High-Throughput Plot-Level Quantitative Phenotyping Using Convolutional Neural Networks on Very High-Resolution Satellite Images
Abstract
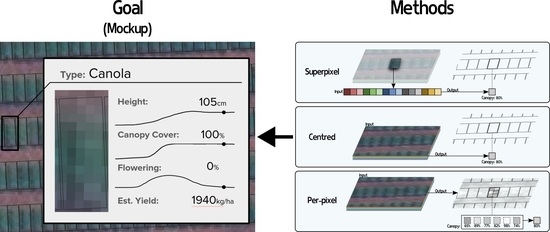
1. Introduction
2. Materials and Methods
2.1. Data
2.1.1. Image Sources
2.1.2. Roseworthy
2.1.3. Obregón
2.1.4. Ground Data Interpolation
2.1.5. Image Preprocessing
2.2. Methods for Per-Plot Prediction
2.2.1. Superpixel
2.2.2. Centred
2.2.3. Per-Pixel
2.3. Training Details
3. Results
3.1. High-Variance Per-Image Evaluation
3.2. High-Variance Image Training
4. Discussion
Limit of Resolvability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Garnett, T.; Appleby, M.C.; Balmford, A.; Bateman, I.J.; Benton, T.G.; Bloomer, P.; Burlingame, B.; Dawkins, M.; Dolan, L.; Fraser, D.; et al. Sustainable Intensification in Agriculture: Premises and Policies. Science 2013, 341, 33–34. [Google Scholar] [CrossRef]
- Campos, H.; Cooper, M.; Habben, J.E.; Edmeades, G.O.; Schussler, J.R. Improving Drought Tolerance in Maize: A View from Industry. Field Crop. Res. 2004, 90, 19–34. [Google Scholar] [CrossRef]
- White, J.W.; Andrade-Sanchez, P.; Gore, M.A.; Bronson, K.F.; Coffelt, T.A.; Conley, M.M.; Feldmann, K.A.; French, A.N.; Heun, J.T.; Hunsaker, D.J.; et al. Field-Based Phenomics for Plant Genetics Research. Field Crop. Res. 2012, 133, 101–112. [Google Scholar] [CrossRef]
- Chapman, S.C.; Merz, T.; Chan, A.; Jackway, P.; Hrabar, S.; Dreccer, M.F.; Holland, E.; Zheng, B.; Ling, T.J.; Jimenez-Berni, J. Pheno-Copter: A Low-Altitude, Autonomous Remote-Sensing Robotic Helicopter for High-Throughput Field-Based Phenotyping. Agronomy 2014, 4, 279–301. [Google Scholar] [CrossRef]
- Sankaran, S.; Marzougui, A.; Hurst, J.P.; Zhang, C.; Schnable, J.C.; Shi, Y. Can High-Resolution Satellite Multispectral Imagery Be Used to Phenotype Canopy Traits and Yield Potential in Field Conditions? Trans. ASABE 2021, 64, 879–891. [Google Scholar] [CrossRef]
- Hedden, P. The Genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Awulachew, M.T. Understanding Basics of Wheat Grain and Flour Quality. J. Health Environ. Res. 2020, 6, 10. [Google Scholar] [CrossRef]
- Fischer, R.; Byerlee, D.; Edmeades, G. Crop Yields and Global Food Security: Will Yield Increase Continue to Feed the World? Number 158 in ACIAR Monograph; Australian Centre for International Agricultural Research: Canberra, Australia, 2014. [Google Scholar]
- Furbank, R.T.; Sirault, X.R.; Stone, E.; Zeigler, R. Plant Phenome to Genome: A Big Data Challenge. In Sustaining Global Food Security: The Nexus of Science and Policy; CSIRO Publishing: Clayton, Australia, 2019; p. 203. [Google Scholar]
- MacDonald, R.B.; Hall, F.G. Global Crop Forecasting. Science 1980, 208, 670–679. [Google Scholar] [CrossRef]
- Zhou, L.; Tucker, C.J.; Kaufmann, R.K.; Slayback, D.; Shabanov, N.V.; Myneni, R.B. Variations in Northern Vegetation Activity Inferred from Satellite Data of Vegetation Index during 1981 to 1999. J. Geophys. Res. Atmos. 2001, 106, 20069–20083. [Google Scholar] [CrossRef]
