Convolutional Neural Network Maps Plant Communities in Semi-Natural Grasslands Using Multispectral Unmanned Aerial Vehicle Imagery
Abstract
1. Introduction
2. Material and Methods
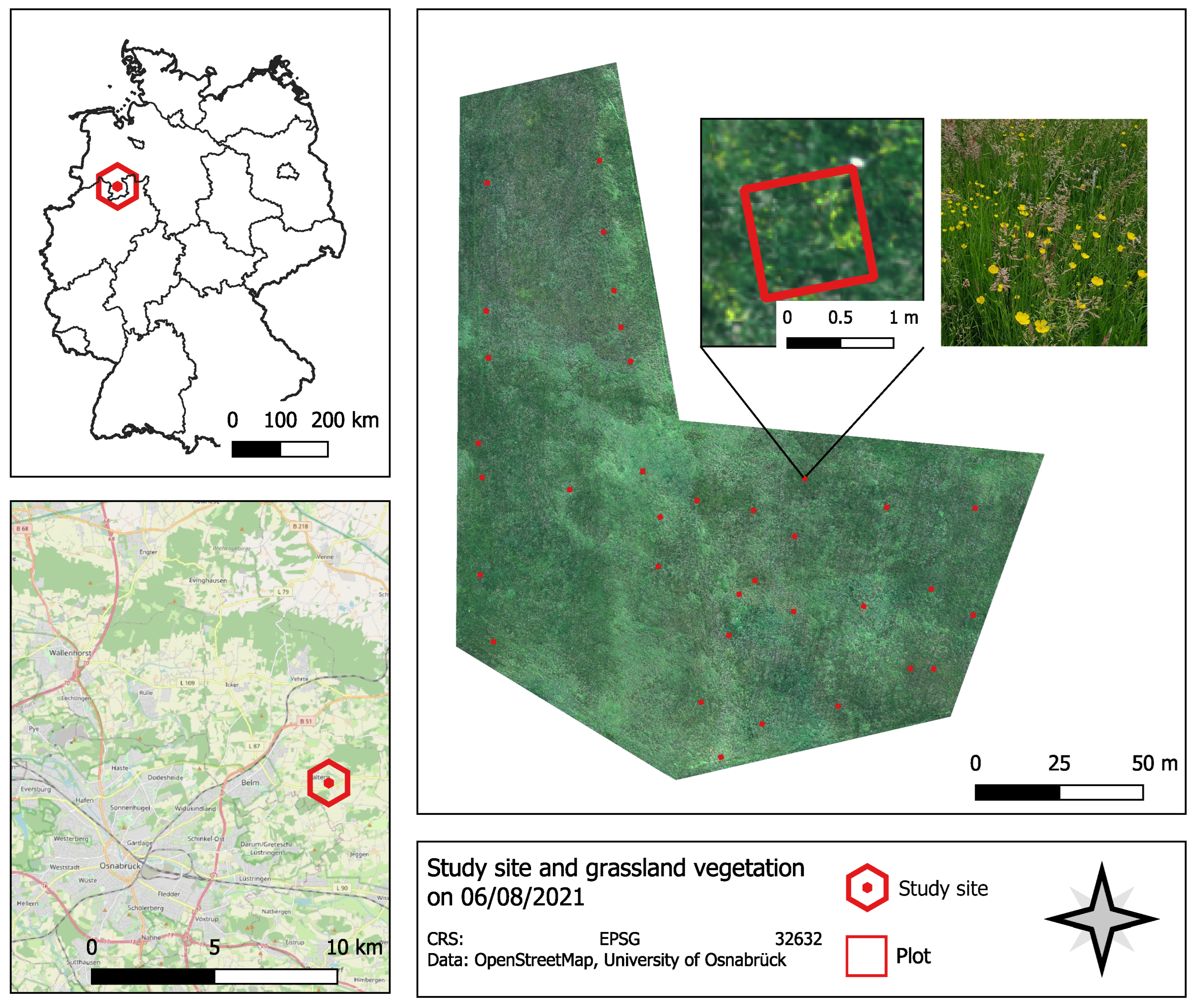
2.1. Study Site
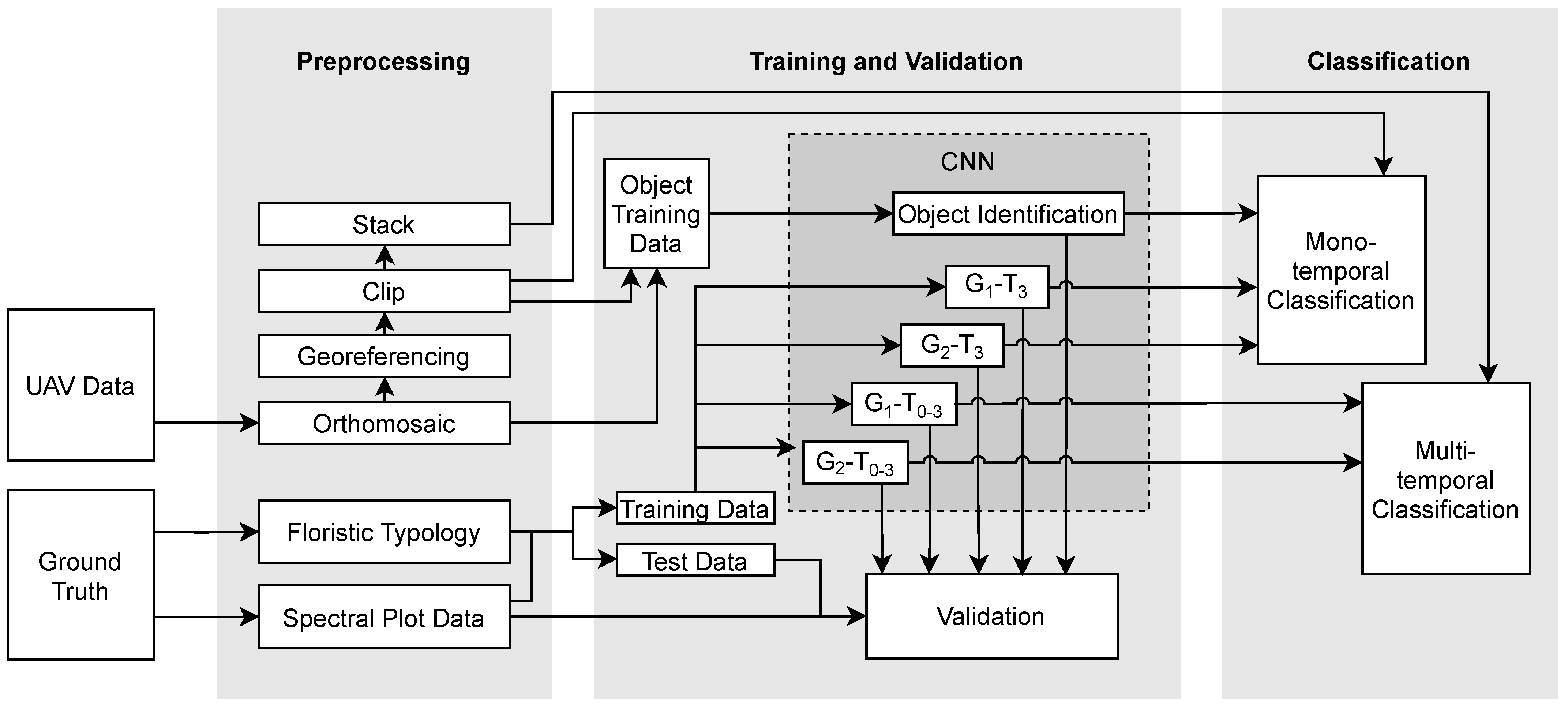
2.2. Data and Preprocessing
2.2.1. UAV Image Data
2.2.2. Vegetation Surveys in the Field
2.3. Methodology
2.3.1. Analysis of Vegetation Data
2.3.2. Training and Test Data
2.3.3. CNN
2.3.4. Classification
2.3.5. Validation Metrics
3. Results
3.1. Floristic Typology
3.2. Phenological Change in Species Spectrum
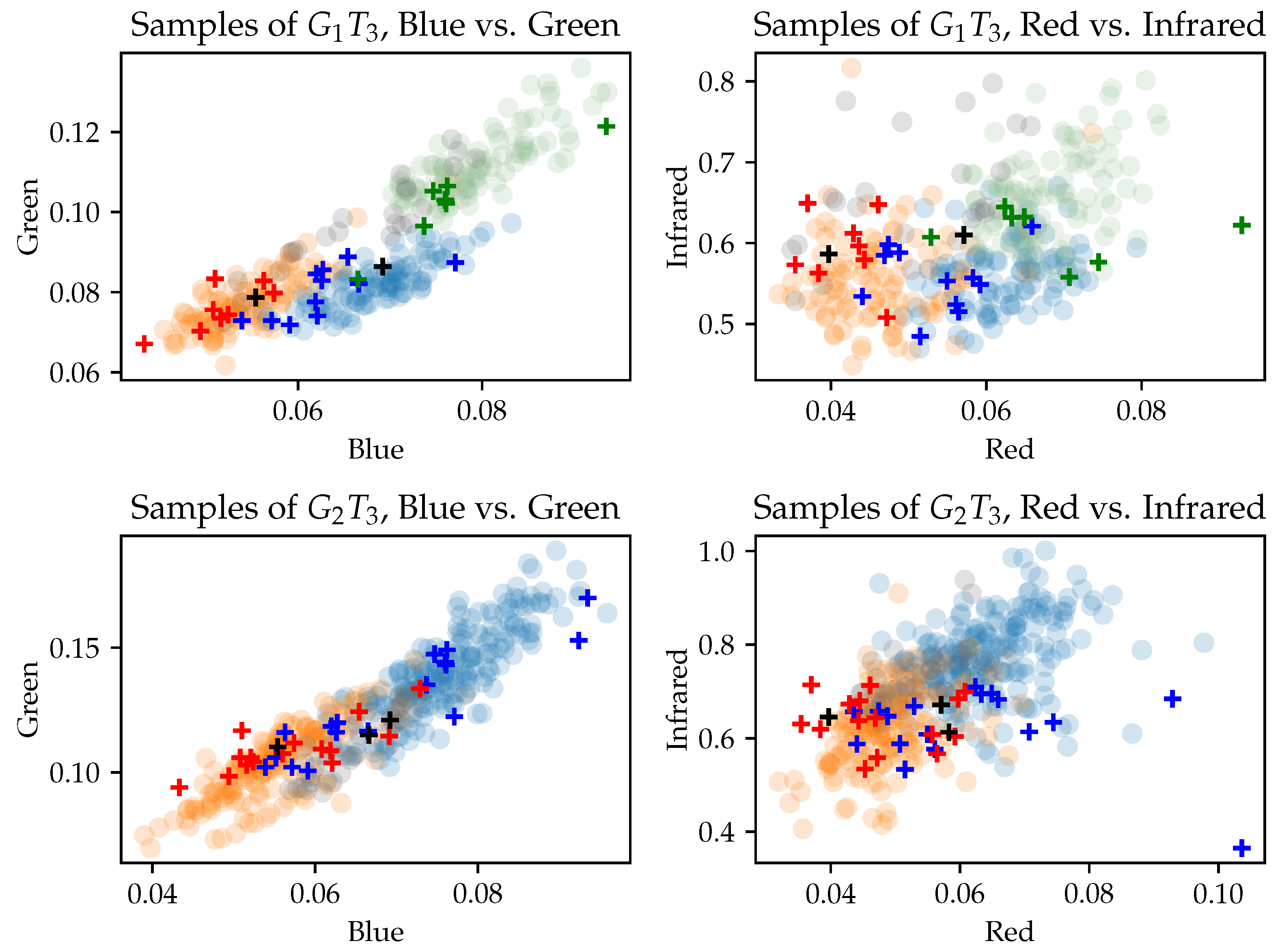
3.3. Separability of Training Data
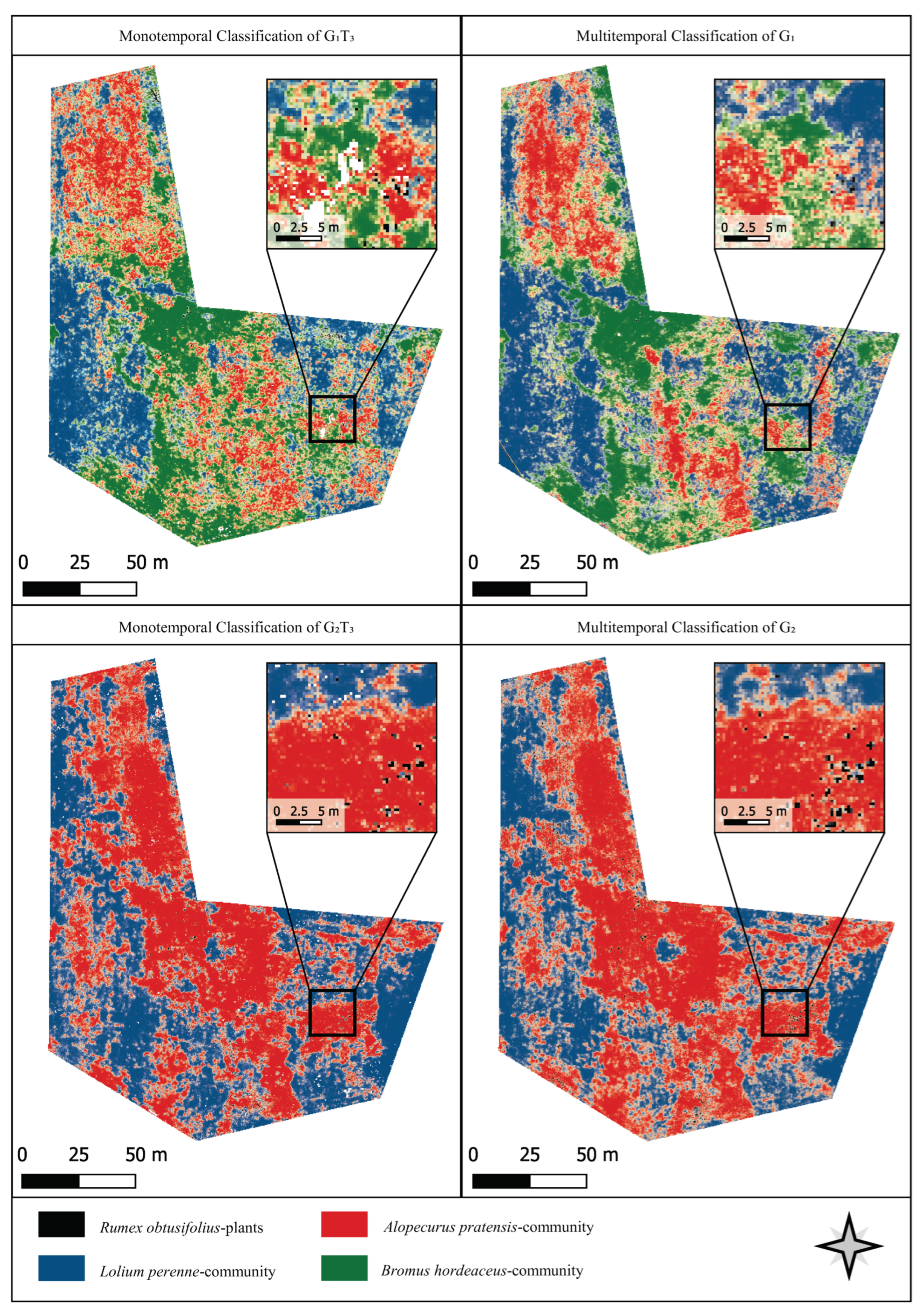
3.4. Classification Results
4. Discussion
4.1. Usability of the Presented Methodology in an Agricultural Context
4.2. Comparison of Mono- and Multitemporal Data for Plant Community Mapping
4.3. CNNs for Plant Community Classification in Grasslands
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNG | Semi-Natural Grasslands |
| UAV | Unmanned Aerial Vehicle |
| CNN | Convolutional Neural Network |
| EIV | Ellenberg Indicator Values |
| VU | Vegetation Unit |
Appendix A
