Land Cover Mapping in a Mangrove Ecosystem Using Hybrid Selective Kernel-Based Convolutional Neural Networks and Multi-Temporal Sentinel-2 Imagery
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Reference Polygons for Classification
2.3. Satellite Imagery
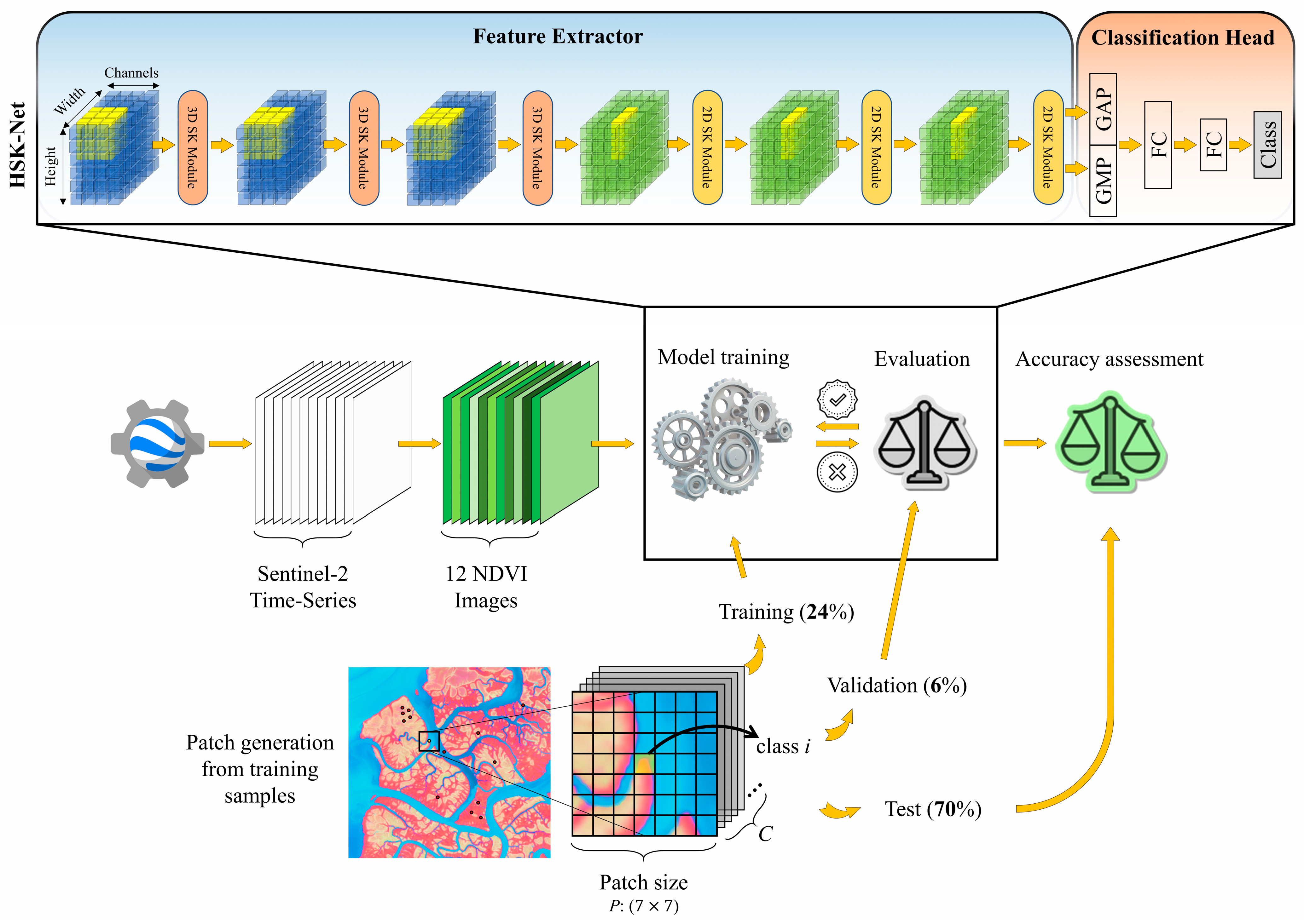
2.4. Methodology
2.4.1. Time-Series NDVI Products
2.4.2. Convolutional Layers
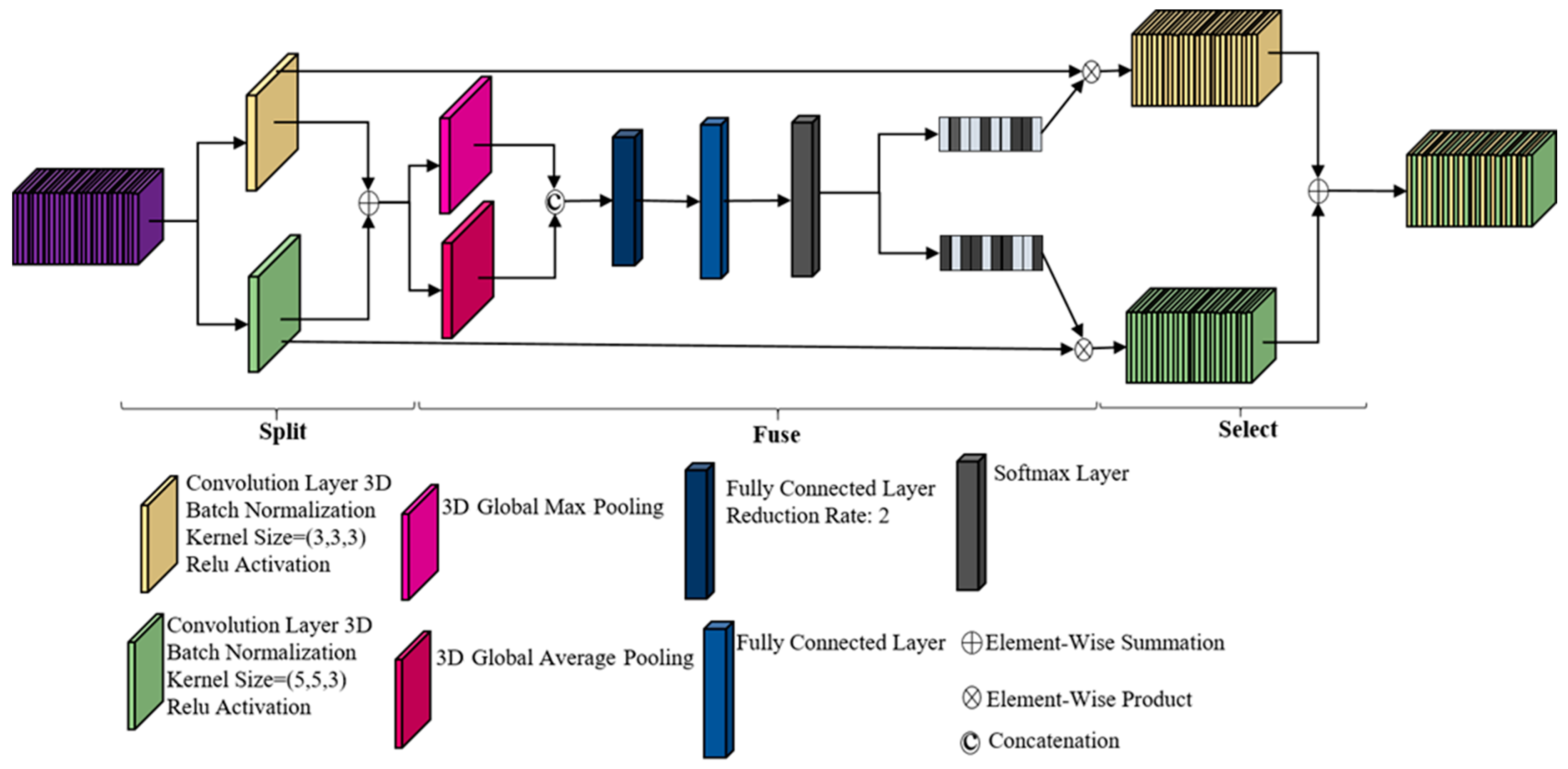
2.4.3. Selective Kernel-Based (SK) Network Modules
Split
Fuse
Select
2.5. Accuracy Assessment
2.6. Implementation
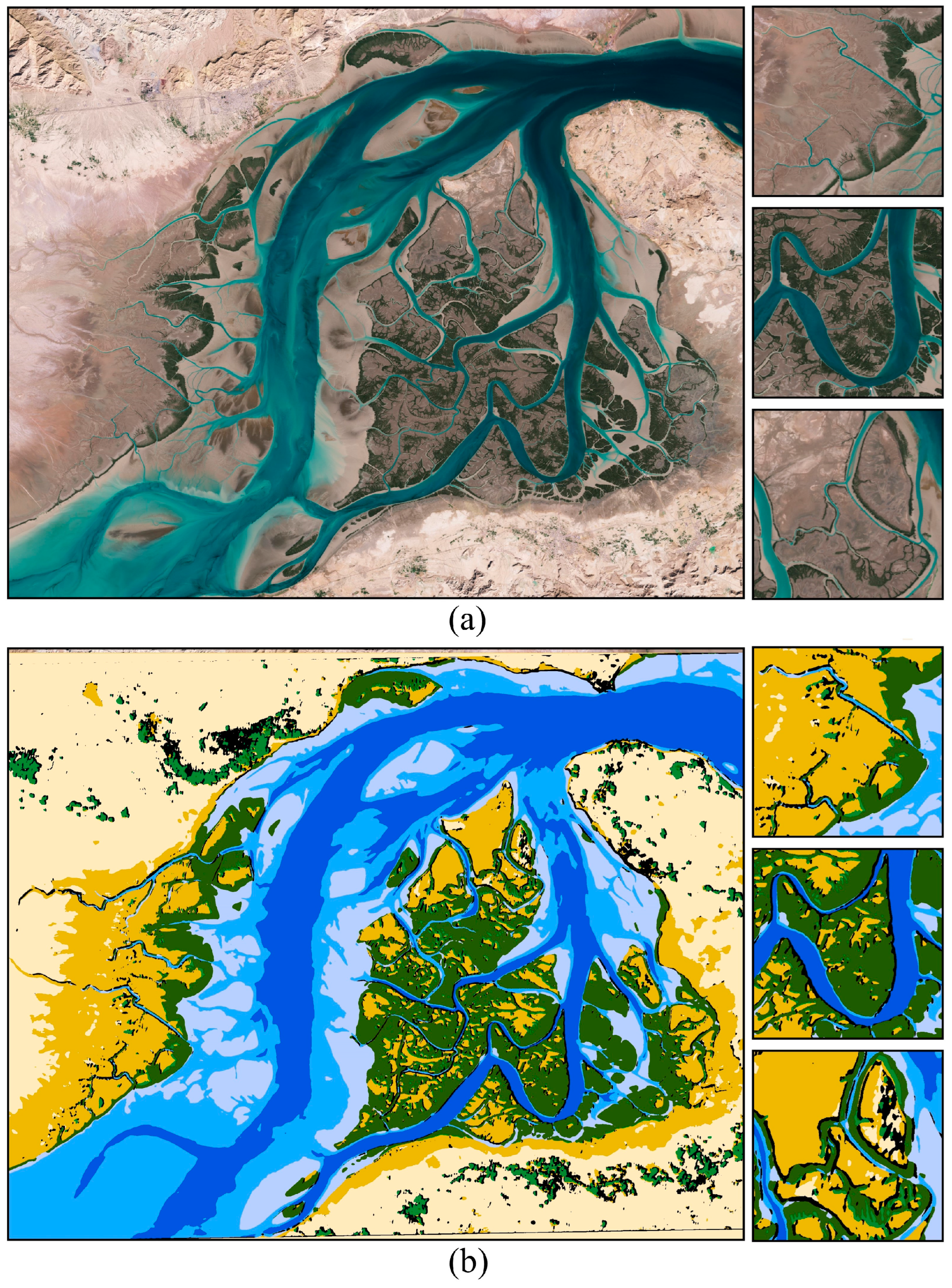
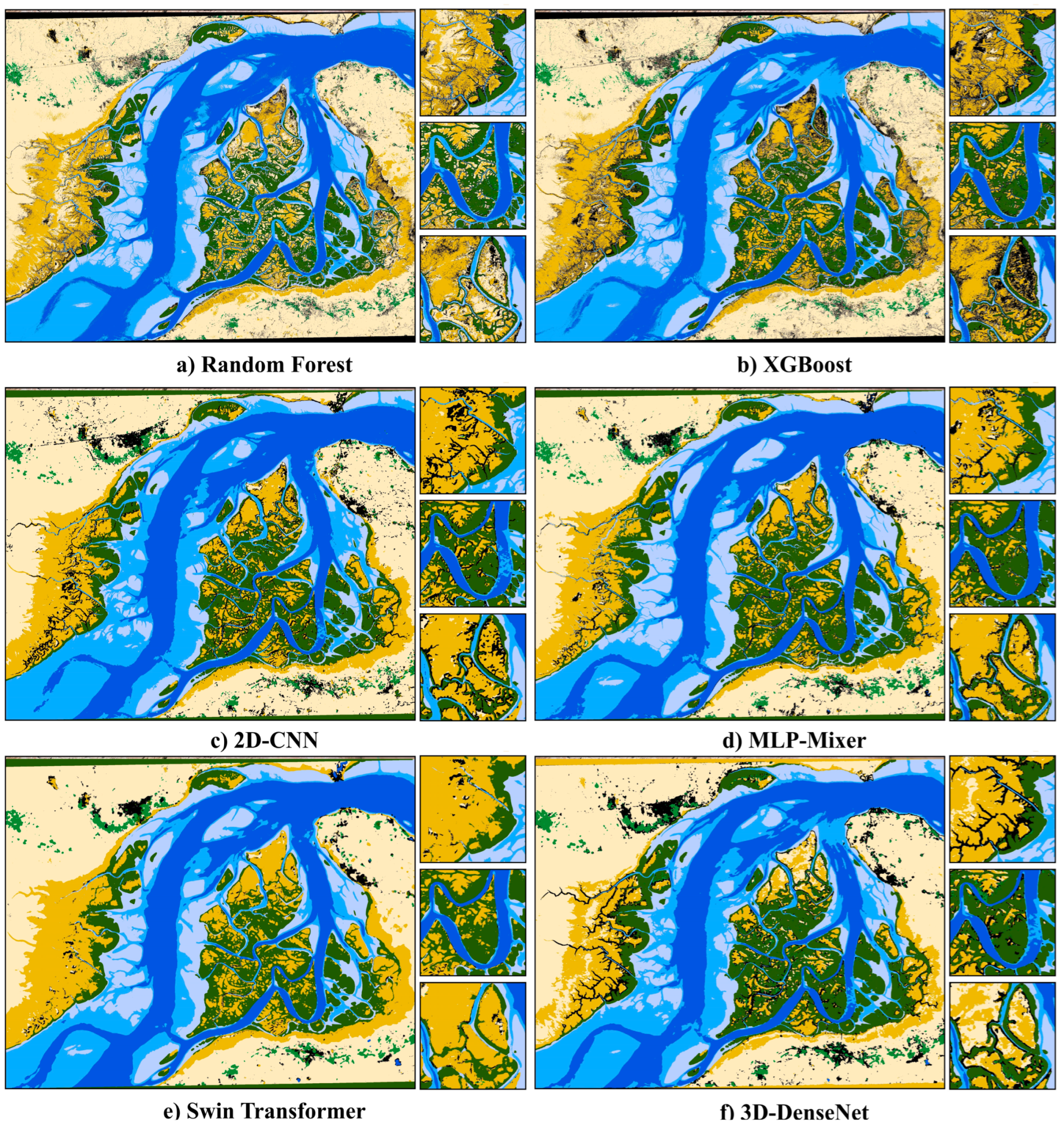
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Getzner, M.; Islam, M.S. Ecosystem Services of Mangrove Forests: Results of a Meta-Analysis of Economic Values. Int. J. Environ. Res. Public Health 2020, 17, 5830. [Google Scholar] [CrossRef] [PubMed]
- Asari, N.; Suratman, M.N.; Mohd Ayob, N.A.; Abdul Hamid, N.H. Mangrove as a Natural Barrier to Environmental Risks and Coastal Protection. In Mangroves: Ecology, Biodiversity and Management; Rastogi, R.P., Phulwaria, M., Gupta, D.K., Eds.; Springer Singapore: Singapore, 2021; pp. 305–322. ISBN 9789811624933. [Google Scholar]
- Hutchison, J.; Spalding, M.; Zu Ermgassen, P. The Role of Mangroves in Fisheries Enhancement. Nat. Conserv. Wetl. Int. 2014, 54, 434. [Google Scholar]
- Arceo-Carranza, D.; Chiappa-Carrara, X.; Chávez López, R.; Yáñez Arenas, C. Mangroves as Feeding and Breeding Grounds. In Mangroves: Ecology, Biodiversity and Management; Rastogi, R.P., Phulwaria, M., Gupta, D.K., Eds.; Springer Singapore: Singapore, 2021; pp. 63–95. ISBN 9789811624933. [Google Scholar]
- Nyanga, C. The Role of Mangroves Forests in Decarbonizing the Atmosphere. In Carbon-Based Material for Environmental Protection and Remediation; Bartoli, M., Frediani, M., Rosi, L., Eds.; IntechOpen: London, UK, 2020. [Google Scholar]