- White, M.A.; De BEURS, K.M.; Didan, K.; Inouye, D.W.; Richardson, A.D.; Jensen, O.P.; O’keefe, J.; Zhang, G.; Nemani, R.R.; Van Leeuwen, W.J.D.; et al. Intercomparison, Interpretation, and Assessment of Spring Phenology in North America Estimated from Remote Sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Kang, Y.; Ozdogan, M.; Zhu, X.; Ye, Z.; Hain, C.; Anderson, M. Comparative Assessment of Environmental Variables and Machine Learning Algorithms for Maize Yield Prediction in the US Midwest. Environ. Res. Lett. 2020, 15, 64005. [Google Scholar] [CrossRef]
- Potopova, V.; Trnka, M.; Hamouz, P.; Soukup, J.; Castravet, T. Statistical Modelling of Drought-Related Yield Losses Using Soil Moisture-Vegetation Remote Sensing and Multiscalar Indices in the South-Eastern Europe. Agric. Water Manag. 2020, 236, 106168. [Google Scholar] [CrossRef]
- Eroglu, O.; Kurum, M.; Boyd, D.; Gurbuz, A.C. High Spatio-Temporal Resolution CYGNSS Soil Moisture Estimates Using Artificial Neural Networks. Remote Sens. 2019, 11, 2272. [Google Scholar] [CrossRef]
- Senanayake, I.P.; Yeo, I.Y.; Walker, J.P.; Willgoose, G.R. Estimating Catchment Scale Soil Moisture at a High Spatial Resolution: Integrating Remote Sensing and Machine Learning. Sci. Total Environ. 2021, 776. [Google Scholar] [CrossRef]
- Sakamoto, T.; Gitelson, A.A.; Arkebauer, T.J. MODIS-based Corn Grain Yield Estimation Model Incorporating Crop Phenology Information. Remote Sens. Environ. 2013, 131, 215–231. [Google Scholar] [CrossRef]
- Waldner, F.; Diakogiannis, F.I.; Batchelor, K.; Ciccotosto-Camp, M.; Cooper-Williams, E.; Herrmann, C.; Mata, G.; Toovey, A. Detect, Consolidate, Delineate: Scalable Mapping of Field Boundaries Using Satellite Images. Remote Sens. 2021, 13, 2197. [Google Scholar] [CrossRef]
- Rahman, M.M.; Robson, A.; Bristow, M. Exploring the Potential of High Resolution WorldView-3 Imagery for Estimating Yield of Mango. Remote Sens. 2018, 10, 1866. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Lotte, R.G.; D’Elia, V.F.; Stamatopoulos, C.; Kim, D.H.; Benjamin, A.R. Accurate Mapping of Brazil Nut Trees (Bertholletia Excelsa) in Amazonian Forests Using WorldView-3 Satellite Images and Convolutional Neural Networks. Ecol. Inform. 2021, 63, 101302. [Google Scholar] [CrossRef]
- Ahlswede, S.; Asam, S.; Roeder, A. Hedgerow Object Detection in Very High-Resolution Satellite Images Using Convolutional Neural Networks. J. Remote Sens. 2021, 15. [Google Scholar] [CrossRef]
- Saralioglu, E.; Gungor, O. Semantic Segmentation of Land Cover from High Resolution Multispectral Satellite Images by Spectral-Spatial Convolutional Neural Network. Geocarto Int. 2022, 37, 657–677. [Google Scholar] [CrossRef]
- Mei, W.; Wang, H.; Fouhey, D.; Zhou, W.; Hinks, I.; Gray, J.M.; Van Berkel, D.; Jain, M. Using Deep Learning and Very-High-Resolution Imagery to Map Smallholder Field Boundaries. Remote Sens. 2022, 14, 3046. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25, pp. 1106–1114. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:cs/1512.03385. [Google Scholar] [CrossRef]
- Sharma, P.; Berwal, Y.P.S.; Ghai, W. Performance Analysis of Deep Learning CNN Models for Disease Detection in Plants Using Image Segmentation. Inf. Process. Agric. 2020, 7, 566–574. [Google Scholar] [CrossRef]
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine Learning in Agriculture: A Comprehensive Updated Review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef]