| Lolium perenne- | Alopecurus pratensis- | Bromus hordeaceus- | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Community | Community | Community | ||||||||||||||
| T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | T1 | T2 | T3 | ||
| Species Group | No. of Plots | 8 | 13 | 19 | 12 | 7 | ||||||||||
| Anthoxanthum odoratum | 38 | 38 | 38 | |||||||||||||
| Ranunculus acris | 38 | 50 | ||||||||||||||
| Veronica chamedrys | 13 | 13 | 13 | |||||||||||||
| Ajuga reptans | 13 | 13 | ||||||||||||||
| Lolium perenne | 100 | 100 | 100 | 100 | 100 | 100 | 25 | 25 | 25 | 25 | 58 | 58 | ||||
| Centaurea jacea | 13 | 13 | 13 | 13 | 13 | |||||||||||
| Galium mollugo | 13 | 13 | 13 | 13 | 13 | 13 | 29 | |||||||||
| Crepis biennis | 31 | 31 | 25 | |||||||||||||
| Agrostis capillaris | 25 | 25 | 25 | |||||||||||||
| Trifolium pratense | 25 | 25 | 25 | |||||||||||||
| Cynosurus cristatus | 38 | 38 | 38 | 44 | 44 | 44 | ||||||||||
| Phleum pratense | 19 | 19 | 0 | |||||||||||||
| Stellaria media | 19 | 13 | ||||||||||||||
| Rumex obtusifolius | 13 | 13 | 13 | |||||||||||||
| Lamium album | 6 | 6 | 13 | |||||||||||||
| Capsella bursa-pastoris | 6 | 6 | 6 | |||||||||||||