- Lee, S.Y.; Primavera, J.H.; Dahdouh-Guebas, F.; McKee, K.; Bosire, J.O.; Cannicci, S.; Diele, K.; Fromard, F.; Koedam, N.; Marchand, C.; et al. Ecological Role and Services of Tropical Mangrove Ecosystems: A Reassessment. Glob. Ecol. Biogeogr. 2014, 23, 726–743. [Google Scholar] [CrossRef]
- Cooke, A.; Lutjeharms, J.R.E.; Vasseur, P. Marine and Coastal Ecosystems. In The Natural History of Madagascar; Goodman, S.M., Benstead, J.P., Eds.; 2003; pp. 179–208. [Google Scholar]
- De Groot, R.S.; Alkemade, R.; Braat, L.; Hein, L.; Willemen, L. Challenges in Integrating the Concept of Ecosystem Services and Values in Landscape Planning, Management and Decision Making. Ecol. Complex. 2010, 7, 260–272. [Google Scholar] [CrossRef]
- Natarajan, P.; Murugesan, A.K.; Govindan, G.; Gopalakrishnan, A.; Kumar, R.; Duraisamy, P.; Balaji, R.; Shyamli, P.S.; Parida, A.K.; Parani, M.; et al. A Reference-Grade Genome Identifies Salt-Tolerance Genes from the Salt-Secreting Mangrove Species Avicennia Marina. Commun. Biol. 2021, 4, 851. [Google Scholar] [CrossRef] [PubMed]
- Adame, M.F.; Roberts, M.E.; Hamilton, D.P.; Ndehedehe, C.E.; Reis, V.; Lu, J.; Griffiths, M.; Curwen, G.; Ronan, M. Tropical Coastal Wetlands Ameliorate Nitrogen Export during Floods. Front. Mar. Sci. 2019, 6, 671. [Google Scholar] [CrossRef]
- Raju, R.D.; Arockiasamy, M. Coastal Protection Using Integration of Mangroves with Floating Barges: An Innovative Concept. J. Mar. Sci. Eng. 2022, 10, 612. [Google Scholar] [CrossRef]
- Rahman, M.M.; Zimmer, M.; Ahmed, I.; Donato, D.; Kanzaki, M.; Xu, M. Co-Benefits of Protecting Mangroves for Biodiversity Conservation and Carbon Storage. Nat. Commun. 2021, 12, 3875. [Google Scholar] [CrossRef] [PubMed]
- Vipriyanti, N.U.; Semadi, I.; Fauzi, A. Developing Mangrove Ecotourism in Nusa Penida Sacred Island, Bali, Indonesia. Environ. Dev. Sustain. 2022, 26, 535–548. [Google Scholar] [CrossRef]
- Arifanti, V.B.; Kauffman, J.B.; Ilman, M.; Tosiani, A.; Novita, N. Contributions of Mangrove Conservation and Restoration to Climate Change Mitigation in Indonesia. Glob. Change Biol. 2022, 28, 4523–4538. [Google Scholar] [CrossRef]
