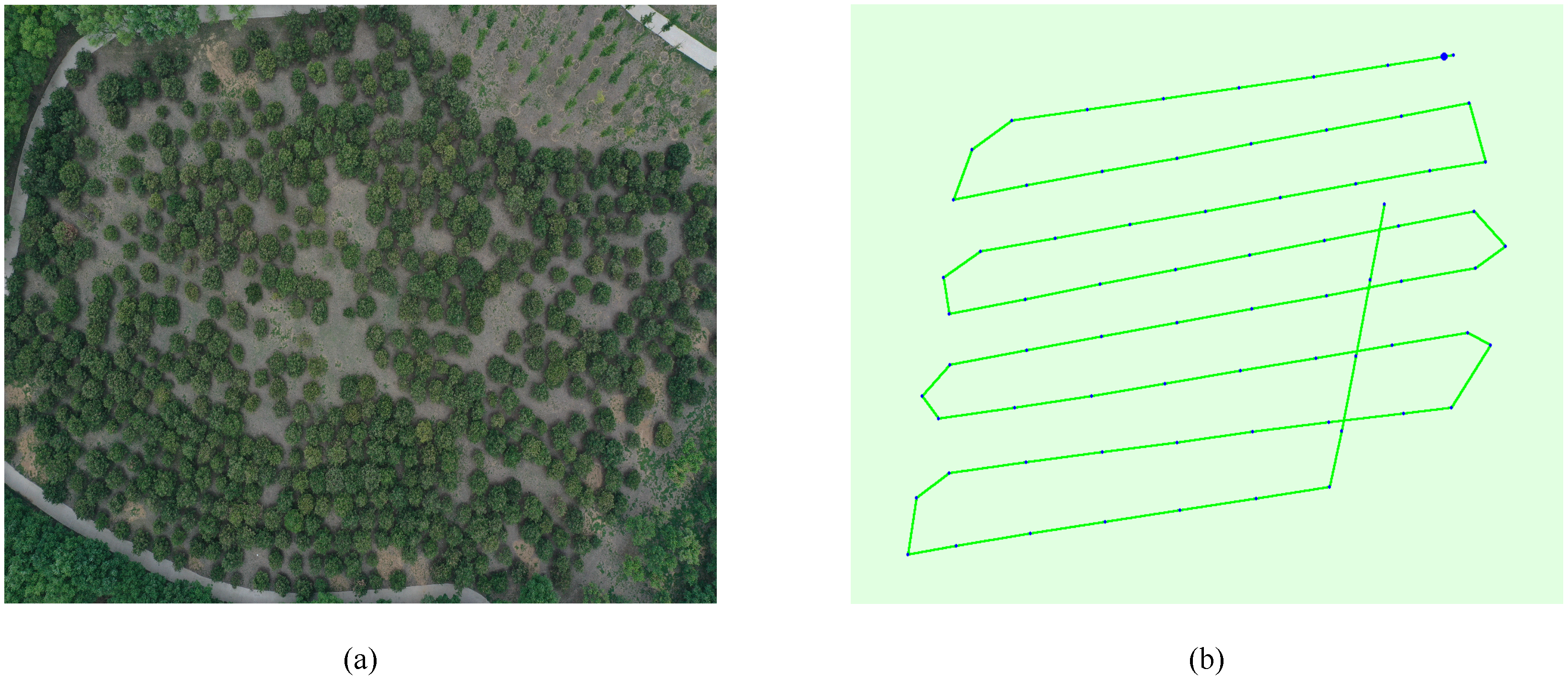
2.1.2. Field Investigation and Data Collection
2.2. Datasets Creation
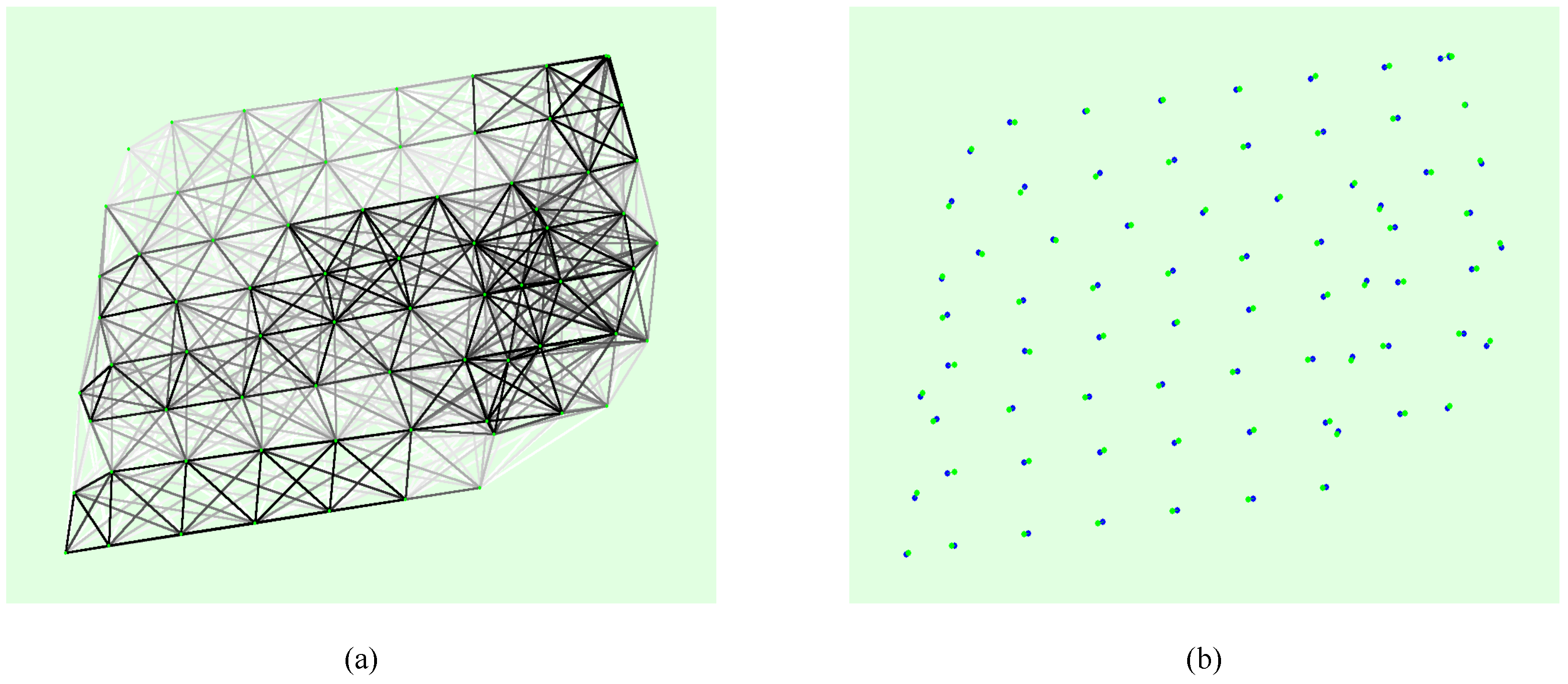
2.2.1. Synthesis of Orthophoto Map
2.2.2. Generating Label Samples
2.3. Evaluation Metrics
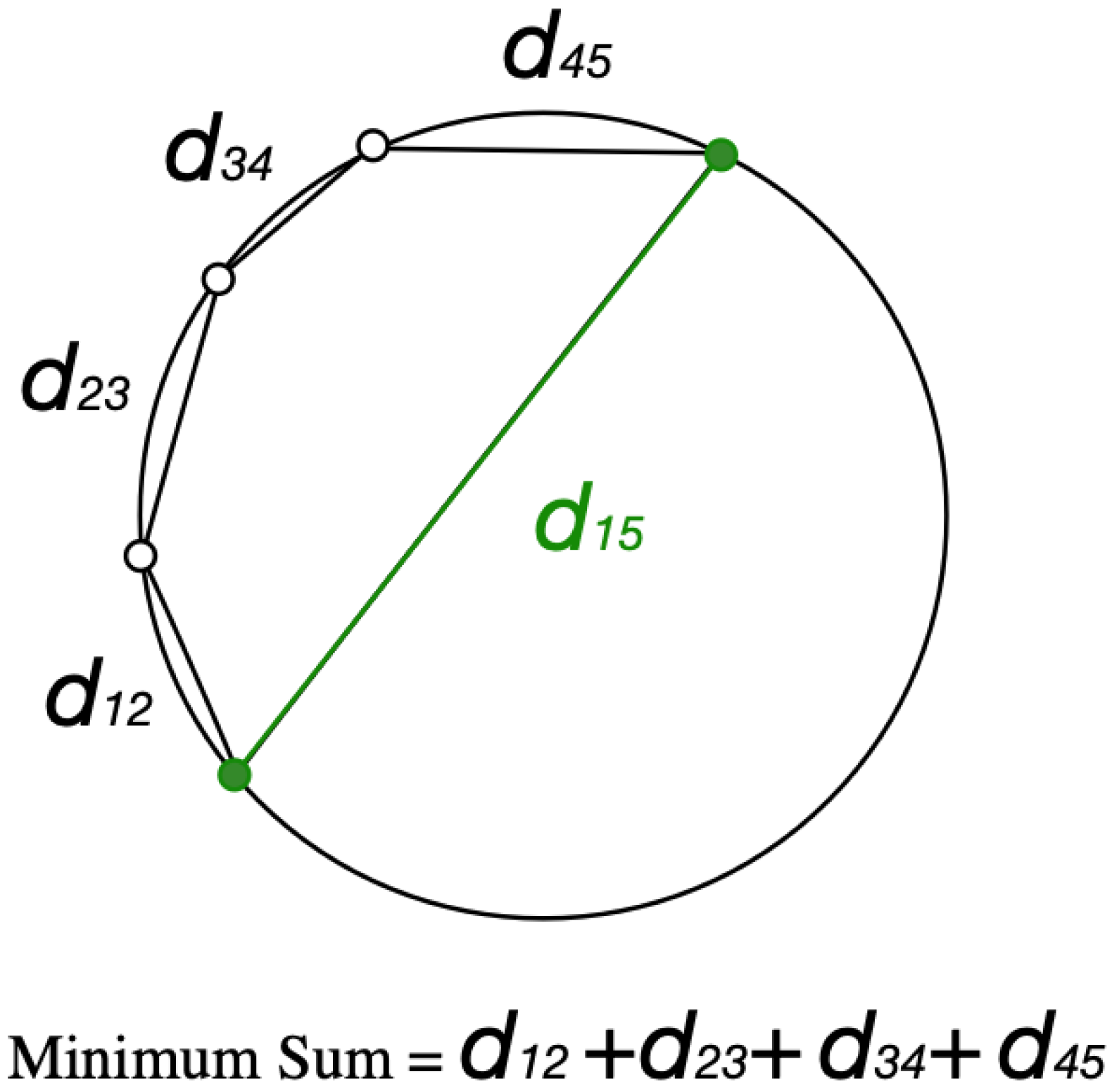
2.3.1. Accuracy Assessment of Individual Tree Detection