| Alopecurus pratensis | 38 | 50 | 63 | 13 | 13 | 13 | 100 | 100 | 100 | 100 | 100 | 100 | 28 | 71 | 71 | |
| Phalaris arundinaea | 38 | 38 | 38 | 8 | 8 | 8 | 14 | 14 | 14 | |||||||
| Cirsium arvense | 13 | 13 | 13 | 14 | 14 | 14 | ||||||||||
| Bromus hordeaceus | 25 | 62 | 62 | 19 | 19 | 19 | 11 | 58 | 58 | 100 | 100 | 100 | ||||
| Other species | Holcus lanatus | 100 | 100 | 100 | 56 | 56 | 56 | 81 | 81 | 81 | 100 | 100 | 100 | 100 | 100 | 100 |
| Poa pratensis | 100 | 100 | 100 | 25 | 25 | 25 | 13 | 13 | 13 | 25 | 58 | 58 | 43 | 43 | 43 | |
| Plantago laneolata | 100 | 100 | 100 | 100 | 100 | 100 | 68 | 65 | 43 | 44 | 8 | 8 | ||||
| Taraxacum officinale agg. | 100 | 100 | 100 | 87 | 68 | 44 | 68 | 62 | 38 | 67 | 41 | 29 | ||||
| Cerastium fontanum | 88 | 100 | 75 | 43 | 56 | 31 | 31 | 31 | 19 | 67 | 58 | 33 | 43 | 71 | 14 | |
| Ranunculus repens | 38 | 50 | 75 | 68 | 62 | 56 | 38 | 31 | 31 | 16 | 29 | 29 | 29 | |||
| Trifolium repens | 63 | 13 | 13 | 38 | 31 | 13 | 19 | 13 | 6 | |||||||
| Rumex acetosa | 63 | 13 | 13 | 43 | 31 | 25 | 19 | 19 | 13 | 8 | 16 | |||||
| Poa trivialis | 100 | 100 | 100 | 25 | 67 | 67 | 100 | 100 | 100 | |||||||