- Hilmi, N.; Chami, R.; Sutherland, M.D.; Hall-Spencer, J.M.; Lebleu, L.; Benitez, M.B.; Levin, L.A. The Role of Blue Carbon in Climate Change Mitigation and Carbon Stock Conservation. Front. Clim. 2021, 3, 710546. [Google Scholar] [CrossRef]
- Pörtner, H.-O.; Roberts, D.C.; Masson-Delmotte, V.; Zhai, P.; Tignor, M.; Poloczanska, E.; Mintenbeck, K.; Nicolai, M.; Okem, A.; Petzold, J.; et al. IPCC Special Report on the Ocean and Cryosphere in a Changing Climate; IPCC Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2019; Volume 1. [Google Scholar]
- Goldberg, L.; Lagomasino, D.; Thomas, N.; Fatoyinbo, T. Global Declines in Human-Driven Mangrove Loss. Glob. Change Biol. 2020, 26, 5844–5855. [Google Scholar] [CrossRef] [PubMed]
- Polidoro, B.A.; Carpenter, K.E.; Collins, L.; Duke, N.C.; Ellison, A.M.; Ellison, J.C.; Farnsworth, E.J.; Fernando, E.S.; Kathiresan, K.; Koedam, N.E.; et al. The Loss of Species: Mangrove Extinction Risk and Geographic Areas of Global Concern. PLoS ONE 2010, 5, e10095. [Google Scholar] [CrossRef] [PubMed]
- Mondal, P.; Dutta, T.; Qadir, A.; Sharma, S. Radar and Optical Remote Sensing for near Real-Time Assessments of Cyclone Impacts on Coastal Ecosystems. Remote Sens. Ecol. Conserv. 2022, 8, 506–520. [Google Scholar] [CrossRef] [PubMed]
- Mafi-Gholami, D.; Zenner, E.K.; Jaafari, A. Mangrove Regional Feedback to Sea Level Rise and Drought Intensity at the End of the 21st Century. Ecol. Indic. 2020, 110, 105972. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Hardy, G.E.S.; Le, T.V.; Nguyen, H.Q.; Nguyen, H.H.; Nguyen, T.V.; Dell, B. Mangrove Forest Landcover Changes in Coastal Vietnam: A Case Study from 1973 to 2020 in Thanh Hoa and Nghe An Provinces. Forests 2021, 12, 637. [Google Scholar] [CrossRef]
- Moschetto, F.A.; Ribeiro, R.B.; De Freitas, D.M. Urban Expansion, Regeneration and Socioenvironmental Vulnerability in a Mangrove Ecosystem at the Southeast Coastal of São Paulo, Brazil. Ocean Coast. Manag. 2021, 200, 105418. [Google Scholar] [CrossRef]