2.3.2. Crown Segmentation Accuracy Metrics
2.3.3. Individual Tree Crown Area and DBH Accuracy Metrics
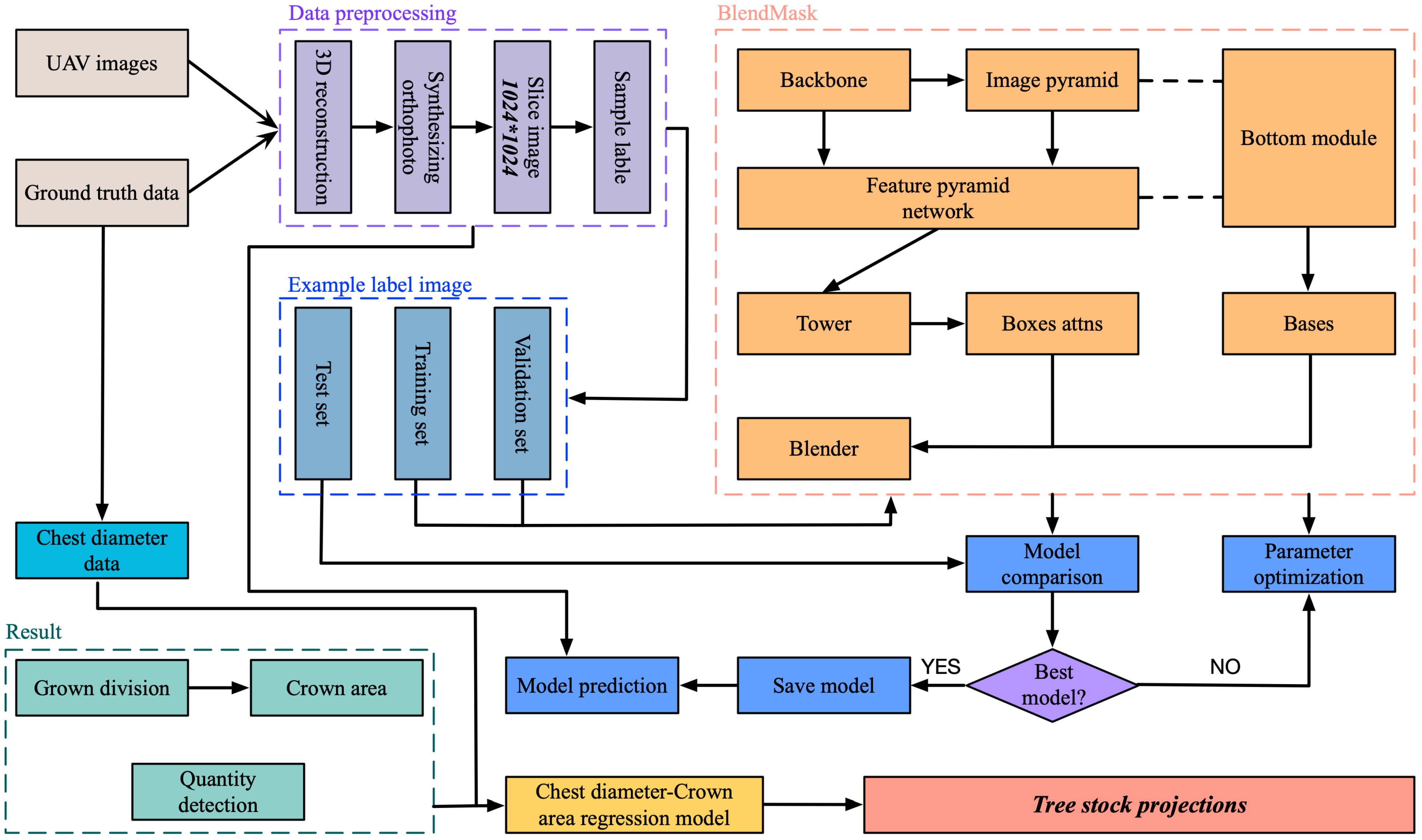
3. Research Methods
3.1. Crown Segmentation Method
3.1.1. Watershed Algorithm
- Convert tree crown images in the dataset to grayscale and classify pixels based on grayscale values, establishing a geodesic distance threshold.