- Darwin, B.; Dharmaraj, P.; Prince, S.; Popescu, D.E.; Hemanth, D.J. Recognition of Bloom/Yield in Crop Images Using Deep Learning Models for Smart Agriculture: A Review. Agronomy 2021, 11, 646. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018. Lecture Notes in Computer Science. pp. 3–11. [Google Scholar] [CrossRef]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep Learning for Cellular Image Analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Thagaard, J.; Stovgaard, E.S.; Vognsen, L.G.; Hauberg, S.; Dahl, A.; Ebstrup, T.; Doré, J.; Vincentz, R.E.; Jepsen, R.K.; Roslind, A.; et al. Automated Quantification of sTIL Density with H&E-Based Digital Image Analysis Has Prognostic Potential in Triple-Negative Breast Cancers. Cancers 2021, 13, 3050. [Google Scholar] [CrossRef]
- Wojke, N.; Bewley, A.; Paulus, D. Simple Online and Realtime Tracking with a Deep Association Metric. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3645–3649. [Google Scholar] [CrossRef]
- Bergmann, P.; Meinhardt, T.; Leal-Taixe, L. Tracking Without Bells and Whistles. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 941–951. [Google Scholar]
- Hall, A.; Victor, B.; He, Z.; Langer, M.; Elipot, M.; Nibali, A.; Morgan, S. The Detection, Tracking, and Temporal Action Localisation of Swimmers for Automated Analysis. Neural Comput. Appl. 2021, 33, 7205–7223. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Rusu, A.A.; Veness, J.; Bellemare, M.G.; Graves, A.; Riedmiller, M.; Fidjeland, A.K.; Ostrovski, G.; et al. Human-Level Control through Deep Reinforcement Learning. Nature 2015, 518, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; van den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the Game of Go with Deep Neural Networks and Tree Search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D Convolutional Neural Networks for Crop Classification with Multi-Temporal Remote Sensing Images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef]
- Debella-Gilo, M.; Gjertsen, A.K. Mapping Seasonal Agricultural Land Use Types Using Deep Learning on Sentinel-2 Image Time Series. Remote Sens. 2021, 13, 289. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Paul, S.; Kumari, M.; Murthy, C.S.; Kumar, D.N. Generating Pre-Harvest Crop Maps by Applying Convolutional Neural Network on Multi-Temporal Sentinel-1 Data. Int. J. Remote Sens. 2022, 42, 6078–6101. [Google Scholar] [CrossRef]
- Chelali, M.; Kurtz, C.; Puissant, A.; Vincent, N. Deep-STaR: Classification of Image Time Series Based on Spatio-Temporal Representations. Comput. Vis. Image Underst. 2021, 208, 103221. [Google Scholar] [CrossRef]
- Roy, D.P.; Wulder, M.A.; Loveland, T.R.; Woodstock, C.E.; Allen, R.G.; Anderson, M.C.; Helder, D.; Irons, J.R.; Johnson, D.M.; Kennedy, R.; et al. Landsat-8: Science and Product Vision for Terrestrial Global Change Research. Remote Sens. Environ. 2014, 145, 154–172. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. J. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Choung, Y.J.; Jung, D. Comparison of Machine and Deep Learning Methods for Mapping Sea Farms Using High-Resolution Satellite Image. J. Coast. Res. 2021, 114, 420–423. [Google Scholar] [CrossRef]