- Ghorbanian, A.; Zaghian, S.; Asiyabi, R.M.; Amani, M.; Mohammadzadeh, A.; Jamali, S. Mangrove Ecosystem Mapping Using Sentinel-1 and Sentinel-2 Satellite Images and Random Forest Algorithm in Google Earth Engine. Remote Sens. 2021, 13, 2565. [Google Scholar] [CrossRef]
- Xue, Z.; Qian, S. Generalized Composite Mangrove Index for Mapping Mangroves Using Sentinel-2 Time Series Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 5131–5146. [Google Scholar] [CrossRef]
- Kuenzer, C.; Bluemel, A.; Gebhardt, S.; Quoc, T.V.; Dech, S. Remote Sensing of Mangrove Ecosystems: A Review. Remote Sens. 2011, 3, 878–928. [Google Scholar] [CrossRef]
- Pham, T.D.; Yokoya, N.; Bui, D.T.; Yoshino, K.; Friess, D.A. Remote Sensing Approaches for Monitoring Mangrove Species, Structure, and Biomass: Opportunities and Challenges. Remote Sens. 2019, 11, 230. [Google Scholar] [CrossRef]
- Maurya, K.; Mahajan, S.; Chaube, N. Remote Sensing Techniques: Mapping and Monitoring of Mangrove Ecosystem—A Review. Complex Intell. Syst. 2021, 7, 2797–2818. [Google Scholar] [CrossRef]
- Bihamta Toosi, N.; Soffianian, A.R.; Fakheran, S.; Pourmanafi, S.; Ginzler, C.T.; Waser, L. Land Cover Classification in Mangrove Ecosystems Based on VHR Satellite Data and Machine Learning—An Upscaling Approach. Remote Sens. 2020, 12, 2684. [Google Scholar] [CrossRef]
- Yang, G.; Huang, K.; Sun, W.; Meng, X.; Mao, D.; Ge, Y. Enhanced Mangrove Vegetation Index Based on Hyperspectral Images for Mapping Mangrove. ISPRS J. Photogramm. Remote Sens. 2022, 189, 236–254. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Qiu, P.; Tan, X.; Zhang, Q. Mapping Mangrove Species Using Combined UAV-LiDAR and Sentinel-2 Data: Feature Selection and Point Density Effects. Adv. Space Res. 2022, 69, 1494–1512. [Google Scholar] [CrossRef]
- Ghorbanian, A.; Ahmadi, S.A.; Amani, M.; Mohammadzadeh, A.; Jamali, S. Application of Artificial Neural Networks for Mangrove Mapping Using Multi-Temporal and Multi-Source Remote Sensing Imagery. Water 2022, 14, 244. [Google Scholar] [CrossRef]