- Identify the pixel with the minimum grayscale value (defaulted as the lowest) and incrementally increase the threshold from the minimum value, designating these as starting points.
- As the plane expands horizontally, it interacts with neighboring pixels, measuring their geodesic distance from the starting point (lowest grayscale). Pixels with distances below the threshold are submerged, while others have dams set, thus categorizing neighboring pixels.
- Use the complementary canopy height model (CHM) distance transform image for segmentation. Utilize the h-minima transform to suppress values smaller than ’H’, generating a marker image for tree tops, followed by reconstruction through erosion.
3.1.2. BlendMask Algorithm
3.2. Calculation of Crown Area
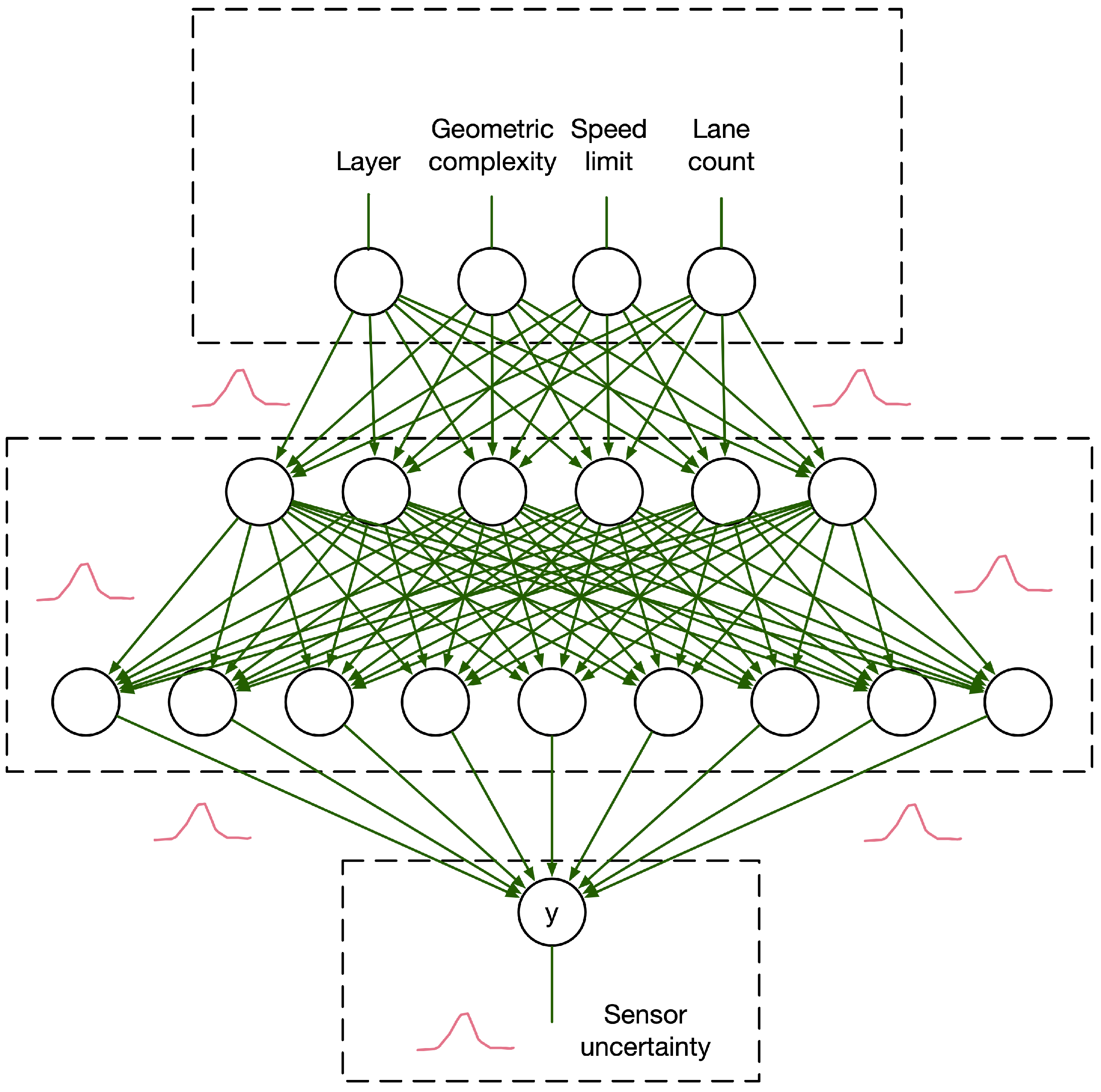
3.3. Crown Area–DBH Model
4. Research Results
4.1. Model Training
4.2. Data Processing and Preprocessing
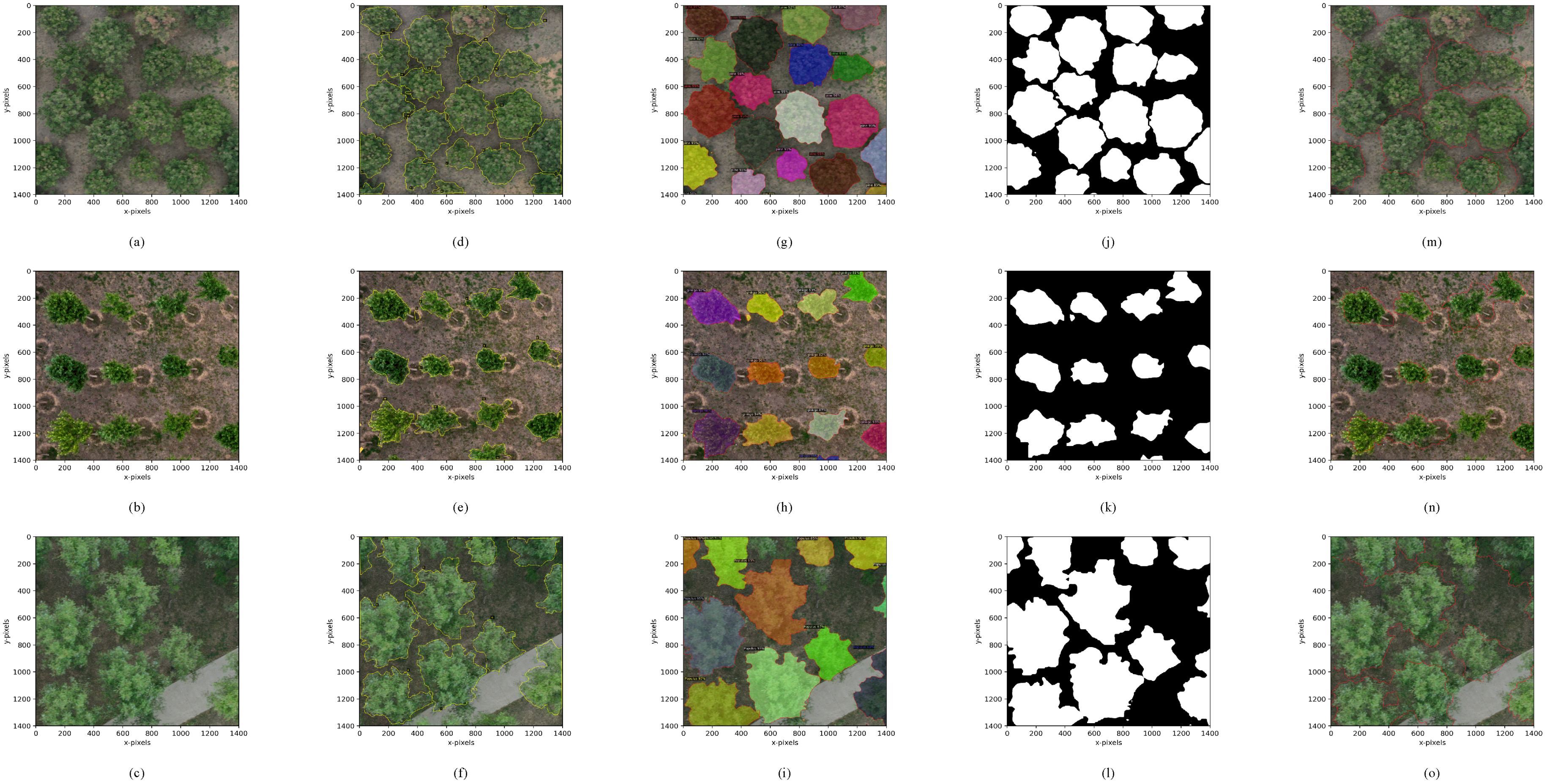
4.2.1. Crown Segmentation of Individual Tree
4.2.2. Testing the Individual Tree Crown Area–DBH Model
4.3. Evaluation of BlendMask’s Performance in Individual Tree Crown Segmentation
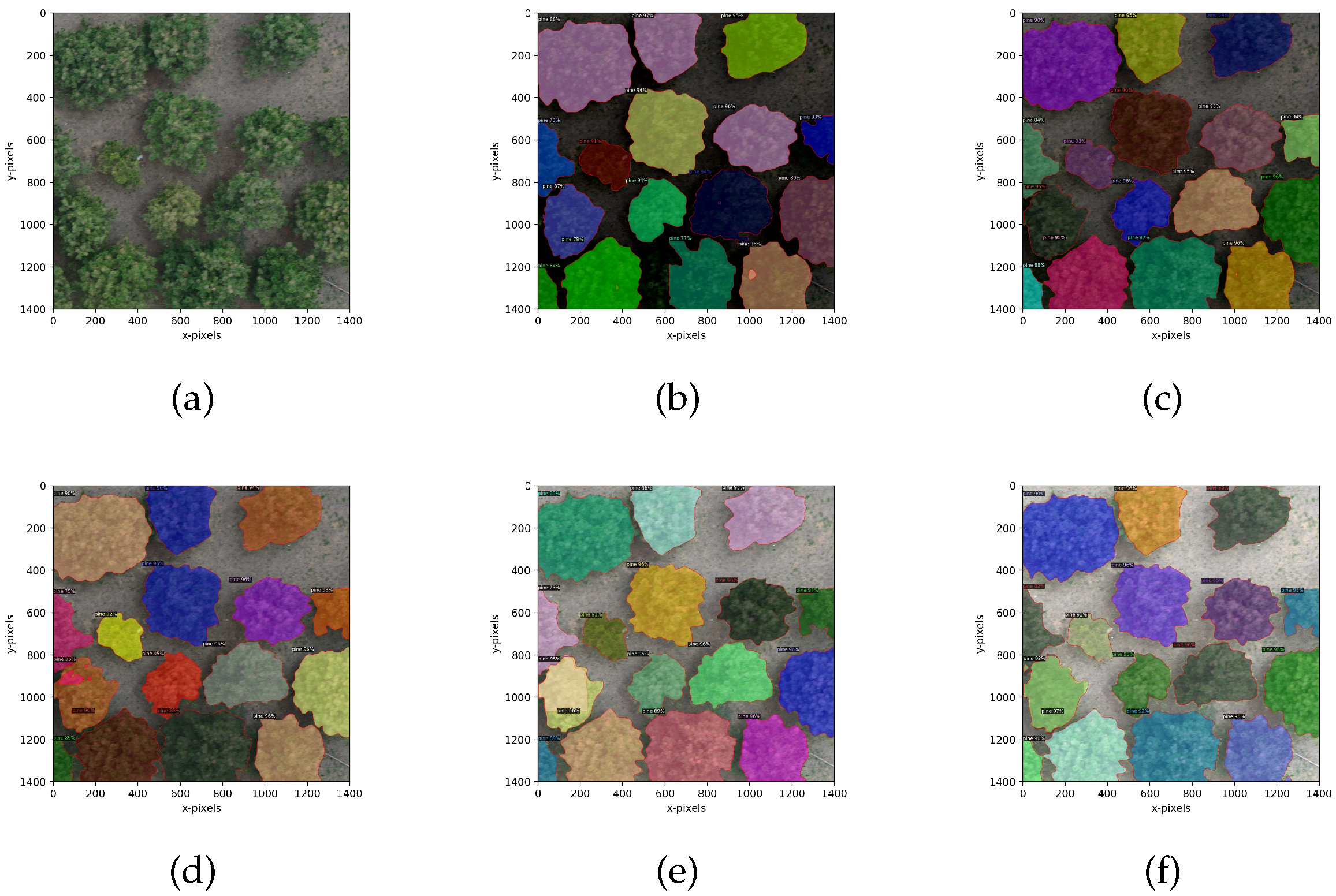
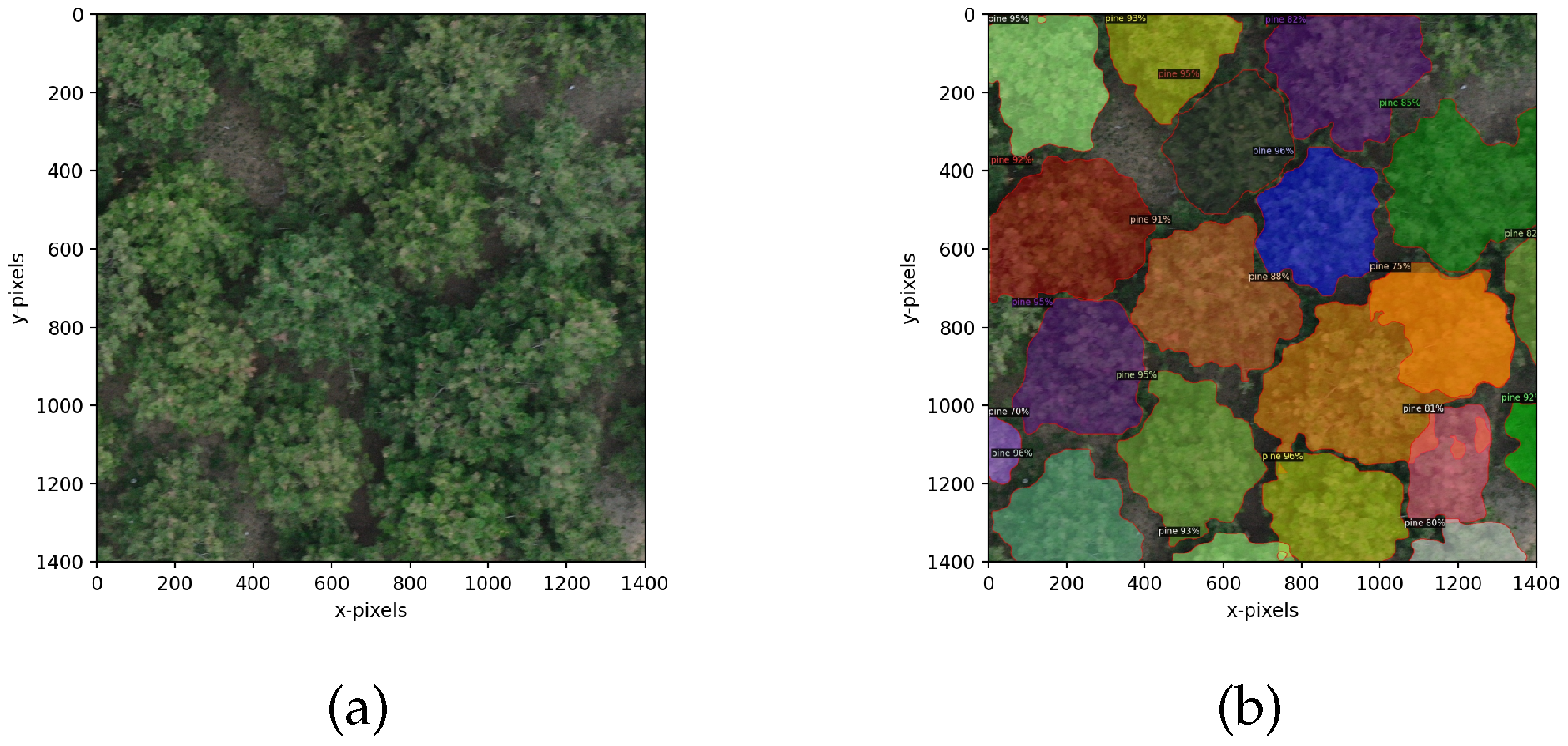
4.3.1. Crown Segmentation Effect
