Abstract
Mapping the spatial and temporal dynamics of species distributions is necessary for biodiversity conservation land-use planning decisions. Recent advances in remote sensing and machine learning have allowed for high-resolution species distribution modeling that can inform landscape-level decision-making. Here we compare the performance of three popular Sentinel-2 (10-m) land cover maps, including dynamic world (DW), European land cover (ELC10), and world cover (WC), in predicting wild bee species richness over southern Norway. The proportion of grassland habitat within 250 m (derived from the land cover maps), along with temperature and distance to sandy soils, were used as predictors in both Bayesian regularized neural network and random forest models. Models using grassland habitat from DW performed best (RMSE = 2.8 ± 0.03; average ± standard deviation across models), followed by ELC10 (RMSE = 2.85 ± 0.03) and WC (RMSE = 2.87 ± 0.02). All satellite-derived maps outperformed a manually mapped Norwegian land cover dataset called AR5 (RMSE = 3.02 ± 0.02). When validating the model predictions of bee species richness against citizen science data on solitary bee occurrences using generalized linear models, we found that ELC10 performed best (AIC = 2278 ± 4), followed by WC (AIC = 2367 ± 3), and DW (AIC = 2376 ± 3). While the differences in RMSE we observed between models were small, they may be significant when such models are used to prioritize grassland patches within a landscape for conservation subsidies or management policies. Partial dependencies in our models showed that increasing the proportion of grassland habitat is positively associated with wild bee species richness, thereby justifying bee conservation schemes that aim to enhance semi-natural grassland habitat. Our results confirm the utility of satellite-derived land cover maps in supporting high-resolution species distribution modeling and suggest there is scope to monitor changes in species distributions over time given the dense time series provided by products such as DW.
1. Introduction
The Anthropocene has heralded an unprecedented loss of biodiversity, primarily due to changes in land use and land cover, climate change, pollution, (over-)exploitation, and biological invasions [1]. In response, governments have established frameworks that address biodiversity loss, including the United Nations’ (UN) Sustainable Development Goals (SDGs, 2030 Agenda) as well as the Aichi biodiversity targets (Strategic Plan for Biodiversity 2011–2020) and the Post-2020 Global Biodiversity Framework of the Convention on Biological Diversity. Recently, the UN established the statistical standards for ecosystem accounting (EA) under the System of Environmental–Economic Accounting (SEEA), which require countries to account for changes in ecosystems over time [2]. To achieve the SDGs, meet biodiversity conservation targets, and to account for ecosystem changes, we require monitoring and evaluation tools that are both globally available and locally relevant [3]. One such tool is species distribution modeling, also known as ecological niche or habitat modeling, which has been widely used to inform decision-making in conservation planning [4].
By modeling and mapping the distribution of species over space and time, we are able to make data-driven decisions about which areas to prioritize for restoration or conservation [5]. The predictive and explanatory power of species distribution models also allows for identifying critical environmental variables (e.g., precipitation, habitat availability) that drive species communities [6]. Insect conservation strategies are a good case in point, whereby mapping and monitoring of insect species richness are prioritized both at the international [7,8] and national levels [9,10]. In Norway, field surveys of bee diversity have been combined with habitat and climate models to create prediction maps that can help determine where wild bee habitat enhancement schemes can be most efficient [11]. However, such priority maps and many species distribution models are currently based on static environmental predictor variables such as land cover maps, whereas management requires more flexible solutions that can detect temporal dynamics in ecosystem conditions and species distributions over time [12].
Recent advances in satellite remote sensing and earth observation have filled data gaps and improved the spatio-temporal transferability of species distribution models [13]. As such, satellite-derived products can capture the environmental processes that underlie the distribution of biodiversity, such as vegetation productivity, water availability, temperature, and perhaps most importantly, land use and land cover. The Sentinel satellites under the Copernicus Programme have been used to produce annually updatable regional and global land cover maps at 10 m resolution, including ELC10 [14], Dynamic World (DW) [15], and World Cover 2020 (WC) [16]. All three products have the capacity to be multi-temporal, but only DW is operationally delivering near real-time land cover maps as new Sentinel-2 scenes become available (every 2–5 days). Due to the novelty of freely available, medium-resolution, high-frequency land cover maps, their use in species distribution modeling is still in its infancy. It is also not clear whether global Sentinel-based land cover maps can replace or improve upon the contribution of regional land cover datasets [17], given that regional maps produced by national mapping agencies through manual methods such as photogrammetry are often more precise and tailored to local conditions.
The aim of this study was to compare Sentinel-based, 10-meter-resolution land cover maps in their ability to predict wild bee species richness distributions across gradients in temperature and habitat availability. To do so, we use a Norwegian land cover dataset called AR5 [18] as a benchmark to evaluate three satellite-based maps, including DW, WC, and ELC10, that are annually updatable. The proportion of semi-natural grassland habitat within 250 m derived from the land cover maps is used as a predictor variable in species distribution models that predict the richness of solitary bee species in southern Norway.
2. Methods
To compare the utility of Sentinel-based land cover maps in species distribution modeling, we chose wild bee species as a model system. We do this for three main reasons, which are elaborated on below: (1) bees are keystone species in grassy ecosystems globally that are good indicators for ecosystem condition; (2) they are experiencing significant local declines in population numbers; and (3) they have a limited home range and are dependent on local resources for survival, so we expect their distribution to respond strongly to landscape-scale land use gradients in the Sentinel-based land cover maps.