| Festuca rubra agg. | 67 | 100 | 100 | 13 | 13 | 13 | 42 | 67 | 67 | 57 | 57 | |||||
| Molinia caerulea | 13 | 16 | 14 | |||||||||||||
| Cardamine pratensis | 50 | 13 | 42 | 8 | ||||||||||||
| Lychnis flos-cuculi | 16 | 16 | ||||||||||||||
Appendix B
| Rumex obtusifolius Plants | Lolium perenne-Community | Alopecurus pratensis-Community | Bromus hordeaceus-Community | |||
|---|---|---|---|---|---|---|
| & | ||||||
| Ellenberg M | 6 | 5.76 | 5.44 | 5.98 | 5.76 | 6.4 |
| Ellenberg R | X | 6.2 | 6.42 | 6.25 | 6.02 | 6.0 |
| Ellenberg N | 9 | 7.17 | 6.41 | 6.68 | 6.37 | 4.97 |
| Forage Value | 2 | 6.26 | 6.59 | 6.67 | 6.4 | 5.26 |
References
- Dengler, J.; Tischew, S. Grasslands of Western and Northern Europe—Between intensification and abandonment. In Grasslands of the World: Diversity, Management and Conservation; Squires, V.S., Dengler, J., Hua, L., Feng, H., Eds.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Leuschner, C.; Ellenberg, H. Ecology of Central European Non-Forest Vegetation: Coastal to Alpine, Natural to Man-Made Habitats: Vegetation Ecology of Central Europe; Springer: Berlin/Heidelberg, Germany, 2018; Volume II. [Google Scholar]