4.3.2. Evaluation of Crown Segmentation Performance
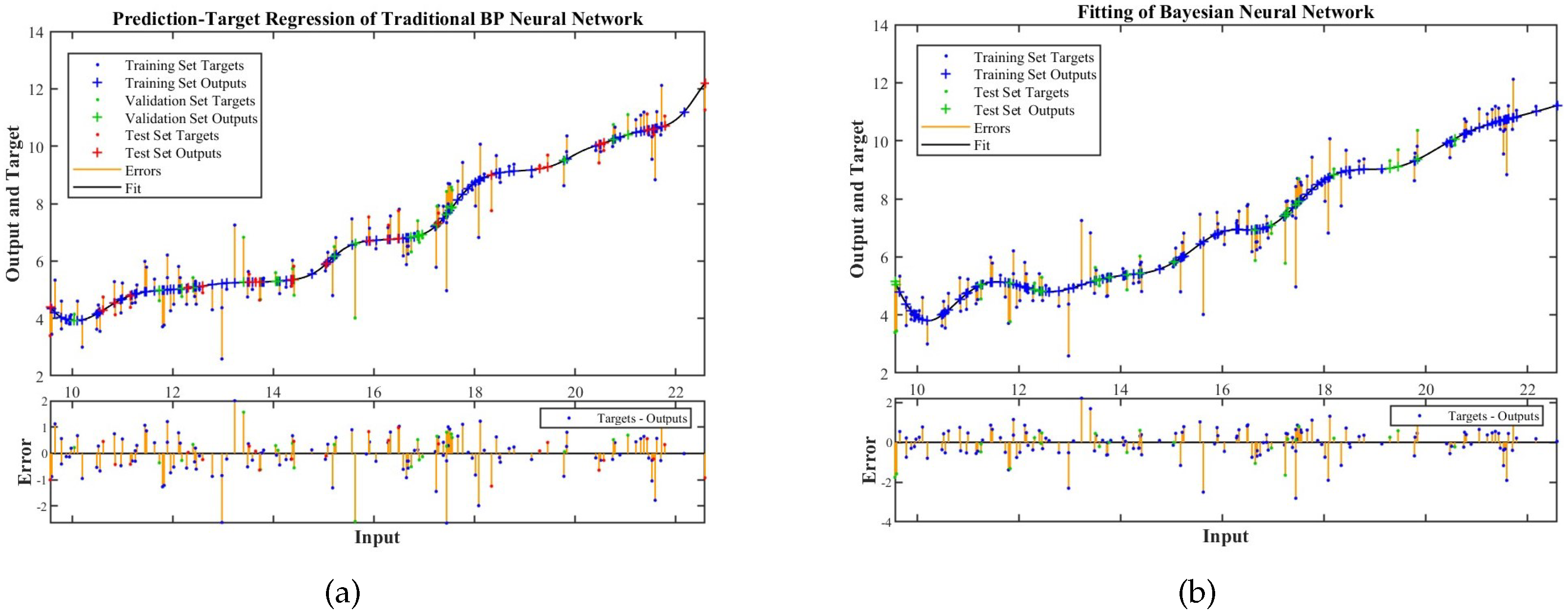
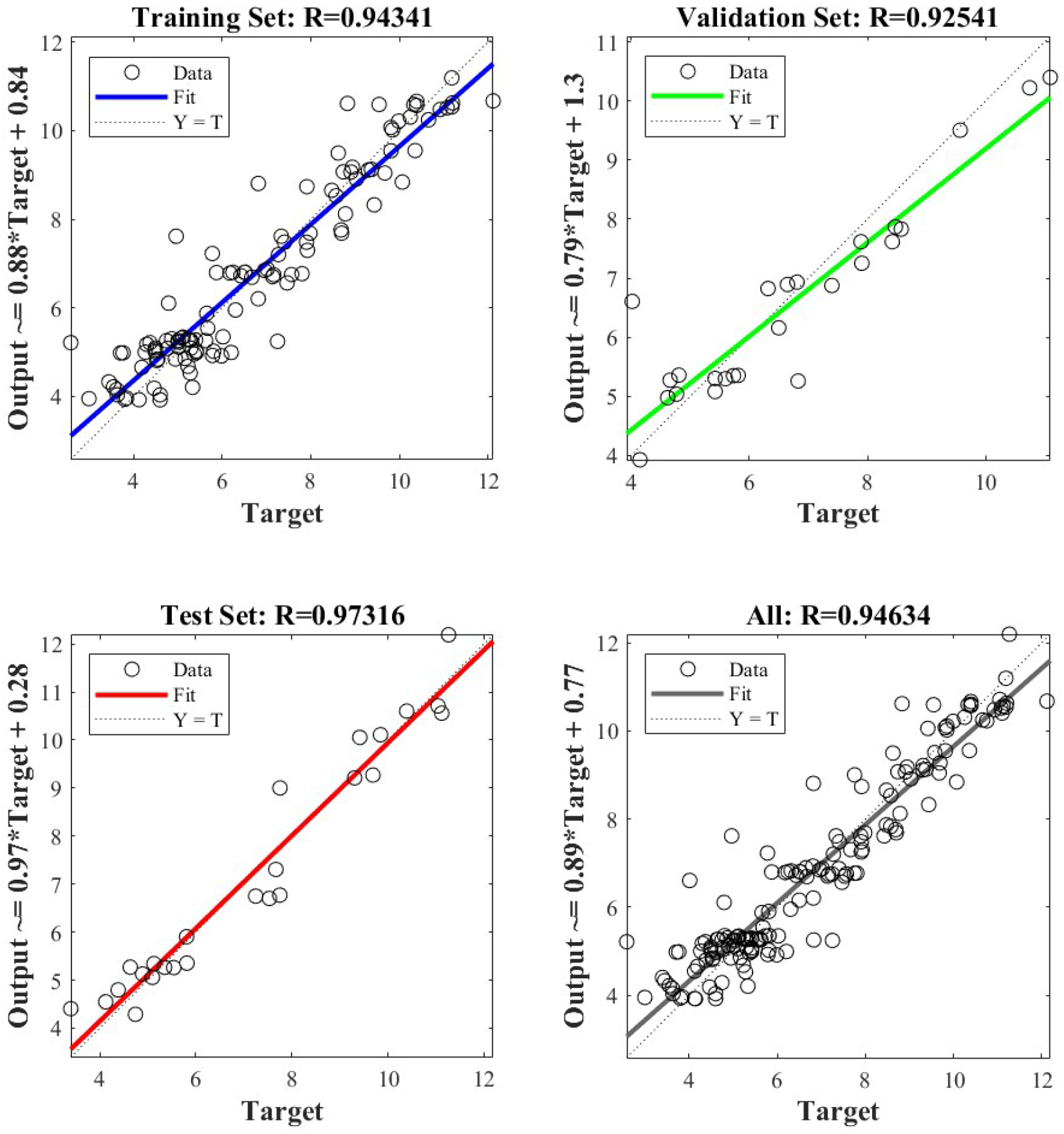
4.4. Performance Evaluation of Crown Area–DBH Model
4.5. Comparative Analysis
4.6. Final Function Validation
5. Discussion
5.1. Comparative Experimentation under Varying Light Intensities
5.2. Analysis of Incorrect Segmentation Cases
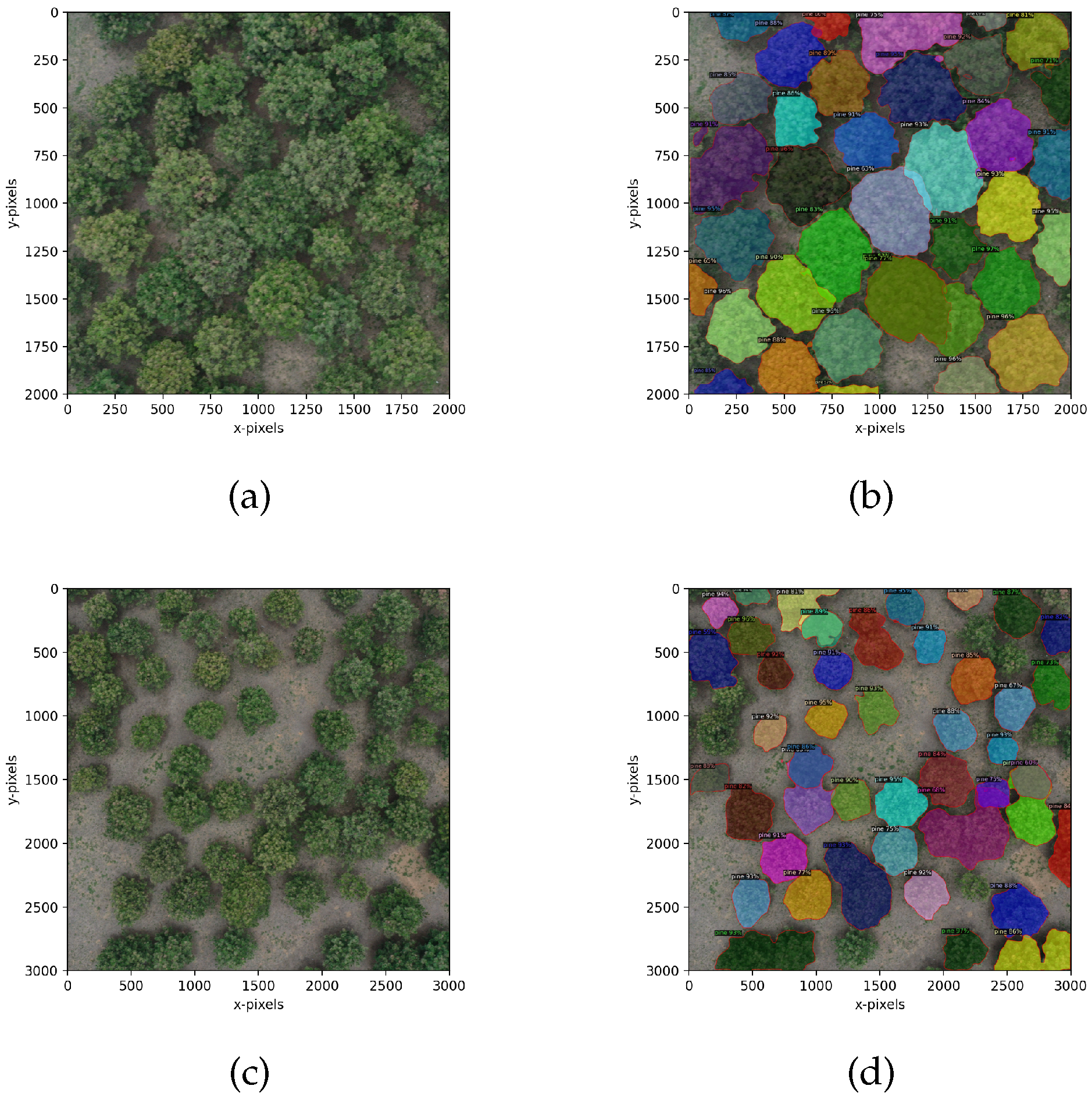
5.3. Image Segmentation Analysis of Different Sizes
6. Conclusions
- BlendMask’s Multi-step Approach: BlendMask utilizes a two-step method for instance segmentation in complex scenes. BlendMask initially extracts the region of interest (ROI) using a pretrained target detector and then performs segmentation of the ROI. Integrating deep learning models, BlendMask delivers more accurate and precise outcomes in handling complex segmentation tasks.
- Robustness to Obstructions and Overlaps: BlendMask effectively handles challenges related to occlusions and overlaps using deep learning models, particularly when distinct objects within a scene overlap or obscure each other. This robustness was beneficial in training Pinus tabulaeformis stand crown information, especially in cases of occluded and intertwined crowns.
