A Physically Interpretable Rice Field Extraction Model for PolSAR Imagery
Abstract
1. Introduction
- (1)
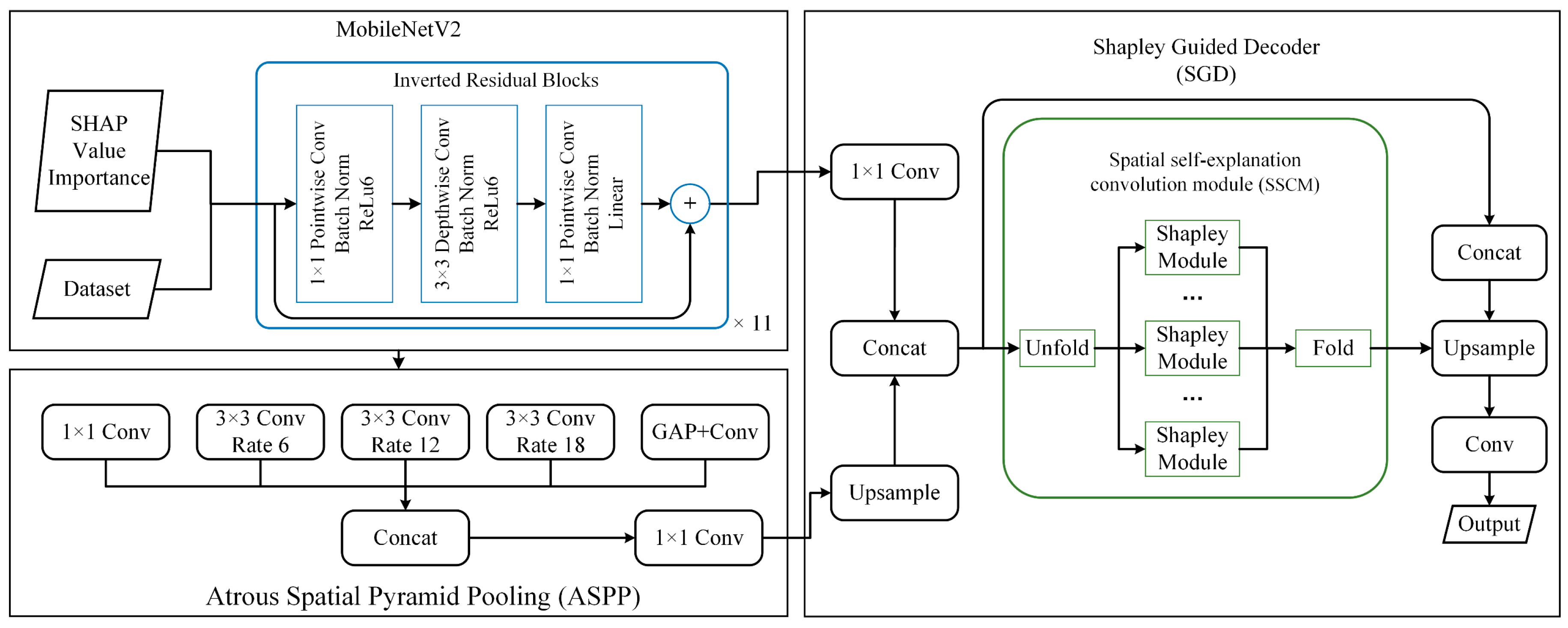
- The SHAP-based Physical Feature Interpretable Module (SPFIM) is proposed for PolSAR data. Physical interpretability refers to the interpretability in the physical feature dimension, i.e., the effect of different physical features on the model outputs can be interpreted. In SPFIM, the LSTM is used to process the feature sequences at the pixel level to obtain the physical characteristics importance weights based on the SHAP value. The physical characteristics importance weights are used to weight the original data to obtain new physical-weighted data, which can increase the physical interpretability of the deep learning method.
- (2)
- The SHAP-guided spatial explanation network (SSEN) is proposed, which contains a spatial self-explanation module SSCM based on the Shapley Module [64] design. The spatial SHAP explanation values of the input features can be calculated and can be input as interlayer features to the neural network along with the abstract features. In such a way, the network is allowed to obtain interpretability in spatial dimensions.