- Schwalbert, R.A.; Amado, T.; Corassa, G.; Pott, L.P.; Prasad, P.V.V.; Ciampitti, I.A. Satellite-Based Soybean Yield Forecast: Integrating Machine Learning and Weather Data for Improving Crop Yield Prediction in Southern Brazil. Agric. For. Meteorol. 2020, 284, 107886. [Google Scholar] [CrossRef]
- Wolanin, A.; Mateo-Garcia, G.; Camps-Valls, G.; Gomez-Chova, L.; Meroni, M.; Duveiller, G.; Liangzhi, Y.; Guanter, L. Estimating and Understanding Crop Yields with Explainable Deep Learning in the Indian Wheat Belt. Environ. Res. Lett. 2020, 15, ab68ac. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Luo, Y.; Cao, J.; Tao, F. Combining Optical, Fluorescence, Thermal Satellite, and Environmental Data to Predict County-Level Maize Yield in China Using Machine Learning Approaches. Remote Sens. 2020, 12, 21. [Google Scholar] [CrossRef]
- Li, A.; Liang, S.; Wang, A.; Qin, J. Estimating Crop Yield from Multi-Temporal Satellite Data Using Multivariate Regression and Neural Network Techniques. Photogramm. Eng. Remote Sens. 2007, 73, 1149–1157. [Google Scholar] [CrossRef]
- Derksen, D.; Inglada, J.; Michel, J. Geometry Aware Evaluation of Handcrafted Superpixel-Based Features and Convolutional Neural Networks for Land Cover Mapping Using Satellite Imagery. Remote Sens. 2020, 12, 513. [Google Scholar] [CrossRef]
- Blaschke, T. Object Based Image Analysis for Remote Sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Haghverdi, A.; Washington-Allen, R.A.; Leib, B.G. Prediction of Cotton Lint Yield from Phenology of Crop Indices Using Artificial Neural Networks. Comput. Electron. Agric. 2018, 152, 186–197. [Google Scholar] [CrossRef]
- Jeong, S.; Ko, J.; Yeom, J.M. Predicting Rice Yield at Pixel Scale through Synthetic Use of Crop and Deep Learning Models with Satellite Data in South and North Korea. Sci. Total Environ. 2022, 802, 149726. [Google Scholar] [CrossRef]
- Gonzalo-Martin, C.; Garcia-Pedrero, A.; Lillo-Saavedra, M.; Menasalvas, E. Deep Learning for Superpixel-Based Classification of Remote Sensing Images. In Proceedings of the GEOBIA 2016: Solutions and Synergies, Enschede, Netherlands, 14–16 September 2016. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitijiang, M.; Bhadra, S.; Maimaitiyiming, M.; Brown, D.R.; Sidike, P.; Fritschi, F.B. Field-Scale Crop Yield Prediction Using Multi-Temporal WorldView-3 and PlanetScope Satellite Data and Deep Learning. ISPRS J. Photogramm. Remote Sens. 2021, 174, 265–281. [Google Scholar] [CrossRef]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A Direct Comparison of Remote Sensing Approaches for High-Throughput Phenotyping in Plant Breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef] [PubMed]
- Pask, A.; Pietragalla, J.; Mullan, D.; Reynolds, M.P. Physiological Breeding II: A Field Guide to Wheat Phenotyping; CIMMYT: Veracruz, Mexico, 2012. [Google Scholar]
- Kuester, M.A. Absolute Radiometric Calibration 2016, DigitalGlobe 2017. Available online: https://dg-cms-uploads-production.s3.amazonaws.com/uploads/document/file/209/ABSRADCAL_FLEET_2016v0_Rel20170606.pdf (accessed on 8 May 2023).
- KOMPSAT-3A Satellite Sensor Satellite Imaging Corp. Available online: https://www.satimagingcorp.com/satellite-sensors/kompsat-3a/ (accessed on 8 May 2023).