- Scalability: BlendMask’s adaptability to large-scale datasets enhances segmentation performance by extracting richer features. It can be applied to various vegetation datasets, aiding in identifying diverse tree crown shapes, sizes, and distributions. This contributes to a comprehensive understanding of forest spatial structures, ecological attributes, and growth patterns.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shiyun, H.G.Z. Evaluation Method of Forest Management Models: A Case Study of Xiaolongshan Forest Area in Gansu Province. Sci. Silvae Sin. 2011, 47, 114. [Google Scholar] [CrossRef]
- Yang, M.; Mou, Y.; Liu, S.; Meng, Y.; Liu, Z.; Li, P.; Xiang, W.; Zhou, X.; Peng, C. Detecting and mapping tree crowns based on convolutional neural network and Google Earth images. Int. J. Appl. Earth Obs. Geoinf. 2022, 108, 102764. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernandez, J.; Corvalan, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B. Selection of Remotely Sensed Data. In Remote Sensing of Forest Environments: Concepts and Case Studies; Wulder, M.A., Franklin, S.E., Eds.; Springer US: Boston, MA, USA, 2003; pp. 13–46. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Yong, X.; Chen, D.; Xia, R.; Ye, B.; Gao, H.; Chen, Z.; Lyu, X. SSCNet: A Spectrum-Space Collaborative Network for Semantic Segmentation of Remote Sensing Images. Remote Sens. 2023, 15, 5610. [Google Scholar] [CrossRef]
- Li, X.; Xu, F.; Lyu, X.; Gao, H.; Tong, Y.; Cai, S.; Li, S.; Liu, D. Dual attention deep fusion semantic segmentation networks of large-scale satellite remote-sensing images. Int. J. Remote Sens. 2021, 42, 3583–3610. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Ferreira, M.P.; de Almeida, D.R.A.; de Almeida Papa, D.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Korpela, I. Individual tree measurements by means of digital aerial photogrammetry. Silva Fennica. Monographs 2004, 3. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhao, K.; Neuenschwander, A.; Lin, C. Satellite lidar vs. small footprint airborne lidar: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ. 2011, 115, 2786–2797. [Google Scholar] [CrossRef]
- Ma, Q.; Su, Y.; Guo, Q. Comparison of Canopy Cover Estimations From Airborne LiDAR, Aerial Imagery, and Satellite Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 4225–4236. [Google Scholar] [CrossRef]
- Dong, T.; Zhang, X.; Ding, Z.; Fan, J. Multi-layered tree crown extraction from LiDAR data using graph-based segmentation. Comput. Electron. Agric. 2020, 170, 105213. [Google Scholar] [CrossRef]
- Riano, D.; Chuvieco, E.; Salas, J.; Aguado, I. Assessment of different topographic corrections in Landsat-TM data for mapping vegetation types (2003). IEEE Trans. Geosci. Remote Sens. 2003, 41, 1056–1061. [Google Scholar] [CrossRef]
- Guo, Y.S.; Liu, Q.S.; Liu, G.H.; Huang, C. Individual Tree Crown Extraction of High Resolution Image Based on Marker-controlled Watershed Segmentation Method. J. Geo-Inf. Sci. 2016, 18, 1259–1266. [Google Scholar]
- Chen, Q.; Baldocchi, D.D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.; Finley, A.O.; Heikkinen, J. Estimating areal means and variances of forest attributes using the k-Nearest Neighbors technique and satellite imagery. Remote Sens. Environ. 2007, 111, 466–480. [Google Scholar] [CrossRef]
- Zhou, S.; Kang, F.; Li, W.; Kan, J.; Zheng, Y.; He, G. Extracting Diameter at Breast Height with a Handheld Mobile LiDAR System in an Outdoor Environment. Sensors 2019, 19, 3212. [Google Scholar] [CrossRef]
- Li, Q.; Lu, W.; Yang, J. A Hybrid Thresholding Algorithm for Cloud Detection on Ground-Based Color Images. J. Atmos. Ocean. Technol. 2011, 28, 1286–1296. [Google Scholar] [CrossRef]
- Chai, G.; Zheng, Y.; Lei, L.; Yao, Z.; Chen, M.; Zhang, X. A novel solution for extracting individual tree crown parameters in high-density plantation considering inter-tree growth competition using terrestrial close-range scanning and photogrammetry technology. Comput. Electron. Agric. 2023, 209, 107849. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Y.F.; Ding, Z.; Liang, R.T.; Xie, Y.; Li, R.; Li, H.; Pan, L.; Sun, Y.J. Individual tree segmentation and biomass estimation based on UAV Digital aerial photograph. J. Mt. Sci. 2023, 20, 724–737. [Google Scholar]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual Tree-Crown Detection in RGB Imagery Using Semi-Supervised Deep Learning Neural Networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Jing, L.; Hu, B.; Noland, T.; Li, J. An individual tree crown delineation method based on multi-scale segmentation of imagery. ISPRS J. Photogramm. Remote Sens. 2012, 70, 88–98. [Google Scholar] [CrossRef]
- Sun, C.; Huang, C.; Zhang, H.; Chen, B.; An, F.; Wang, L.; Yun, T. Individual Tree Crown Segmentation and Crown Width Extraction From a Heightmap Derived From Aerial Laser Scanning Data Using a Deep Learning Framework. Front. Plant Sci. 2022, 13, 914974. [Google Scholar] [CrossRef]
- Huang, Y.X.; Fang, L.M.; Huang, S.Q.; Gao, H.L.; Yang, L.B.; Lou, X.L. Research on Crown Extraction Based on Improved Faster R-CNN Model. For. Resour. Wanagement 2021, 1, 173. [Google Scholar]
- Xu, X.; Zhou, Z.; Tang, Y.; Qu, Y. Individual tree crown detection from high spatial resolution imagery using a revised local maximum filtering. Remote Sens. Environ. 2021, 258, 112397. [Google Scholar] [CrossRef]
- G. Braga, J.R.; Peripato, V.; Dalagnol, R.; P. Ferreira, M.; Tarabalka, Y.; O. C. Aragão, L.E.; F. de Campos Velho, H.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, S.; Xu, A.J.; Chen, B. Passive measurement method of tree diameter at breast height using a smartphone. Comput. Electron. Agric. 2019, 163, 104875. [Google Scholar] [CrossRef]
- Hao, Y.; Widagdo, F.R.A.; Liu, X.; Quan, Y.; Dong, L.; Li, F. Individual Tree Diameter Estimation in Small-Scale Forest Inventory Using UAV Laser Scanning. Remote Sens. 2021, 13, 24. [Google Scholar] [CrossRef]
- de Almeida, C.T.; Galvao, L.S.; Ometto, J.P.H.B.; Jacon, A.D.; de Souza Pereira, F.R.; Sato, L.Y.; Lopes, A.P.; de Alencastro Graça, P.M.L.; de Jesus Silva, C.V.; Ferreira-Ferreira, J.; et al. Combining LiDAR and hyperspectral data for aboveground biomass modeling in the Brazilian Amazon using different regression algorithms. Remote Sens. Environ. 2019, 232, 111323. [Google Scholar] [CrossRef]
- Adhikari, A.; Montes, C.R.; Peduzzi, A. A Comparison of Modeling Methods for Predicting Forest Attributes Using Lidar Metrics. Remote Sens. 2023, 15, 1284. [Google Scholar] [CrossRef]
- Tian, X.; Sun, S.; Mola-Yudego, B.; Cao, T. Predicting individual tree growth using stand-level simulation, diameter distribution, and Bayesian calibration. Ann. For. Sci. 2020, 77, 57. [Google Scholar] [CrossRef]