- Kumar, T.; Mandal, A.; Dutta, D.; Nagaraja, R.; Dadhwal, V.K. Discrimination and Classification of Mangrove Forests Using EO-1 Hyperion Data: A Case Study of Indian Sundarbans. Geocarto. Int. 2019, 34, 415–442. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Johnson, B.A. Deep Learning in Remote Sensing Applications: A Meta-Analysis and Review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- de Souza Moreno, G.M.; de Carvalho Júnior, O.A.; de Carvalho, O.L.F.; Andrade, T.C. Deep Semantic Segmentation of Mangroves in Brazil Combining Spatial, Temporal, and Polarization Data from Sentinel-1 Time Series. Ocean Coast. Manag. 2023, 231, 106381. [Google Scholar] [CrossRef]
- Guo, Y.; Liao, J.; Shen, G. Mapping Large-Scale Mangroves along the Maritime Silk Road from 1990 to 2015 Using a Novel Deep Learning Model and Landsat Data. Remote Sens. 2021, 13, 245. [Google Scholar] [CrossRef]
- Guo, M.; Yu, Z.; Xu, Y.; Huang, Y.; Li, C. ME-Net: A Deep Convolutional Neural Network for Extracting Mangrove Using Sentinel-2A Data. Remote Sens. 2021, 13, 1292. [Google Scholar] [CrossRef]
- Jamaluddin, I.; Thaipisutikul, T.; Chen, Y.-N.; Chuang, C.-H.; Hu, C.-L. MDPrePost-Net: A Spatial-Spectral-Temporal Fully Convolutional Network for Mapping of Mangrove Degradation Affected by Hurricane Irma 2017 Using Sentinel-2 Data. Remote Sens. 2021, 13, 5042. [Google Scholar] [CrossRef]
- Toosi, N.B.; Soffianian, A.R.; Fakheran, S.; Waser, L.T. Mapping Disturbance in Mangrove Ecosystems: Incorporating Landscape Metrics and PCA-Based Spatial Analysis. Ecol. Indic. 2022, 136, 108718. [Google Scholar] [CrossRef]
- Alipour-Fard, T.; Paoletti, M.E.; Haut, J.M.; Arefi, H.; Plaza, J.; Plaza, A. Multibranch Selective Kernel Networks for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2020, 18, 1089–1093. [Google Scholar] [CrossRef]
- Alireza, S.M.; Mansour, J.B. Satellite based assessment of the area and changes in the Mangrove ecosystem of the QESHM island, Iran. J. Environ. Res. Develop. 2012, 7, 1052–1060. [Google Scholar]