- Thuillier, G.; Hersé, M.; Labs, D.; Foujols, T.; Peetermans, W.; Gillotay, D.; Simon, P.; Mandel, H. The Solar Spectral Irradiance from 200 to 2400 Nm as Measured by the SOLSPEC Spectrometer from the Atlas and Eureca Missions. Sol. Phys. 2003, 214, 1–22. [Google Scholar] [CrossRef]
- GDAL/OGR Contributors. GDAL/OGR Geospatial Data Abstraction Software Library; Open Source Geospatial Foundation: Beaverton, ON, USA, 2023. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Representations by Back-Propagating Errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Phan, H. Huyvnphan/PyTorch_CIFAR10. Zenodo; CERN: Meyrin, Switzerland, 2021. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:cs/1706.05587. [Google Scholar] [CrossRef]
- Hussein, M.S.; Hanafy, M.E.; Mahmoud, T.A. Characterization of the Sources of Degradation in Remote Sensing Satellite Images. In Proceedings of the 2019 International Conference on Innovative Trends in Computer Engineering (ITCE), Aswan, Egypt, 2–4 February 2019; pp. 29–34. [Google Scholar] [CrossRef]
- Camacho, F.; Fuster, B.; Li, W.; Weiss, M.; Ganguly, S.; Lacaze, R.; Baret, F. Crop Specific Algorithms Trained over Ground Measurements Provide the Best Performance for GAI and fAPAR Estimates from Landsat-8 Observations. Remote Sens. Environ. 2021, 260, 112453. [Google Scholar] [CrossRef]








| Method | Model | Flowering | Canopy Cover | Green | Height | Average |
|---|---|---|---|---|---|---|
| Superpixel | RF | 0.825 ± 0.008 | 0.991 ± 0.003 | 0.982 ± 0.004 | 0.963 ± 0.005 | 0.940 ± 0.005 |
| MLP | 0.858 ± 0.006 | 0.994 ± 0.001 | 0.985 ± 0.001 | 0.969 ± 0.001 | 0.952 ± 0.002 | |
| Centred | VGG-A | 0.880 ± 0.009 | 0.989 ± 0.002 | 0.985 ± 0.001 | 0.973 ± 0.003 | 0.957 ± 0.004 |
| ResNet18 | 0.888 ± 0.015 | 0.993 ± 0.000 | 0.986 ± 0.001 | 0.975 ± 0.001 | 0.960 ± 0.004 | |
| ResNet50 | 0.886 ± 0.010 | 0.989 ± 0.002 | 0.983 ± 0.003 | 0.969 ± 0.003 | 0.957 ± 0.004 | |
| DenseNet161 | 0.863 ± 0.017 | 0.991 ± 0.003 | 0.983 ± 0.002 | 0.970 ± 0.004 | 0.952 ± 0.007 | |
| Per-pixel | UNet++ | 0.871 ± 0.029 | 0.994 ± 0.001 | 0.986 ± 0.002 | 0.974 ± 0.002 | 0.956 ± 0.008 |
| DeepLabv3 | 0.824 ± 0.008 | 0.994 ± 0.001 | 0.983 ± 0.002 | 0.966 ± 0.002 | 0.941 ± 0.003 | |
| Hypothetical use avg per img | 0.782 ± 0.010 | 0.991 ± 0.002 | 0.978 ± 0.003 | 0.952 ± 0.003 | 0.926 ± 0.005 | |
| Method | Model | Biomass | NDVI | Average |
|---|---|---|---|---|
| Superpixel | RF | 0.834 ± 0.130 | 0.899 ± 0.031 | 0.867 ± 0.080 |
| MLP | 0.823 ± 0.140 | 0.948 ± 0.010 | 0.885 ± 0.075 | |
| Centred | VGG-A | 0.863 ± 0.121 | 0.884 ± 0.154 | 0.873 ± 0.137 |
| ResNet18 | 0.855 ± 0.123 | 0.949 ± 0.017 | 0.902 ± 0.070 | |
| ResNet50 | 0.832 ± 0.135 | 0.866 ± 0.142 | 0.849 ± 0.138 | |