2. Materials and Methods
2.1. Study Area
2.2. Study Data
2.3. Methods
2.3.1. Physical Features Extraction
2.3.2. SHAP-Based Physical Feature Interpretable Module
2.3.3. SHAP-Guided Spatial Explanation Network
2.3.4. Accuracy Assessment
3. Experiments and Results
3.1. Physical Interpretability of SSEN
3.2. Spatial Interpretability of SSEN
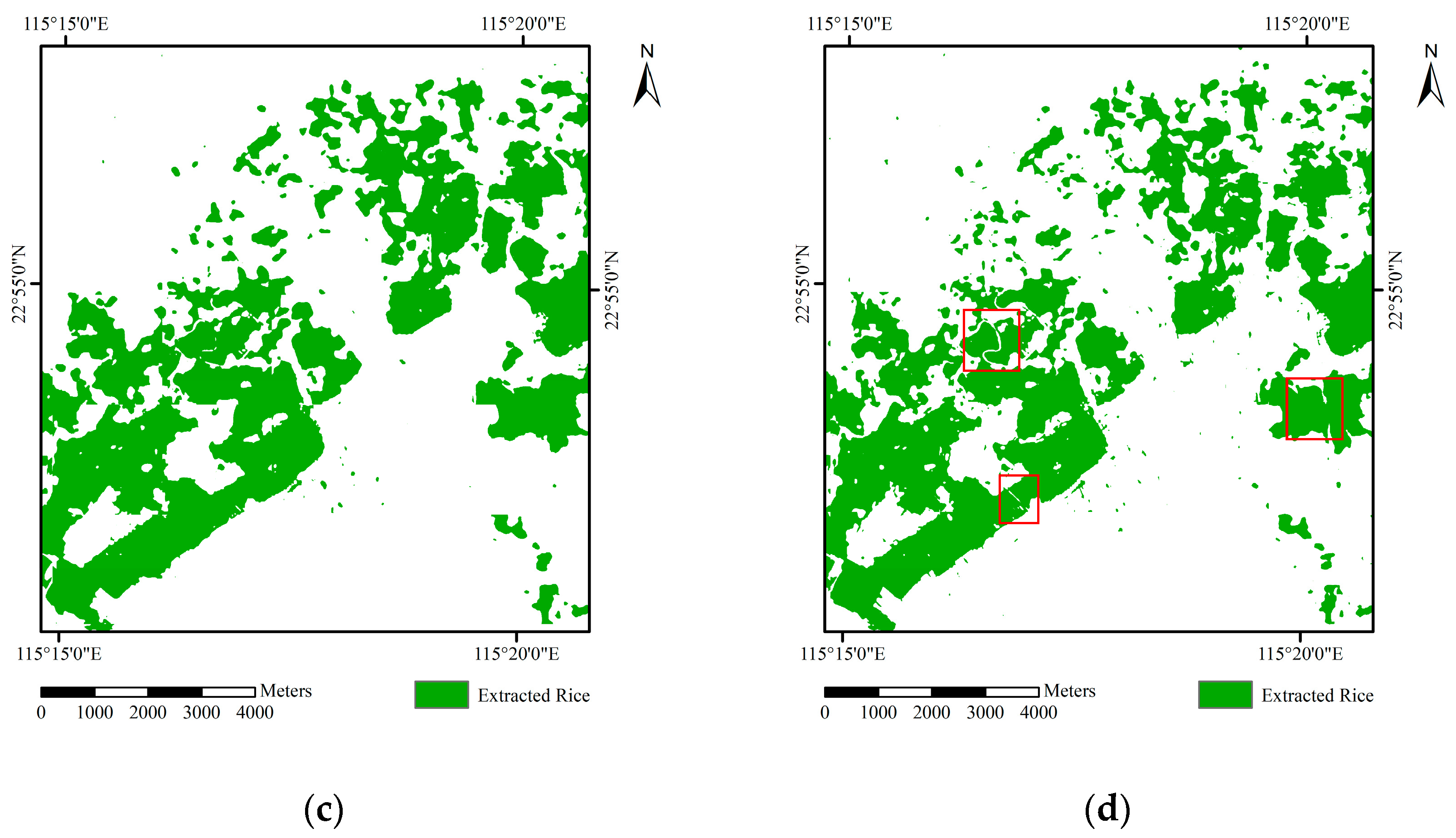
3.3. Comparison of Different Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for global nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef]
- FAO. FAO Rice Market Monitor. 2018, Volume 21. Available online: http://www.fao.org/economic/est/publications/rice-publications/rice-market-monitor-rmm/en/ (accessed on 30 September 2022).