Insects in general and wild bees in particular are key components of many ecosystems and provide important contributions to people [19,20]. For example, the economic value of pollinating insects is considerable, estimated at EUR 153 billion globally [21]. Although pollination is often associated with domestic insects such as the honey bee (Apis mellifera), a large and diverse community of wild bee species contributes significantly to the pollination of wild plants and crops [22,23]. Insect abundance, biomass [24], and diversity [25] are declining in some regions due to urbanization, deforestation, climate change, pesticide use, and invasive species [26]. The same is true for bee species [27]. Although the diversity of wild bees is predominantly driven by temperature at global and regional scales, habitat and landscape characteristics are important drivers at local scales [28,29]. Bees are central place foragers that travel back and forth from a nesting site to collect resources [30], and therefore, the abundance and diversity of wild bees correlate to the diversity and abundance of floral resources (vascular plants) in the landscape [28,31]. Land cover types such as semi-natural grasslands, including hay and flower meadows, provide ample floral [32] and non-floral resources to wild bees [33]. Non-floral resources include nesting sites, and although some wild bee species nest in dead wood, the majority use sandy soil sediments as nest sites [34]. Consequently, wild bee diversity can be predicted by the availability of suitable land cover types for resources (i.e., semi-natural grasslands) and for nests (i.e., sandy soils), in combination with climatic variables [29].
2.1. Solitary Bee Surveys
We surveyed solitary bee communities in 72 traditionally managed (mowed) semi-natural grasslands distributed along a climatic gradient from south-eastern Norway to mid-Norway (Figure 1A). In 2019, we sampled 32 semi-natural grasslands in south-eastern Norway [11], adding another 20 study sites in 2020 [29]. To capture potential influences of climatic conditions on solitary bee diversity at regional scales, solitary bee communities were sampled in another set of 20 semi-natural grasslands in mid-Norway in 2021. All surveys were conducted using pan traps, which are an efficient method for surveying wild bee communities [35], in particular when the aim is to survey the solitary as opposed to social bee species [36]. Each pan trap consisted of three white plastic soup bowls, coated with fluorescent yellow or blue, or left white, mounted on a fence pole at the height of the surrounding vegetation [11,29]. We deployed 3 traps per site in 2019 and 2 traps per site for sites sampled in 2020 and 2021. The number of traps per site was reduced from 3 to 2 after 2019 to allow more sites to be sampled. This resulted in 176 samples (the sum of traps across all sites) distributed across 72 study sites. Within sites, traps were always placed at least 20 m apart to avoid inter-trap competition [37].
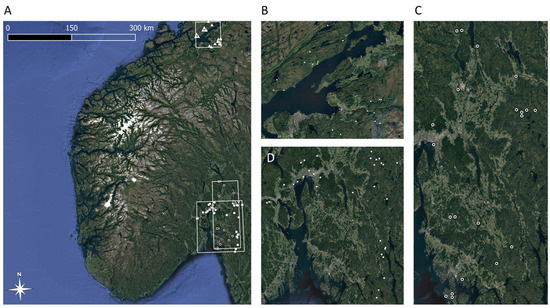
Figure 1.
Extent of study area with solitary bee sampling locations and individual survey extents (A). Location of study sites in Trøndelag County, sampled in 2021 (B). Location of study sites in Oslo, Viken, and Innlandet counties, sampled in 2020 (C). Locations of study sites in Oslo, Viken, and Innlandet, sampled in 2019 (D).
In all years, sites were sampled in May, June, and July at similar times of day and on days with weather conditions that are optimal for bee activity (i.e., low wind and temperatures above 15 °C). For each sampling period, we activated the pan traps at a site by mounting the bowls and filling them with water and a drop of detergent. Sampled bee specimens were collected after 48 h. In 2019, sampling was initiated on 13 May, 21 June, 9 July, and 23 July [11]. In 2020, sampling was initiated on 13 May, 25 May, 14 June, and 16 July [29]. In 2021, sampling began on 29 May, 22 June, and 24 July. Collected bees were stored in 96% laboratory ethanol prior to pinning and identification. Voucher specimens are stored in the entomological collections at the Norwegian Institute for Nature Research. We tallied the number of solitary bee species sampled per trap across the season.
2.2. Land Cover Maps
Data pre-processing and extraction of land cover maps took place in the Google Earth Engine [38]. The AR5 map obtained from the Norwegian Institute of Bioeconomy Research [18] is provided as a vector map at 1:5000 scale, which we rasterized at 5 m resolution. AR5 is updated every 5–8 years and therefore represents a mosaic of years across the country. WC and ELC10 are available in Google Earth Engine for the years 2020 and 2018, respectively. However, DW is provided as a collection of classified Sentinel-2 images with less than 35% cloud cover. To generate an annual land cover composite for 2020 comparable with WC and ELC10, we calculated the mode predicted land cover class in the image band named “label” across all DW images during June, July, and August. For all land cover maps, we used a focal mean function to calculate the proportion of grassland habitat within 250 m of each pixel in the study area (Figure 1) at 10 m resolution. We used the 250-m radius because solitary bee diversity has been shown to respond strongly to habitat availability at this spatial scale [39]. To isolate grassland pixels, we used the “grassland” class from WC and ELC10 and the “grass” class from DW, which are both defined as areas dominated by natural herbaceous vegetation, including grasslands, prairies, steppes, savannahs, and pastures. For AR5, we used the “innmarksbeite” and “åpent fastmark” classes, which are defined as open ecosystems dominated by herbaceous vegetation and often used for extensive grazing [18]. We used a radius of 250 m as solitary bee species richness has previously been shown to respond to habitat area at this spatial scale [39].