- Sharifi, N.; Danehkar, A.; Robati, M.; Khorasani, N.A.; Rajaee, T. Developing Decision Algorithm for Determination of Protection Zones in Protected Areas (Case Study: Hara Protected Area). Int. J. Environ. Sci. Technol. 2021, 18, 2237–2250. [Google Scholar] [CrossRef]
- Vahidi, F.; Fatemi, S.M.R.; Danehkar, A.; Mashinchian Moradi, A.; Mousavi Nadushan, R. Patterns of Mollusks (Bivalvia and Gastropoda) Distribution in Three Different Zones of Harra Biosphere Reserve, the Persian Gulf, Iran. Iran. J. Fish. Sci. 2021, 20, 1336–1353. [Google Scholar]
- Bunting, P.; Rosenqvist, A.; Lucas, R.M.; Rebelo, L.-M.; Hilarides, L.; Thomas, N.; Hardy, A.; Itoh, T.; Shimada, M.; Finlayson, C.M. The Global Mangrove Watch—A New 2010 Global Baseline of Mangrove Extent. Remote Sens. 2018, 10, 1669. [Google Scholar] [CrossRef]
- Murray, N.J.; Phinn, S.R.; DeWitt, M.; Ferrari, R.; Johnston, R.; Lyons, M.B.; Clinton, N.; Thau, D.; Fuller, R.A. The Global Distribution and Trajectory of Tidal Flats. Nature 2019, 565, 222–225. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Seydi, S.T.; Arefi, H. A Comparison of Deep Learning-Based Super-Resolution Frameworks for SENTINEL-2 Imagery in Urban Areas. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2023, 10, 1021–1026. [Google Scholar] [CrossRef]
- Seydi, S.T.; Hasanolu, M.; Chanussot, J. A Novel Deep Siamese Framework for Burned Area Mapping Leveraging Mixture of Experts. Eng. Appl. Artif. Intell. 2024, 133, 108280. [Google Scholar] [CrossRef]
- Seydi, S.T.; Hasanlou, M. Binary Hyperspectral Change Detection Based on 3D Convolution Deep Learning. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, 43, 1629–1633. [Google Scholar] [CrossRef]
- Seydi, S.T.; Hasanlou, M.; Chanussot, J. DSMNN-Net: A Deep Siamese Morphological Neural Network Model for Burned Area Mapping Using Multispectral Sentinel-2 and Hyperspectral PRISMA Images. Remote Sens. 2021, 13, 5138. [Google Scholar] [CrossRef]