- Plantureux, S.; Peeters, A.; McCracken, D. Biodiversity in intensive grasslands: Effect of management, improvement and challenges. Agron. Res. 2005, 3, 153–164. [Google Scholar]
- Wesche, K.; Krause, B.; Culmsee, H.; Leuschner, C. Fifty years of change in Central European grassland vegetation: Large losses in species richness and animal-pollinated plants. Biol. Conserv. 2012, 150, 76–85. [Google Scholar] [CrossRef]
- European Parliament and the Council. OJ L 347/608; Regulation (EU) No 1307/2013 of the European Parliament and of the Council of 17 December 2013 Establishing Rules for Direct Payments to Farmers Under Support Schemes Within the Framework of the Common Agricultural Policy and Repealing Council Regulation (EC) No 637/2008 and Council Regulation (EC) No 73/2009; European Parliament and the Council: Brussels, Belgium, 2013. [Google Scholar]
- Niedersächsisches Ministerium für Ernährung, Landwirtschaft und Verbraucherschutz (ML). Merkblatt zu den Besonderen Förderbestimmungen GL 1—Extensive Bewirtschaftung von Dauergrünland GL 11—Grundförderung. Available online: https://www.ml.niedersachsen.de/download/85100/GL_12_-_Merkblatt_Zusatzfoerderung_nicht_vollstaendig_barrierefrei_.pdf (accessed on 7 October 2021).