2.3. Modeling
Data modeling and visualization were performed in R [40]. As in [29], we included predictor variables related to climatic conditions, habitat availability, and distances to high-quality nesting substrates for below ground nesting bees, which account for the vast majority of solitary bees. In [29], the spatial variation in climatic conditions was estimated using a digital elevation model (DEM) together with latitude. Here, we used the average temperature for the warmest quarter (June, July, and August), during the current 30-year climate reference period (1990–2021), calculated from daily estimates and interpolated station measurements at a 1 km resolution from the Norwegian Meteorological Institute’s database [41]. We used the average temperature during the warmest quarter instead of the annual mean temperature because high winter temperatures along the coast result in annual mean temperatures not reflecting the gradient in ambient temperature experienced by bees during their main activity periods in Norway (spring to autumn). Using modeled climate data instead of DEMs to estimate the effects of temperature on bee diversity enables one to project changes in bee diversity as a function of future climate scenarios. In [29] we used a habitat suitability model to estimate the potential habitat area surrounding each pan trap at a 60-m radius. In contrast, in the current study, we use the proportion of pixels identified as “grassland” by satellite-derived land cover maps as estimates of habitat availability within a 250-m radius surrounding each trap. A benefit of using satellite-derived grassland classifications instead of estimates of habitat suitability maps that may partly rely on existing land cover products is that one reduces the number of modeling steps required to produce maps of pollinator diversity from remote sensing data. In addition, we used the geographic distance to areas with soils mapped as having a high water infiltration capacity by the Norwegian geological survey [42], as such areas are typically located on sandy soils, which is the preferred nesting substrate for the majority of Norwegian soil nesting bees.
We ran and compared eight models of solitary bee species richness that all followed the general formula:
Solitary bee species richness (survey year + average temperature during the warmest quarter + Distance to sandy soils + proportion of grassland within a 250-m radius).
The eight models differed in terms of the data source (map) used to estimate the proportion of grassland around each pan trap and the type of model used to predict solitary bee species richness. The survey year was included as a categorical variable to account for inter-annual variation in bee species richness as well as for annual differences in climatic conditions, which could influence the number of species sampled in a trap. For each land cover map, we trained a Random Forest (RF) regression model [43] and a Bayesian Regularized Neural Network (BRNN) model [44] in Caret [45] in R. BRNN models were tuned by selecting the number of neurons (1, 2, or 3) that resulted in the lowest root mean square error (RMSE) following 25 bootstrap resamples of the training data. RF models were tuned by selecting the mtry (number of parameters tested at each node) and split-rule variance or extra trees that resulted in the lowest RMSE, following 25 bootstrap resamples. We adopted leave-one-out cross-validation (LOOCV) to assess model predictive performance due to the small number of study sites. Small training datasets can result in large variances in model performance when using traditional training-testing splits (e.g., 70% training and 30% testing) before model fitting, compared to LOOCV [46]. In LOOCV, each model was iteratively trained and tuned on data from 71 study sites and then used to predict solitary bee species richness in pan traps from the one held-out site. Predictive performance is calculated across all LOOCV iterations. Following this nested cross-validation procedure ensures independence between the data used for tuning the models and the data used for final validations.
For each land cover-map model prediction, we evaluated the predictive power in terms of the Pearson correlation coefficient (Cor), the root-mean-square deviation (RMSE), and the mean absolute error (MAE) between observed values of solitary bee species richness and the solitary bee species richness predicted by the model. As an alternative form of model validation, we also tested the ability of model predictions of bee species richness to predict the variance in occurrence of solidary bee records obtained from citizen science data. We first downloaded all post-2015 solitary bee species observations from the Global Information Biodiversity Facility (GBIF) that intersected our study area (xmin: 9.836, xmax: 12.719, ymin: 58.909, ymax: 63.962) and had a GPS error of less than 50 m (n = 2111). To reduce spatial bias in the data—i.e., that some areas have been surveyed more frequently or intensively than others—we only included one occurrence per square kilometer. We randomly sampled 10,000 pseudo-absences from within our study area and used binomial generalized linear models to quantify how well the predicted species richness scores explained the variance in the GBIF presence-absence data. We used the Akaike information criterion (AIC) to compare the explanatory capacity of the different models (δAIC > 2). Finally, we used partial dependency plots through the R-package pdp [47] as a means to visualize the estimated marginal effect of the proportion of grassland, using the different land cover maps, on solitary bee species richness.
In summary, our modeling workflow included 16 unique models for combinations of reference data (survey, GBIF), model type (RF, BRNN), and land cover dataset (AR5, DW, ELC10, WC). Each model was iterated 100 times in order to estimate the mean and standard deviation in model performance metrics.
3. Results
We did not find that models using the proportion of grassland estimated from the vector-based Norwegian land cover map (AR5) outperformed models where grassland had been estimated from satellite-derived land use models with 10 m resolution. Specifically, satellite-derived maps exhibited an average RMSE of 2.87 ± 0.03 (±standard deviation), whereas the Norwegian AR5 map produced models with a RMSE of 3.02 ± 0.02 (Figure 2). Similarly, for the AR5-based BRNN models, the correlation coefficient between observed and predicted solitary bee species richness (SR) was slightly lower (Figure 2A) than for the satellite-based models (Figure 2B–D). This was also the case for the Random Forest (RF) models (Figure 2E–H).
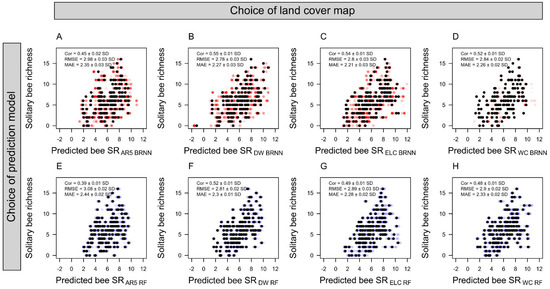
Figure 2.
Performance of Bayesian regularized neural network (BRNN) and random forest (RF) models for predicting solitary bee species richness (SR). Data points represent model predictions against observed SR following a leave-one-out cross-validation procedure. The average and standard deviation of correlation coefficients (Cor), root-mean and mean-absolute error (RMSE, MAE) from 100 iterations of each leave-one-out cross validation are reported for models trained with land cover data from (A,E) a Norwegian map (AR5), (B,F) dynamic world (DW), (C,G) European land cover (ELC10), and (D,H) world cover (WC). Red and blue points show the predicted and observed solitary bee species richness for each pan trap across the 100 leave-one-out iterations, and black points show the average predicted bee species richness and the associated observed species richness for each specific trap.