- Tolstikhin, I.O.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Steiner, A.; Keysers, D.; Uszkoreit, J. Mlp-Mixer: An All-Mlp Architecture for Vision. Adv. Neural Inf. Process. Syst. 2021, 34, 24261–24272. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Li, G.; Zhang, C.; Lei, R.; Zhang, X.; Ye, Z.; Li, X. Hyperspectral Remote Sensing Image Classification Using Three-Dimensional-Squeeze-and-Excitation-DenseNet (3D-SE-DenseNet). Remote Sens. Lett. 2020, 11, 195–203. [Google Scholar] [CrossRef]
- Zhang, C.; Li, G.; Du, S.; Tan, W.; Gao, F. Three-Dimensional Densely Connected Convolutional Network for Hyperspectral Remote Sensing Image Classification. J. Appl. Remote Sens. 2019, 13, 016519. [Google Scholar] [CrossRef]


| Class | All Patches | Training | Validation | Test |
|---|---|---|---|---|
| Mangrove | 4870 | 1198 | 302 | 3370 |
| Tidal Zone | 4419 | 1111 | 245 | 3063 |
| Deep Water | 4074 | 1002 | 333 | 2739 |
| Shallow Water | 3575 | 879 | 214 | 2482 |
| Mudflat | 3856 | 1126 | 248 | 3170 |
| Urban | 4070 | 724 | 159 | 3187 |
| Barren | 5926 | 1412 | 311 | 4203 |
| Vegetation | 3554 | 503 | 111 | 2940 |
| Total | 34,344 | 7955 | 1923 | 25,154 |
| Algorithm | Hyperparameters | General Hyperparameters |
|---|---|---|
| Random Forest | n_estimators = 128, max_depth = 10 | - |
| XGBoost | learning_rate = 0.1, n_estimators = 250, min_child_weight = 1, gamma = 0, subsample = 0.8, colsample_bytree = 0.8, nthread = 4 | |
| 2D-CNN | dropout_rate = 0.1 | optimizers = Adam, learning_rate = 1 × 10−3, epochs = 500, batch_size = 550, loss function = Categorical-Crossentropy, kernel_initializer = Glorot |
| MLP-Mixer | num_blocks = 4, window_size = 5, stem_width = 128, mlp_dim = 512, dropout = 0.1, tokens_mlp_dim = 512 | |
| Swin Transformer | num_heads = 4, window_size = 4, shift_size = 2, embed_dim = 128, mlp_dim = 256, dropout = 0.1 | |
| 3D-DenseNet | Dense_blocks = 3, transition_block = 2, compression rate at transition layers = 0.5, number of building blocks = 6 | |
| Proposed HSK-CNN | SK blocks = 2, reduction_rate = 0.5, dropout_rate = 0.5 |