- Gyawali, A.; Aalto, M.; Peuhkurinen, J.; Villikka, M.; Ranta, T. Comparison of Individual Tree Height Estimated from LiDAR and Digital Aerial Photogrammetry in Young Forests. Sustainability 2022, 14, 3720. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G.E. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C., Bottou, L., Weinberger, K., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Graves, A.; Mohamed, A.R.; Hinton, G. Speech recognition with deep recurrent neural networks. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 6645–6649. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches. arXiv 2014, arXiv:1409.1259. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Graves, A. Supervised Sequence Labelling with Recurrent Neural Networks. Ph.D. Thesis, Technical University of Munich, Munich, Germany, 2012. Volume 385. [Google Scholar] [CrossRef]
- Shi, J.Q.; Feng, Z.K.; Liu, J. Design and experiment of high precision forest resource investigation system based on UAV remote sensing images. Nongye Gongcheng Xuebao/Transactions Chin. Soc. Agric. Eng. 2017, 33, 82–90. [Google Scholar]
- Brede, B.; Terryn, L.; Barbier, N.; Bartholomeus, H.; Bartolo, R.; Calders, K.; Derroire, G.; Moorthy, S.; Lau Sarmiento, A.; Levick, S.; et al. Non-destructive estimation of individual tree biomass: Allometric models, terrestrial and UAV laser scanning. Remote Sens. Environ. 2022, 280, 113180. [Google Scholar] [CrossRef]
- Bucksch, A.; Lindenbergh, R.; Abd Rahman, M.; Menenti, M. Breast Height Diameter Estimation From High-Density Airborne LiDAR Data. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1056–1060. [Google Scholar] [CrossRef]
- Heo, H.K.; Lee, D.K.; Park, J.H.; Thorne, J. Estimating the heights and diameters at breast height of trees in an urban park and along a street using mobile LiDAR. Landsc. Ecol. Eng. 2019, 15, 253–263. [Google Scholar] [CrossRef]
- Zhang, B.; Yuan, J.; Shi, B.; Chen, T.; Li, Y.; Qiao, Y. Uni3D: A Unified Baseline for Multi-dataset 3D Object Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023. [Google Scholar]
- Chen, H.; Sun, K.; Tian, Z.; Shen, C.; Huang, Y.; Yan, Y. BlendMask: Top-Down Meets Bottom-Up for Instance Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Lahivaara, T.; Seppanen, A.; Kaipio, J.P.; Vauhkonen, J.; Korhonen, L.; Tokola, T.; Maltamo, M. Bayesian Approach to Tree Detection Based on Airborne Laser Scanning Data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2690–2699. [Google Scholar] [CrossRef]
- Huang, H.; Li, X.; Chen, C. Individual Tree Crown Detection and Delineation From Very-High-Resolution UAV Images Based on Bias Field and Marker-Controlled Watershed Segmentation Algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2253–2262. [Google Scholar] [CrossRef]


| Step | Type | Specific Settings |
|---|---|---|
| Image scale | 1/2 | |
| Densification of point cloud | Point cloud density | best |
| Minimum matching number | 3 | |
| Matching window size | 7 × 7 pixels | |
| Configuration | Medium resolution | |
| Three-dimensional grid | Sampling density distribution | 1 |
| Texture color source | Visible color | |
| Texture mapping | Texture compression quality | 75% JPEG image quality |
| Maximum texture size | 8192 | |
| Texture sharpening | Enabled |
| Hardware | Attribute |
|---|---|
| CPU | E5-12640 |
| GPU | RTX 6000 24GB |
| SSD | 1T SSD |
| Memory | 32GB |
| Total Sample Number | Mean DBH (cm) | Maximum Breast Diameter (cm) | Minimum DBH (cm) | Average Crown Area (m2) | Maximum Crown Area (m2) | Minimum Crown Area (m2) |
|---|---|---|---|---|---|---|
| 164 | 15.5892 | 22.5781 | 9.5670 | 6.8217 | 12.1171 | 2.5853 |
| / | h = 0.5 | h = 1.0 | h = 1.5 | h = 2.0 |
|---|---|---|---|---|
| b=3 | 0.696 | 0.715 | 0.721 | 0.707 |
| b=5 | 0.675 | 0.689 | 0.695 | 0.686 |
| b=7 | 0.661 | 0.647 | 0.653 | 0.656 |
| b=9 | 0.624 | 0.612 | 0.625 | 0.611 |
| Model Weight | Average Precision | Mean Average Precision IOU = 0.5 | Mean Average Precision IOU = 0.75 |
|---|---|---|---|
| BlendMask | 0.724 | 0.893 | 0.745 |
| Watershed algorithm | 0.685 | 0.763 | 0.674 |
| / | Relative Error RE | Average Absolute Error MAE | Root Mean Square Error RMSE |
|---|---|---|---|
| Crown area | 0.05653 | 0.3290 | 0.4563 |
| DBH | 0.03308 | 92.18 | 106.4 |
| Target Training Times | Learning Rate | Minimum Error of Training Target | Additional Momentum Factor | Minimum Performance Gradient |
|---|---|---|---|---|
| 10000 | 0.001 | 0.000001 | 0.95 | 0.00001 |
| Model | Training Set/ | Test Set/ | All/ | RMSE |
|---|---|---|---|---|
| Traditional BP neural network | 0.96523 | 0.90999 | 0.9456 | 0.74516 |
| Bayesian neural network | 0.9488 | 0.95628 | 0.94775 | 0.72602 |
| Model | Training Set | Test Set | All |
|---|---|---|---|
| BlendMask + Bayesian neural network | 0.7855 | 0.7926 | 0.7862 |
| BlendMask + raditional BP neural network | 0.69882 | 0.65883 | 0.6846 |
| BlendMask + raditional BP neural network | 0.69882 | 0.65883 | 0.6846 |
| Watershed + traditional BP neural network | 0.6499 | 0.6551 | 0.6492 |
| Sample Area | The Tree Number | MDTBH/cm | CDTBH/cm | DTBH Error/cm |
|---|---|---|---|---|
| No.8 | 9.56 | 9.43 | 0.13 | |
| No.15 | 9.98 | 9.94 | 0.04 | |
| Pinus tabulaeformis | No.29 | 10.1 | 10.20 | 0.10 |
| No.41 | 10.84 | 10.72 | 0.12 | |
| No.53 | 11.79 | 11.92 | 0.13 | |
| No.13 | 17.28 | 17.41 | 0.13 | |
| No.18 | 18.53 | 18.67 | 0.14 | |
| Ginkgo biloba | No.25 | 18.81 | 18.69 | 0.12 |
| No.31 | 19.32 | 19.23 | 0.09 | |
| No.44 | 19.45 | 19.25 | 0.20 | |
| No.4 | 23.32 | 23.42 | 0.10 | |
| No.11 | 23.81 | 23.78 | 0.03 | |
| Populus nigra varitalica | No.19 | 24.54 | 24.85 | 0.31 |
| No.23 | 24.98 | 24.72 | 0.26 | |
| No.29 | 25.10 | 25.01 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Su, M.; Sun, Y.; Pan, W.; Cui, H.; Jin, S.; Zhang, L.; Wang, P. Tree Crown Segmentation and Diameter at Breast Height Prediction Based on BlendMask in Unmanned Aerial Vehicle Imagery. Remote Sens. 2024, 16, 368. https://doi.org/10.3390/rs16020368
Xu J, Su M, Sun Y, Pan W, Cui H, Jin S, Zhang L, Wang P. Tree Crown Segmentation and Diameter at Breast Height Prediction Based on BlendMask in Unmanned Aerial Vehicle Imagery. Remote Sensing. 2024; 16(2):368. https://doi.org/10.3390/rs16020368
Chicago/Turabian StyleXu, Jie, Minbin Su, Yuxuan Sun, Wenbin Pan, Hongchuan Cui, Shuo Jin, Li Zhang, and Pei Wang. 2024. "Tree Crown Segmentation and Diameter at Breast Height Prediction Based on BlendMask in Unmanned Aerial Vehicle Imagery" Remote Sensing 16, no. 2: 368. https://doi.org/10.3390/rs16020368
APA StyleXu, J., Su, M., Sun, Y., Pan, W., Cui, H., Jin, S., Zhang, L., & Wang, P. (2024). Tree Crown Segmentation and Diameter at Breast Height Prediction Based on BlendMask in Unmanned Aerial Vehicle Imagery. Remote Sensing, 16(2), 368. https://doi.org/10.3390/rs16020368