- Johansen, M.; Lund, P.; Weisbjerg, M. Feed intake and milk production in dairy cows fed different grass and legume species: A meta-analysis. Animal 2018, 12, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Sturm, P.; Zehm, A.; Baumbauch, H.; von Brackel, W.; Verbücheln, G.; Stock, M.; Zimmermann, F. Grünlandtypen; Quelle & Meyer Verlag: Wiebelsheim, Germany, 2018. [Google Scholar]
- Gräßler, J.; von Borstel, U. Fructan content in pasture grasses. Pferdeheilkunde 2005, 21, 75. [Google Scholar] [CrossRef]
- van Eps, A.; Pollitt, C. Equine laminitis induced with oligofructose. Equine Vet. J. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, D.; Belesky, D.; Lewis, G. Abiotic stresses in endophytic grasses. In Neotyphodium in Cool-Season Grasses; Blackwell Publishing: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Bourke, C.A.; Hunt, E.; Watson, R. Fescue-associated oedema of horses grazing on endophyte-inoculated tall fescue grass (Festuca arundinacea) pastures. Aust. Vet. J. 2009, 87, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, J.; Bullock, J.M.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.; Smith, H.; Lindborg, R. Grasslands—More important for ecosystem services than you might think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- Le Clec’h, S.; Finger, R.; Buchmann, N.; Gosal, A.S.; Hörtnagl, L.; Huguenin-Elie, O.; Jeanneret, P.; Lüscher, A.; Schneider, M.K.; Huber, R. Assessment of spatial variability of multiple ecosystem services in grasslands of different intensities. J. Environ. Manag. 2019, 251, 109372. [Google Scholar] [CrossRef]
- Lavorel, S.; Grigulis, K.; Lamarque, P.; Colace, M.P.; Garden, D.; Girel, J.; Pellet, G.; Douzet, R. Using plant functional traits to understand the landscape distribution of multiple ecosystem services. J. Ecol. 2011, 99, 135–147. [Google Scholar] [CrossRef]
- Díaz, S.; Lavorel, S.; de Bello, F.; Quétier, F.; Grigulis, K.; Robson, T.M. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl. Acad. Sci. USA 2007, 104, 20684–20689. [Google Scholar] [CrossRef]
- Smith, T.; Huston, M. A theory of the spatial and temporal dynamics of plant communities. In Progress in Theoretical Vegetation Science; Springer: Berlin/Heidelberg, Germany, 1990; pp. 49–69. [Google Scholar] [CrossRef]
- Dierschke, H. Pflanzensoziologie: Grundlagen und Methoden; 55 Tabellen; Eugen Ulmer KG: Darmstadt, Germany, 1994. [Google Scholar]
- Mueller-Dombois, D.; Ellenberg, H. Aims and Methods of Vegetation Ecology; The Blackburn Press: West Caldwell, NJ, USA, 2003. [Google Scholar]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, Á.; Székely, B.; Pfeifer, N. Categorizing grassland vegetation with full-waveform airborne laser scanning: A feasibility study for detecting Natura 2000 habitat types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef]
- Reinermann, S.; Asam, S.; Kuenzer, C. Remote sensing of grassland production and management—A review. Remote Sens. 2020, 12, 1949. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Wood, D.J.; Preston, T.M.; Powell, S.; Stoy, P.C. Multiple UAV Flights across the Growing Season Can Characterize Fine Scale Phenological Heterogeneity within and among Vegetation Functional Groups. Remote Sens. 2022, 14, 1290. [Google Scholar] [CrossRef]
- Rapinel, S.; Mony, C.; Lecoq, L.; Clement, B.; Thomas, A.; Hubert-Moy, L. Evaluation of Sentinel-2 time-series for mapping floodplain grassland plant communities. Remote Sens. Environ. 2019, 223, 115–129. [Google Scholar] [CrossRef]
- Fauvel, M.; Lopes, M.; Dubo, T.; Rivers-Moore, J.; Frison, P.L.; Gross, N.; Ouin, A. Prediction of plant diversity in grasslands using Sentinel-1 and-2 satellite image time series. Remote Sens. Environ. 2020, 237, 111536. [Google Scholar] [CrossRef]
- Cruzan, M.B.; Weinstein, B.G.; Grasty, M.R.; Kohrn, B.F.; Hendrickson, E.C.; Arredondo, T.M.; Thompson, P.G. Small unmanned aerial vehicles (micro-UAVs, drones) in plant ecology. Appl. Plant Sci. 2016, 4, 1600041. [Google Scholar] [CrossRef]