| DenseNet161 | 0.857 ± 0.123 | 0.949 ± 0.014 | 0.903 ± 0.068 | |
| Per-pixel | UNet++ | 0.820 ± 0.146 | 0.956 ± 0.020 | 0.888 ± 0.083 |
| DeepLabv3 | 0.837 ± 0.140 | 0.709 ± 0.179 | 0.773 ± 0.160 | |
| Hypothetical use avg per img | 0.843 ± 0.139 | 0.952 ± 0.005 | 0.898 ± 0.072 | |
| Method | Model | Flowering (3, 4) | Canopy Cover (2) | Green (2, 3, 4) | Height (3, 4, 5) |
|---|---|---|---|---|---|
| Superpixel | RF | 0.268 ± 0.203 | −0.156 ± 0.000 | 0.146 ± 0.178 | 0.237 ± 0.138 |
| MLP | 0.364 ± 0.123 | 0.250 ± 0.000 | 0.262 ± 0.087 | 0.327 ± 0.129 | |
| Centred | VGG-A | 0.442 ± 0.055 | 0.364 ± 0.000 | 0.296 ± 0.034 | 0.401 ± 0.100 |
| ResNet18 | 0.473 ± 0.002 | 0.208 ± 0.000 | 0.337 ± 0.054 | 0.468 ± 0.084 | |
| ResNet50 | 0.470 ± 0.062 | 0.290 ± 0.000 | 0.219 ± 0.089 | 0.325 ± 0.141 | |
| DenseNet161 | 0.362 ± 0.055 | 0.083 ± 0.000 | 0.235 ± 0.021 | 0.358 ± 0.070 | |
| Per-pixel | UNet++ | 0.401 ± 0.032 | 0.258 ± 0.000 | 0.342 ± 0.098 | 0.433 ± 0.056 |
| DeepLabv3 | 0.166 ± 0.028 | 0.274 ± 0.000 | 0.171 ± 0.087 | 0.252 ± 0.073 |
| Model | Subset | Flowering (3, 4) | Canopy Cover (2) | Green (3, 4, 5) | Height (2, 3, 4) |
|---|---|---|---|---|---|
| ResNet18 | 0.473 ± 0.002 | 0.208 ± 0.000 | 0.337 ± 0.054 | 0.468 ± 0.084 | |
| ResNet18 | ✓ | 0.500 ± 0.055 | 0.089 ± 0.000 | 0.328 ± 0.075 | 0.455 ± 0.095 |
| UNet++ | 0.401 ± 0.032 | 0.258 ± 0.000 | 0.342 ± 0.098 | 0.433 ± 0.056 | |
| UNet++ | ✓ | 0.414 ± 0.053 | 0.287 ± 0.000 | 0.289 ± 0.087 | 0.294 ± 0.023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Victor, B.; Nibali, A.; Newman, S.J.; Coram, T.; Pinto, F.; Reynolds, M.; Furbank, R.T.; He, Z. High-Throughput Plot-Level Quantitative Phenotyping Using Convolutional Neural Networks on Very High-Resolution Satellite Images. Remote Sens. 2024, 16, 282. https://doi.org/10.3390/rs16020282
Victor B, Nibali A, Newman SJ, Coram T, Pinto F, Reynolds M, Furbank RT, He Z. High-Throughput Plot-Level Quantitative Phenotyping Using Convolutional Neural Networks on Very High-Resolution Satellite Images. Remote Sensing. 2024; 16(2):282. https://doi.org/10.3390/rs16020282
Chicago/Turabian StyleVictor, Brandon, Aiden Nibali, Saul Justin Newman, Tristan Coram, Francisco Pinto, Matthew Reynolds, Robert T. Furbank, and Zhen He. 2024. "High-Throughput Plot-Level Quantitative Phenotyping Using Convolutional Neural Networks on Very High-Resolution Satellite Images" Remote Sensing 16, no. 2: 282. https://doi.org/10.3390/rs16020282
APA StyleVictor, B., Nibali, A., Newman, S. J., Coram, T., Pinto, F., Reynolds, M., Furbank, R. T., & He, Z. (2024). High-Throughput Plot-Level Quantitative Phenotyping Using Convolutional Neural Networks on Very High-Resolution Satellite Images. Remote Sensing, 16(2), 282. https://doi.org/10.3390/rs16020282