Among the satellite-derived models (Figure 2B–D,F–H), there were only marginal differences in their performances as predictors of solitary bee SR. Models using grassland habitat from DW performed best (RMSE = 2.8 ± 0.03; averaged across RF and BRNN models), followed by ELC10 (RMSE = 2.85 ± 0.03) and WC (RMSE = 2.87 ± 0.02). Grassland habitat was ranked the third most important predictor variable in BRNN and RF models (Figure 3). The mean summer temperature and sampling year were the most important predictors in the models. All models, independent of data source or model type, produced partial dependence plots that corroborated a positive association between grassland habitat and solitary bee SR (Figure 4). The association was less distinct in the AR5 model (Figure 4A) compared to the satellite-based models (Figure 4B–D).
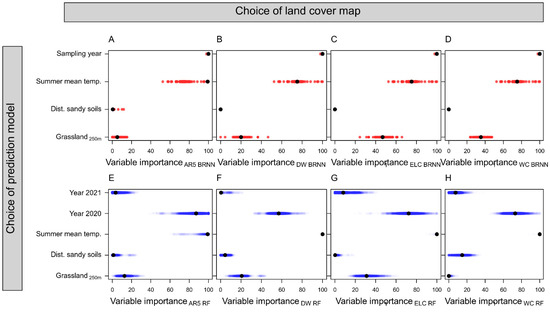
Figure 3.
Variable importance plots showing the scaled relative importance of sampling year (3 levels), summer mean temperature, distance to sandy soils, and grassland proportion within 250 m for predicting solitary bee species richness. For the sampling year, the year 2019 was used as the reference year and does therefore not appear in the figures. Variable importance is derived from Bayesian regularized neural network (BRNN; A–D) and random forest (RF; E–H) models for predicting solitary bee species richness (SR). Models were trained with land cover data from (A,E) a Norwegian map (AR5), (B,F) dynamic world (DW), (C,G) European land cover (ELC10), and (D,H) world cover (WC).
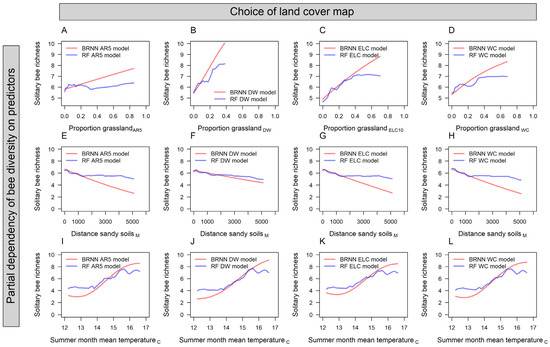
Figure 4.
Partial dependence plots from Bayesian regularized neural network (BRNN) and random forest (RF) models of bee species richness. Partial dependencies are shown for the spatial predictors: proportion of grassland habitat within 250 m, derived from (A) a Norwegian map (AR5), (B) dynamic world (DW), (C) European land cover (ELC), and (D) world cover (WC); (E–H) distance to sandy soils; and (I–L) mean temperature during the summer months.
When validating the model predictions of bee SR against citizen science data on solitary bee occurrences using generalized linear models (Figure 5), we found that the order of performance was changed. We found that ELC10 performed best (AIC = 2278 ± 4), followed by WC (AIC = 2367 ± 3), and DW (AIC = 2376 ± 3). The BRNN models performed slightly better than RF models, both in terms of leave-one-out cross-validation (Figure 2) and in terms of explaining the occurrence of solitary bees from GBIF (Figure 5). Therefore, we used the BRNN models to generate wall-to-wall prediction maps of bee SR over the study region (Figure 6). A qualitative visual comparison shows that the broad-scale spatial patterns of bee SR predictions are similar between models, with bee SR increasing with north-south temperature gradients and along populated valleys where extensive grazing practices have established semi-natural grassland patches. At the landscape scale (Figure 7), AR5 predictions show less spatial variation than the satellite-derived maps. All models appear to pick up the grassland habitat adjacent to the runways at Gardermoen International Airport; however, ELC10 appears to pick up the most habitat in the agricultural landscapes southwest of the airport (Figure 7C).
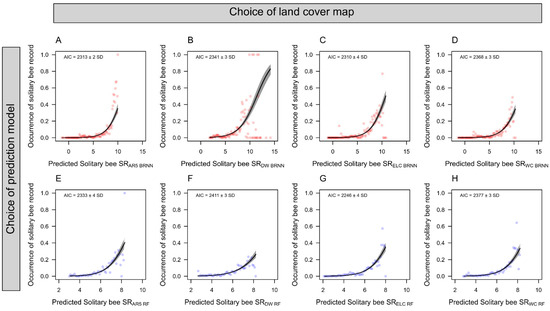
Figure 5.
Explanatory power of bee species richness (SR) predictions from Bayesian regularized neural network (BRNN) and random forest (RF) models. Effects plots are shown along with AIC scores from binomial generalized linear models of solitary bee species occurrence data from GBIF as a function of predicted bee SR from models trained with land cover data from (A,E) a Norwegian map (AR5), (B,F) dynamic world (DW), (C,G) European land cover (ELC10), and (D,H) world cover (WC). Points in the figures show the mean occurrence of solitary bee records calculated within bins of one decimal. Individual GLMs were run for each of the 100 spatially filtered datasets of solitary bee records in order to calculate the average AIC and its standard deviation.
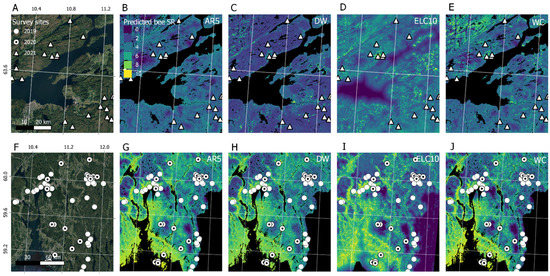
Figure 6.