| Evaluation Metrics | |||||
|---|---|---|---|---|---|
| OA (%) | CKC | MCC | BA | ||
| Models | Random forest | 85 | 0.83 | 0.83 | 0.85 |
| XGBoost | 87 | 0.85 | 0.85 | 0.87 | |
| 2D-CNN | 91 | 0.90 | 0.90 | 0.91 | |
| MLP-Mixer | 92 | 0.91 | 0.91 | 0.92 | |
| Swin Transformer | 93 | 0.92 | 0.92 | 0.92 | |
| 3D-DenseNet | 90 | 0.89 | 0.89 | 0.90 | |
| Proposed HSK-CNN | 94 | 0.93 | 0.93 | 0.94 | |
| Convolutional Layer Structure | OA (%) | CKC | MCC | BA |
|---|---|---|---|---|
| Without 2D SK | 92 | 0.90 | 0.91 | 0.92 |
| Without 3D SK | 90 | 0.89 | 0.89 | 0.90 |
| With 2D and 3D SK | 94 | 0.93 | 0.93 | 0.94 |
| Patch Size (Pixels) | OA (%) | CKC | MCC | BA |
|---|---|---|---|---|
| 7 × 7 | 91 | 0.90 | 0.90 | 0.91 |
| 9 × 9 | 94 | 0.93 | 0.93 | 0.93 |
| 11 × 11 | 94 | 0.93 | 0.93 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seydi, S.T.; Ahmadi, S.A.; Ghorbanian, A.; Amani, M. Land Cover Mapping in a Mangrove Ecosystem Using Hybrid Selective Kernel-Based Convolutional Neural Networks and Multi-Temporal Sentinel-2 Imagery. Remote Sens. 2024, 16, 2849. https://doi.org/10.3390/rs16152849
Seydi ST, Ahmadi SA, Ghorbanian A, Amani M. Land Cover Mapping in a Mangrove Ecosystem Using Hybrid Selective Kernel-Based Convolutional Neural Networks and Multi-Temporal Sentinel-2 Imagery. Remote Sensing. 2024; 16(15):2849. https://doi.org/10.3390/rs16152849
Chicago/Turabian StyleSeydi, Seyd Teymoor, Seyed Ali Ahmadi, Arsalan Ghorbanian, and Meisam Amani. 2024. "Land Cover Mapping in a Mangrove Ecosystem Using Hybrid Selective Kernel-Based Convolutional Neural Networks and Multi-Temporal Sentinel-2 Imagery" Remote Sensing 16, no. 15: 2849. https://doi.org/10.3390/rs16152849
APA StyleSeydi, S. T., Ahmadi, S. A., Ghorbanian, A., & Amani, M. (2024). Land Cover Mapping in a Mangrove Ecosystem Using Hybrid Selective Kernel-Based Convolutional Neural Networks and Multi-Temporal Sentinel-2 Imagery. Remote Sensing, 16(15), 2849. https://doi.org/10.3390/rs16152849