- Gholizadeh, H.; Gamon, J.A.; Townsend, P.A.; Zygielbaum, A.I.; Helzer, C.J.; Hmimina, G.Y.; Yu, R.; Moore, R.M.; Schweiger, A.K.; Cavender-Bares, J. Detecting prairie biodiversity with airborne remote sensing. Remote Sens. Environ. 2019, 221, 38–49. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Species classification using Unmanned Aerial Vehicle (UAV)-acquired high spatial resolution imagery in a heterogeneous grassland. ISPRS J. Photogramm. Remote Sens. 2017, 128, 73–85. [Google Scholar] [CrossRef]
- Weisberg, P.J.; Dilts, T.E.; Greenberg, J.A.; Johnson, K.N.; Pai, H.; Sladek, C.; Kratt, C.; Tyler, S.W.; Ready, A. Phenology-based classification of invasive annual grasses to the species level. Remote Sens. Environ. 2021, 263, 112568. [Google Scholar] [CrossRef]
- Wijesingha, J.; Astor, T.; Schulze-Brüninghoff, D.; Wengert, M.; Wachendorf, M. Predicting forage quality of grasslands using UAV-borne imaging spectroscopy. Remote Sens. 2020, 12, 126. [Google Scholar] [CrossRef]
- Pecina, M.V.; Bergamo, T.F.; Ward, R.; Joyce, C.; Sepp, K. A novel UAV-based approach for biomass prediction and grassland structure assessment in coastal meadows. Ecol. Indic. 2021, 122, 107227. [Google Scholar] [CrossRef]
- Valente, J.; Doldersum, M.; Roers, C.; Kooistra, L. Detecting Rumex Obtusifolius weed plants in grasslands from UAV RGB imagery using deep learning. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 4, 179–185. [Google Scholar] [CrossRef]
- Lam, O.H.Y.; Dogotari, M.; Prüm, M.; Vithlani, H.N.; Roers, C.; Melville, B.; Zimmer, F.; Becker, R. An open source workflow for weed mapping in native grassland using unmanned aerial vehicle: Using Rumex obtusifolius as a case study. Eur. J. Remote Sens. 2021, 54, 71–88. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef] [PubMed]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sens. Ecol. Conserv. 2020, 6, 472–486. [Google Scholar] [CrossRef]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef]
- Niedersächsisches Landesamt für Bergbau, Energie und Geologie (LBEG). Bodenübersichtskarte im Maßstab 1:50 000 (BÜK50); LBEG: Hannover, Germany, 1999. [Google Scholar]
- Meteostat. Belm. Available online: https://meteostat.net/en/station/D0342 (accessed on 7 October 2021).
- Reichelt, G.; Wilmanns, O. Vegetationsgeographie; Westermann: Braunschweig, Germany, 1973. [Google Scholar]
- Jäger, E.J. Rothmaler-Exkursionsflora von Deutschland. Gefäßpflanzen: Grundband, 21st ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Ellenberg, H.; Weber, H.E.; Düll, R.; Wirth, V.; Werner, W.; Paulißen, D. Zeigerwerte von Pflanzen in Mitteleuropa, 3, durch gesehene Aufl. Scr. Geobot. 2001, 18, 1–261. [Google Scholar]
- Dierschke, H.; Briemle, G. Kulturgrasland, 2nd ed.; Eugen Ulmer KG: Darmstadt, Germany, 2008. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Pawara, P.; Okafor, E.; Schomaker, L.; Wiering, M. Data augmentation for plant classification. In Proceedings of the International Conference on Advanced Concepts for Intelligent Vision Systems, Antwerp, Belgium, 18–21 September 2017; pp. 615–626. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.E.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.E.; Jackel, L.D. Handwritten digit recognition with a back-propagation network. In Proceedings of the Advances in Neural Information Processing Systems, Denver, CO, USA, 26–29 November 1990; pp. 396–404. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014, arXiv:1412.6806. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Zhang, Z.; Sabuncu, M. Generalized cross entropy loss for training deep neural networks with noisy labels. Adv. Neural Inf. Process. Syst. 2018, 31, 8778–8788. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Thakkar, V.; Tewary, S.; Chakraborty, C. Batch Normalization in Convolutional Neural Networks—A comparative study with CIFAR-10 data. In Proceedings of the 2018 5th International Conference on Emerging Applications of Information Technology (EAIT), West Bengal, India, 12–13 January 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Phaisangittisagul, E. An analysis of the regularization between L2 and dropout in single hidden layer neural network. In Proceedings of the 2016 7th International Conference on Intelligent Systems, Modelling and Simulation (ISMS), Bangkok, Thailand, 25–27 January 2016; pp. 174–179. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Elsäßer, M.; Engel, S.; Roßberg, R.; Thumm, U. Unkräuter im Grünland. Erkennen - Bewerten - Handeln, 2nd ed.; DLG-Verlag: Frankfurt am Main, Germany, 2018. [Google Scholar]