Solitary bee species richness (SR) prediction maps for Trøndelag county (A–E) and Oslo and Innlandet county (F–J) from Bayesian regularized neural network (BRNN) models trained on grassland habitat data from (B,G) a Norwegian map (AR5), (C,H) dynamic world (DW), (D,I) European land cover (ELC10), and (E,J) world cover (WC). Each map is overlaid with the bee survey sites.
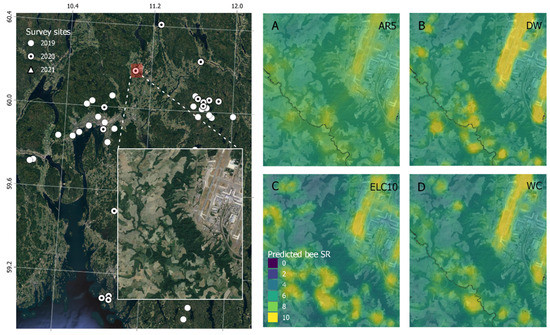
Figure 7.
Solitary bee species richness (SR) prediction maps over the landscape surrounding Gardermoen international airport (left panel) from Bayesian regularized neural network (BRNN) models trained on grassland habitat data from (A) a Norwegian map (AR5), (B) dynamic world (DW), (C) European land cover (ELC10), and (D) world cover (WC).
4. Discussion
Species distribution modeling that incorporates high-resolution satellite data is still in its infancy [13], yet the application to solitary bee species richness presented in this study confirms its potential. Our results show that the Sentinel satellite-based land cover maps outperformed a regional manually digitized land cover map over southern Norway (AR5) in predicting solitary bee species richness (Figure 2). While the use of satellite imagery to map vegetation types or individual forest species is common [48], using derived products to predict habitat suitability for animal or plant species that are not directly visible in satellite images is far less common [11,49]. Here we show that the availability of grassland habitat within 250 m, measured from satellite data, is positively associated with solitary bee species richness (Figure 4) and is therefore predictive of solitary bee occurrences at regional scales (Figure 5).
We found larger differences in the predictive capacity of grassland habitat derived from manually mapped versus satellite-derived land cover maps than differences between the satellite-based maps themselves (Figure 2). The AR5 map uses a minimum mapping unit of 2000 m2, which results in very small grassland fragments being subsumed into a broader land cover class [18]. For example, road verges or small urban parks will be classified as “built” in the AR5 maps. However, such small grassland patches can harbor significant floral resources for bees, and the fact that AR5 does not map these areas may therefore be why AR5 was less predictive of species richness than satellite-based maps. Furthermore, AR5 is not as up-to-date as the satellite-based maps and may misrepresent the conditions on the ground during 2019, 2020, and 2021, when the field surveys were conducted. In contrast, ELC10 and WC use a minimum mapping unit of 100 m2, while DW uses 2500 m2. At the landscape scale, it is evident that predictions of bee species richness with ELC10 were more spatially heterogeneous than AR5 or DW (Figure 6), probably due to its smaller minimum mapping unit and ability to detect smaller grassland patches. This may have contributed to its greater predictive capacity compared to AR5. This also explains why, when validating our models using citizen science data from GBIF, ELC10 outperformed DW. In contrast to the survey dataset, GBIF data are spatially clustered and biased toward urban landscapes, which are easily accessible but also have complex landscapes. Due to ELC10’s small minimum mapping unit, it captures the landscape complexity more than DW does and is able to predict the GBIF bee SR better.
The accuracies of the solitary bee species richness models presented here are arguably high enough to make data-driven management decisions at the landscape scale. The average RMSE of 2.87 means that one can at least distinguish very species-rich areas (maximum species richness of 16 in our study) from species-poor areas (zero species). The differences in model accuracy between satellite-based grassland maps were marginal (RMSE difference of 0.04); however, when visualized at a landscape scale (Figure 6), small nuances may have important implications for management and decision-making. For instance, a ranking or prioritization of grassland patches for receiving conservation subsidies based on the maps in Figure 6 may yield different results depending on the data source used. Therefore, post-stratified accuracy assessment of species distribution models in specific landscapes may be necessary before they can be adopted in practice [50].
Based on several limitations identified in our study, we outline avenues for further research on the integration of high-resolution satellite data in species distribution modeling. Firstly, it is not necessary to use derived products such as land cover maps, as we have carried out here. Instead, one can use the spectral signatures themselves from a satellite image to calibrate distribution models [13], although this would not allow ecologists or policymakers to relate species-rich areas to specific land use types. Secondly, maps with a higher thematic resolution than those used in this study would produce more detailed species distribution maps [51]. For example, all four maps tested here contained a single broad category for grassland, without distinguishing between intensively managed grasslands and extensively managed grasslands, such as the mowed meadows from which we sampled solitary bees [11,29]. Therefore, measuring aspects of ecosystem condition or use, such as grassland use intensity [52], might further improve the accuracy of species distribution models. Thirdly, we did not explore how accurately satellite-based prediction models can detect real changes in species distributions over time because we did not implement a field survey design that was comprehensive enough to capture changes in species ranges over time. However, we know from earlier studies that changes in land cover that are detectable from space are direct drivers of species range shifts [53]. To this end, DW is probably the most suitable Sentinel-based land cover dataset to quantify land cover and use dynamics [54] because of its continuous updates and delivery and would be well-suited to such dynamic species distribution modeling. This also strengthens the call for investment in long-term biodiversity monitoring programs so that satellite-based distribution models can be calibrated and validated with in situ data [55].