- Klapp, E.; Opitz von Boberfeld, W. Taschenbuch der Gräser, 12th ed.; Eugen Ulmer KG: Darmstadt, Germany, 2013. [Google Scholar]
- Stilmant, D.; Bodson, B.; Vrancken, C.; Losseau, C. Impact of cutting frequency on the vigour of Rumex obtusifolius. Grass Forage Sci. 2010, 65, 147–153. [Google Scholar] [CrossRef]
| Growth | ID | Date | Time of Flight | No. of Plots | Weather Conditions | Wind Speed |
|---|---|---|---|---|---|---|
| Growth 1 | 05/03/2021 | 10:58 a.m.–11:24 a.m. | 0 | closed cloud cover | 2 m/s | |
| Growth 1 | 05/12/2021 | 2:47 p.m.–3:11 p.m. | 30 | closed cloud cover | 5.7 m/s | |
| Growth 1 | 05/28/2021 | 1:59 p.m.–2:28 p.m. | 30 | sunny with a few clouds | 6.4 m/s | |
| Growth 1 | 06/08/2021 | 2:06 p.m.–2:30 p.m. | 29 | closed cloud cover | 2.9 m/s | |
| Growth 2 | 06/25/2021 | 12:45 a.m.–1:22 p.m. | 0 | sunny and cloudless | 3.9 m/s | |
| Growth 2 | 07/12/2021 | 11:22 a.m.–11:47 a.m. | 35 | sunny and cloudless | 1.6 m/s | |
| Growth 2 | 07/27/2021 | 11:22 a.m.–11:49 a.m. | 35 | first sunny, then cloudy | 2.2 m/s | |
| Growth 2 | 08/06/2021 | 11:13 a.m.–11:44 a.m. | 35 | closed cloud cover | 2.2 m/s |
| Layer | Parameter |
|---|---|
| Input | 64 × 64 × 5 |
| Conv2D_1 | Filter: 32, Kernel: 3 × 3, Strides: 2 × 2, Activation: ReLU |
| BatchNormalization | - |
| Dropout | 0.1 |
| Conv2D_2 | Filter: 128, Kernel: 3 × 3, Strides: 2 × 2, Activation: ReLU |
| BatchNormalization | - |
| Dropout | 0.1 |
| Reshape | - |
| FullyConnected_1 | Dense: 64, Activation: ReLU |
| BatchNormalization | - |
| Dropout | 0.2 |
| FullyConnected_2 | Dense: n, Activation: Softmax |
| Precision in % | OA | |||||||||
| Rumex obtusifolius plants | Lolium perenne-community | Alopecurus pratensis-community | Bromus hordeaceus-community | |||||||
| 87.72 | 100 | 97.98 | 81.81 | 99.00 | 80.00 | 96.51 | 83.33 | 97.06 | 82.75 | |
| 96.25 | 33.33 | 95.51 | 78.95 | 97.11 | 81.82 | 96.01 | 71.43 | |||
| 83.33 | 0.00 | 93.94 | 72.73 | 95.81 | 70.00 | 87.34 | 83.33 | 91.14 | 68.97 | |
| 96.62 | 100 | 95.12 | 94.11 | 97.68 | 86.67 | 95.72 | 88.57 | |||
| Recall in % | ||||||||||
| 98.04 | 71.42 | 96.37 | 100 | 94.75 | 75 | 98.04 | 100 | 97.06 | 82.75 | |
| 95.06 | 33.33 | 97.50 | 78.95 | 94.39 | 69.23 | 96.01 | 71.43 | |||
| 79.71 | 0.00 | 96.44 | 100 | 83.40 | 58.33 | 99.07 | 71.43 | 91.14 | 68.97 | |
| 98.85 | 66.67 | 97.82 | 84.21 | 92.32 | 100 | 95.72 | 88.57 | |||
| 97.71 | ||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pöttker, M.; Kiehl, K.; Jarmer, T.; Trautz, D. Convolutional Neural Network Maps Plant Communities in Semi-Natural Grasslands Using Multispectral Unmanned Aerial Vehicle Imagery. Remote Sens. 2023, 15, 1945. https://doi.org/10.3390/rs15071945
Pöttker M, Kiehl K, Jarmer T, Trautz D. Convolutional Neural Network Maps Plant Communities in Semi-Natural Grasslands Using Multispectral Unmanned Aerial Vehicle Imagery. Remote Sensing. 2023; 15(7):1945. https://doi.org/10.3390/rs15071945
Chicago/Turabian StylePöttker, Maren, Kathrin Kiehl, Thomas Jarmer, and Dieter Trautz. 2023. "Convolutional Neural Network Maps Plant Communities in Semi-Natural Grasslands Using Multispectral Unmanned Aerial Vehicle Imagery" Remote Sensing 15, no. 7: 1945. https://doi.org/10.3390/rs15071945
APA StylePöttker, M., Kiehl, K., Jarmer, T., & Trautz, D. (2023). Convolutional Neural Network Maps Plant Communities in Semi-Natural Grasslands Using Multispectral Unmanned Aerial Vehicle Imagery. Remote Sensing, 15(7), 1945. https://doi.org/10.3390/rs15071945