5. Conclusions
The proliferation of high-resolution earth observation data and derived land cover products provides scope for mapping biodiversity distributions with models that are both locally relevant for decision making and scalable to the globe. Here we found that globally available Sentinel-based land cover maps can improve upon manually digitized regional land cover maps for predicting the richness of solitary bee species in southern Norway. The differences in predictive performance between DW, WC, and ELC10 were marginal; however, at the landscape scale, the smaller minimum mapping units of WC and ELC10 allow them to resolve smaller habitat patches, which are reflected in the landscape variations in predicted species richness. Furthermore, the rich time series provided by maps such as DW (from 2015 to present) offer unique opportunities to model short-term changes in species distributions in response to land use changes if paired with in-situ temporal monitoring data. We conclude that the use of satellite-derived land cover maps can facilitate high-resolution species distribution models that can guide decision-making relevant to landscape ecology. To this end, future modeling efforts should be aimed at those species that perform key roles in ecosystems, are indicators of ecosystem status, and support nature’s contribution to people.
Author Contributions
Conceptualization, M.A.K.S., Z.S.V. and R.E.R.; methodology, M.A.K.S., Z.S.V. and R.E.R.; formal analysis, M.A.K.S., Z.S.V. and R.E.R.; data curation, M.A.K.S., G.M.K. and Z.S.V.; writing—original draft preparation, Z.S.V., R.E.R. and M.A.K.S.; writing—review and editing, Z.S.V., R.E.R., M.A.K.S., G.M.K., G.M.R. and M.S.N.; project administration, M.A.K.S.; funding acquisition, M.A.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
Insect sampling from semi-natural grasslands in 2019 and 2020 was funded by the Norwegian Agricultural Agency (Klima-og Miljøprogrammet: POLLILAND, grant number 2018/72806). Sampling in 2021 was financed by The Research Council of Norway, project no. 160022/F40 NINA basic funding. This study was financed by the Norwegian Agricultural Agency (Klima- og Miljøprogrammet: POLLILAND-MIDT, grant number 2021/40219).
Data Availability Statement
The data and code to reproduce this analysis are archived on Zenodo: 10.5281/zenodo.7762665.
Acknowledgments
We thank Mikaela E.G.P. Olsen, Solveig Haug, Jonas Lystrup Andresen, April McKay, Stian Brønner, and Ida Elise Løvall Rastad for operating the traps installed in the semi-natural grasslands.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johnson, C.N.; Balmford, A.; Brook, B.W.; Buettel, J.C.; Galetti, M.; Guangchun, L.; Wilmshurst, J.M. Biodiversity Losses and Conservation Responses in the Anthropocene. Science 2017, 356, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Edens, B.; Maes, J.; Hein, L.; Obst, C.; Siikamaki, J.; Schenau, S.; Javorsek, M.; Chow, J.; Chan, J.Y.; Steurer, A.; et al. Establishing the SEEA Ecosystem Accounting as a Global Standard. Ecosyst. Serv. 2022, 54, 101413. [Google Scholar] [CrossRef]
- Schmeller, D.S.; Böhm, M.; Arvanitidis, C.; Barber-Meyer, S.; Brummitt, N.; Chandler, M.; Chatzinikolaou, E.; Costello, M.J.; Ding, H.; García-Moreno, J.; et al. Building Capacity in Biodiversity Monitoring at the Global Scale. Biodivers. Conserv. 2017, 26, 2765–2790. [Google Scholar] [CrossRef]
- Villero, D.; Pla, M.; Camps, D.; Ruiz-Olmo, J.; Brotons, L. Integrating Species Distribution Modelling into Decision-Making to Inform Conservation Actions. Biodivers. Conserv. 2017, 26, 251–271. [Google Scholar] [CrossRef]
- Guisan, A.; Tingley, R.; Baumgartner, J.B.; Naujokaitis-Lewis, I.; Sutcliffe, P.R.; Tulloch, A.I.T.; Regan, T.J.; Brotons, L.; McDonald-Madden, E.; Mantyka-Pringle, C.; et al. Predicting Species Distributions for Conservation Decisions. Ecol. Lett. 2013, 16, 1424–1435. [Google Scholar] [CrossRef]
- McShea, W.J. What Are the Roles of Species Distribution Models in Conservation Planning? Environ. Conserv. 2014, 41, 93–96. [Google Scholar] [CrossRef]
- Harvey, J.A.; Heinen, R.; Armbrecht, I.; Basset, Y.; Baxter-Gilbert, J.H.; Bezemer, T.M.; Böhm, M.; Bommarco, R.; Borges, P.A.; Cardoso, P. International Scientists Formulate a Roadmap for Insect Conservation and Recovery. Nat. Ecol. Evol. 2020, 4, 174–176. [Google Scholar] [CrossRef]
- Senapathi, D.; Goddard, M.A.; Kunin, W.E.; Baldock, K.C. Landscape Impacts on Pollinator Communities in Temperate Systems: Evidence and Knowledge Gaps. Funct. Ecol. 2017, 31, 26–37. [Google Scholar] [CrossRef]
- Norwegian Ministries. National Pollinator Strategy A Strategy for Viable Populations of Wild Bees and Other Pollinating Insects; Norwegian Government Security and Service Organisation: Oslo, Norway, 2018; p. 67.
- IPBES. The Assessment Report of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services on Pollinators, Pollination and Food Production; Potts, S.G., Imperatriz-Fonseca, V.L., Ngo, H.T., Eds.; Secretariat of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services: Bonn, Germany, 2016; p. 552. [Google Scholar] [CrossRef]
- Sydenham, M.A.K.; Venter, Z.S.; Eldegard, K.; Moe, S.R.; Steinert, M.; Staverløkk, A.; Dahle, S.; Skoog, D.I.J.; Hanevik, K.A.; Skrindo, A.; et al. High Resolution Prediction Maps of Solitary Bee Diversity Can Guide Conservation Measures. Landsc. Urban Plan. 2022, 217, 104267. [Google Scholar] [CrossRef]
- Zurell, D.; Thuiller, W.; Pagel, J.; Cabral, J.S.; Münkemüller, T.; Gravel, D.; Dullinger, S.; Normand, S.; Schiffers, K.H.; Moore, K.A.; et al. Benchmarking Novel Approaches for Modelling Species Range Dynamics. Glob. Change Biol. 2016, 22, 2651–2664. [Google Scholar] [CrossRef]
- Randin, C.F.; Ashcroft, M.B.; Bolliger, J.; Cavender-Bares, J.; Coops, N.C.; Dullinger, S.; Dirnböck, T.; Eckert, S.; Ellis, E.; Fernández, N.; et al. Monitoring Biodiversity in the Anthropocene Using Remote Sensing in Species Distribution Models. Remote Sens. Environ. 2020, 239, 111626. [Google Scholar] [CrossRef]
- Venter, Z.S.; Sydenham, M.A.K. Continental-Scale Land Cover Mapping at 10 m Resolution Over Europe (ELC10). Remote Sens. 2021, 13, 2301. [Google Scholar] [CrossRef]
- Brown, C.F.; Brumby, S.P.; Guzder-Williams, B.; Birch, T.; Hyde, S.B.; Mazzariello, J.; Czerwinski, W.; Pasquarella, V.J.; Haertel, R.; Ilyushchenko, S.; et al. Dynamic World, Near Real-Time Global 10 m Land Use Land Cover Mapping. Sci. Data 2022, 9, 251. [Google Scholar] [CrossRef]
- Zanaga, D.; Van De Kerchove, R.; De Keersmaecker, W.; Souverijns, N.; Brockmann, C.; Quast, R.; Wevers, J.; Grosu, A.; Paccini, A.; Vergnaud, S.; et al. ESA WorldCover 10 m 2020 V100. Zenodo 2021. [Google Scholar] [CrossRef]
- Tulbure, M.G.; Hostert, P.; Kuemmerle, T.; Broich, M. Regional Matters: On the Usefulness of Regional Land-Cover Datasets in Times of Global Change. Remote Sens. Ecol. Conserv. 2022, 8, 272–283. [Google Scholar] [CrossRef]
- Bjørdal, I.; Bjørkelo, K. AR5 Klassifikasjonssystem: Klassifikasjon Av Arealressurser. In Håndbok Fra Skog Og Landskap; The Norwegian Institute of Bioeconomy Research: Ås, Norway, 2006. [Google Scholar]
- Noriega, J.A.; Hortal, J.; Azcárate, F.M.; Berg, M.P.; Bonada, N.; Briones, M.J.I.; Del Toro, I.; Goulson, D.; Ibanez, S.; Landis, D.A.; et al. Research Trends in Ecosystem Services Provided by Insects. Basic Appl. Ecol. 2018, 26, 8–23. [Google Scholar] [CrossRef]
- Prather, C.M.; Pelini, S.L.; Laws, A.; Rivest, E.; Woltz, M.; Bloch, C.P.; Del Toro, I.; Ho, C.; Kominoski, J.; Newbold, T.S. Invertebrates, Ecosystem Services and Climate Change. Biol. Rev. 2013, 88, 327–348. [Google Scholar] [CrossRef]
- Gallai, N.; Salles, J.-M.; Settele, J.; Vaissière, B.E. Economic Valuation of the Vulnerability of World Agriculture Confronted with Pollinator Decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Losey, J.E.; Vaughan, M. The Economic Value of Ecological Services Provided by Insects. Bioscience 2006, 56, 311–323. [Google Scholar] [CrossRef]
- Smith, T.J.; Saunders, M.E. Honey Bees: The Queens of Mass Media, despite Minority Rule among Insect Pollinators. Insect Conserv. Divers. 2016, 9, 384–390. [Google Scholar] [CrossRef]
- Hallmann, C.A.; Sorg, M.; Jongejans, E.; Siepel, H.; Hofland, N.; Schwan, H.; Stenmans, W.; Müller, A.; Sumser, H.; Hörren, T. More than 75 Percent Decline over 27 Years in Total Flying Insect Biomass in Protected Areas. PLoS ONE 2017, 12, e0185809. [Google Scholar] [CrossRef] [PubMed]
- Seibold, S.; Gossner, M.M.; Simons, N.K.; Blüthgen, N.; Müller, J.; Ambarlı, D.; Ammer, C.; Bauhus, J.; Fischer, M.; Habel, J.C. Arthropod Decline in Grasslands and Forests Is Associated with Landscape-Level Drivers. Nature 2019, 574, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.L.; Grames, E.M.; Forister, M.L.; Berenbaum, M.R.; Stopak, D. Insect Decline in the Anthropocene: Death by a Thousand Cuts. Proc. Natl. Acad. Sci. USA 2021, 118, e2023989118. [Google Scholar] [CrossRef] [PubMed]
- Zattara, E.E.; Aizen, M.A. Worldwide Occurrence Records Suggest a Global Decline in Bee Species Richness. One Earth 2021, 4, 114–123. [Google Scholar] [CrossRef]
- Orr, M.C.; Hughes, A.C.; Chesters, D.; Pickering, J.; Zhu, C.-D.; Ascher, J.S. Global Patterns and Drivers of Bee Distribution. Curr. Biol. 2021, 31, 451–458. [Google Scholar] [CrossRef]
- Sydenham, M.A.K.; Eldegard, K.; Venter, Z.S.; Evju, M.; Åström, J.; Rusch, G.M. Priority Maps for Pollinator Habitat Enhancement Schemes in Semi-Natural Grasslands. Landsc. Urban Plan. 2022, 220, 104354. [Google Scholar] [CrossRef]
- Westrich, P. Habitat Requirements of Central European Bees and the Problems of Partial Habitats; Academic Press Limited: Cambridge, MA, USA, 1996; Volume 18, pp. 1–16. [Google Scholar]
- Woodard, S.H.; Jha, S. Wild Bee Nutritional Ecology: Predicting Pollinator Population Dynamics, Movement, and Services from Floral Resources. Curr. Opin. Insect Sci. 2017, 21, 83–90. [Google Scholar] [CrossRef]
- Carrié, R.; Lopes, M.; Ouin, A.; Andrieu, E. Bee Diversity in Crop Fields Is Influenced by Remotely-Sensed Nesting Resources in Surrounding Permanent Grasslands. Ecol. Indic. 2018, 90, 606–614. [Google Scholar] [CrossRef]
- Requier, F.; Leonhardt, S.D. Beyond Flowers: Including Non-Floral Resources in Bee Conservation Schemes. J. Insect Conserv. 2020, 24, 5–16. [Google Scholar] [CrossRef]
- Antoine, C.M.; Forrest, J.R. Nesting Habitat of Ground-nesting Bees: A Review. Ecol. Entomol. 2021, 46, 143–159. [Google Scholar] [CrossRef]
- O’Connor, R.S.; Kunin, W.E.; Garratt, M.P.; Potts, S.G.; Roy, H.E.; Andrews, C.; Jones, C.M.; Peyton, J.M.; Savage, J.; Harvey, M.C. Monitoring Insect Pollinators and Flower Visitation: The Effectiveness and Feasibility of Different Survey Methods. Methods Ecol. Evol. 2019, 10, 2129–2140. [Google Scholar] [CrossRef]
- Hutchinson, L.A.; Oliver, T.H.; Breeze, T.D.; O’Connor, R.S.; Potts, S.G.; Roberts, S.P.; Garratt, M.P. Inventorying and Monitoring Crop Pollinating Bees: Evaluating the Effectiveness of Common Sampling Methods. Insect Conserv. Divers. 2022, 15, 299–311. [Google Scholar] [CrossRef]
- Droege, S.; Tepedino, V.J.; LeBuhn, G.; Link, W.; Minckley, R.L.; Chen, Q.; Conrad, C. Spatial Patterns of Bee Captures in North American Bowl Trapping Surveys. Insect Conserv. Divers. 2010, 3, 15–23. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Steffan-Dewenter, I.; Münzenberg, U.; Bürger, C.; Thies, C.; Tscharntke, T. Scale-dependent Effects of Landscape Context on Three Pollinator Guilds. Ecology 2002, 83, 1421–1432. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing 2021; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Lussana, C.; Tveito, O.; Uboldi, F. Three-dimensional Spatial Interpolation of 2 m Temperature over Norway. Q. J. R. Meteorol. Soc. 2018, 144, 344–364. [Google Scholar] [CrossRef]
- Geological Survey of Norway Løsmasser WMS. Available online: https://kartkatalog.geonorge.no/metadata/norges-geologiske-undersokelse/losmasser-wms/aa780848-5de8-4562-8f35-3d5c80ea8b48/ (accessed on 24 October 2022).
- Wright, M.N.; Wager, S.; Probst, P. R Package, Version 0.12; Ranger: A Fast Implementation of Random Forests; R Core Team: Vienna, Austria, 2020; Volume 1.
- Rodriguez, P.P.; Gianola, D. R Package, Version 0.6; BRNN: Bayesian Regularization for Feed-Forward Neural Networks; R Core Team: Vienna, Austria, 2016.
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Singh, V.; Pencina, M.; Einstein, A.J.; Liang, J.X.; Berman, D.S.; Slomka, P. Impact of Train/Test Sample Regimen on Performance Estimate Stability of Machine Learning in Cardiovascular Imaging. Sci. Rep. 2021, 11, 14490. [Google Scholar] [CrossRef]
- Greenwell, B.M. Pdp: An R Package for Constructing Partial Dependence Plots. R J. 2017, 9, 421. [Google Scholar] [CrossRef]
- Nkhwanana, N.; Adam, E.; Ramoelo, A. Assessing the Utility of Sentinel-2 MSI in Mapping an Encroaching Serephium Plumosum in South African Rangeland. Appl. Geomat. 2022, 14, 435–449. [Google Scholar] [CrossRef]
- De Simone, W.; Allegrezza, M.; Frattaroli, A.R.; Montecchiari, S.; Tesei, G.; Zuccarello, V.; Di Musciano, M. From Remote Sensing to Species Distribution Modelling: An Integrated Workflow to Monitor Spreading Species in Key Grassland Habitats. Remote Sens. 2021, 13, 1904. [Google Scholar] [CrossRef]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good Practices for Estimating Area and Assessing Accuracy of Land Change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Marshall, L.; Beckers, V.; Vray, S.; Rasmont, P.; Vereecken, N.J.; Dendoncker, N. High Thematic Resolution Land Use Change Models Refine Biodiversity Scenarios: A Case Study with Belgian Bumblebees. J. Biogeogr. 2021, 48, 345–358. [Google Scholar] [CrossRef]
- Griffiths, P.; Nendel, C.; Pickert, J.; Hostert, P. Towards National-Scale Characterization of Grassland Use Intensity from Integrated Sentinel-2 and Landsat Time Series. Remote Sens. Environ. 2020, 238, 111124. [Google Scholar] [CrossRef]
- Coops, N.C.; Waring, R.H.; Plowright, A.; Lee, J.; Dilts, T.E. Using Remotely-Sensed Land Cover and Distribution Modeling to Estimate Tree Species Migration in the Pacific Northwest Region of North America. Remote Sens. 2016, 8, 65. [Google Scholar] [CrossRef]
- Venter, Z.S.; Barton, D.N.; Chakraborty, T.; Simensen, T.; Singh, G. Global 10 mL and Use Land Cover Datasets: A Comparison of Dynamic World, World Cover and Esri Land Cover. Remote Sens. 2022, 14, 4101. [Google Scholar] [CrossRef]
- White, E.R. Minimum Time Required to Detect Population Trends: The Need for Long-Term Monitoring Programs. BioScience 2019, 69, 40–46. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).