- McNairn, H.; Brisco, B. The application of C-band polarimetric SAR for agriculture: A review. Can. J. Remote Sens. 2004, 30, 525–542. [Google Scholar] [CrossRef]
- Chen, J.; Han, Y.; Zhang, J. Mapping rice crop fields using C band polarimetric SAR data. In Proceedings of the 2014 The Third International Conference on Agro-Geoinformatics, Beijing, China, 11–14 August 2014; pp. 1–4. [Google Scholar]
- Li, K.; Brisco, B.; Yun, S.; Touzi, R. Polarimetric decomposition with RADARSAT-2 for rice mapping and monitoring. Can. J. Remote Sens. 2012, 38, 169–179. [Google Scholar] [CrossRef]
- Wu, F.; Wang, C.; Zhang, H.; Zhang, B.; Tang, Y. Rice crop monitoring in South China with RADARSAT-2 quad-polarization SAR data. IEEE Geosci. Remote Sens. Lett. 2010, 8, 196–200. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, B.; Zhang, H.; Wang, C.; Tang, Y. Analysis of rice growth using multi-temporal RADARSAT-2 quad-pol SAR images. Intell. Autom. Soft Comput. 2012, 18, 997–1007. [Google Scholar] [CrossRef]
- Wang, H.; Magagi, R.; Goïta, K.; Trudel, M.; McNairn, H.; Powers, J. Crop phenology retrieval via polarimetric SAR decomposition and Random Forest algorithm. Remote Sens. Environ. 2019, 231, 111234. [Google Scholar] [CrossRef]
- Lopez-Sanchez, J.M.; Vicente-Guijalba, F.; Ballester-Berman, J.D.; Cloude, S.R. Polarimetric Response of Rice Fields at C-Band: Analysis and Phenology Retrieval. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2977–2993. [Google Scholar] [CrossRef]
- Xu, J.; Li, Z.; Tian, B.; Huang, L.; Chen, Q.; Fu, S. Polarimetric analysis of multi-temporal RADARSAT-2 SAR images for wheat monitoring and mapping. Int. J. Remote Sens. 2014, 35, 3840–3858. [Google Scholar] [CrossRef]
- He, Z.; Li, S.; Zhai, P.; Deng, Y. Mapping Rice Planting Area Using Multi-Temporal Quad-Pol Radarsat-2 Datasets and Random Forest Algorithm. In Proceedings of the IGARSS 2020-2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 4653–4656. [Google Scholar]
- Srikanth, P.; Ramana, K.; Deepika, U.; Chakravarthi, P.K.; Sai, M.S. Comparison of various polarimetric decomposition techniques for crop classification. J. Indian Soc. Remote Sens. 2016, 44, 635–642. [Google Scholar] [CrossRef]
- Lopez-Sanchez, J.; Ballester-Berman, J.; Hajnsek, I. First Results of Rice Monitoring Practices in Spain by Means of Time Series of TerraSAR-X Dual-Pol Images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 412–422. [Google Scholar] [CrossRef]
- Corcione, V.; Nunziata, F.; Mascolo, L.; Migliaccio, M. A study of the use of COSMO-SkyMed SAR PingPong polarimetric mode for rice growth monitoring. Int. J. Remote Sens. 2016, 37, 633–647. [Google Scholar] [CrossRef]
- Lopez-Sanchez, J. Rice phenology monitoring by means of SAR polarimetry at X-band. IEEE Trans. Geosci. Remote Sens. 2012, 50, 2695–2709. [Google Scholar] [CrossRef]
- Hoang, H.K.; Bernier, M.; Duchesne, S.; Tran, Y.M. Rice mapping using RADARSAT-2 dual-and quad-pol data in a complex land-use Watershed: Cau River Basin (Vietnam). IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3082–3096. [Google Scholar] [CrossRef]
- Ohki, M.; Shimada, M. Large-area land use and land cover classification with quad, compact, and dual polarization SAR data by PALSAR-2. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5550–5557. [Google Scholar] [CrossRef]
- Valcarce-Diñeiro, R.; Arias-Pérez, B.; Lopez-Sanchez, J.M.; Sánchez, N. Multi-temporal dual-and quad-polarimetric synthetic aperture radar data for crop-type mapping. Remote Sens. 2019, 11, 1518. [Google Scholar] [CrossRef]
- Dey, S.; Bhattacharya, A.; Ratha, D.; Mandal, D.; McNairn, H.; Lopez-Sanchez, J.M.; Rao, Y.S. Novel clustering schemes for full and compact polarimetric SAR data: An application for rice phenology characterization. ISPRS J. Photogramm. Remote Sens. 2020, 169, 135–151. [Google Scholar] [CrossRef]
- Dey, S.; Mandal, D.; Robertson, L.D.; Banerjee, B.; Kumar, V.; McNairn, H.; Bhattacharya, A.; Rao, Y. In-season crop classification using elements of the Kennaugh matrix derived from polarimetric RADARSAT-2 SAR data. Int. J. Appl. Earth Obs. Geoinf. 2020, 88, 102059. [Google Scholar] [CrossRef]
- Dey, S.; Bhogapurapu, N.; Bhattacharya, A.; Mandal, D.; Lopez-Sanchez, J.M.; McNairn, H.; Frery, A.C. Rice phenology mapping using novel target characterization parameters from polarimetric SAR data. Int. J. Remote Sens. 2021, 42, 5515–5539. [Google Scholar] [CrossRef]
- Yonezawa, C. An Attempt to Extract Paddy Fields Using Polarimetric Decomposition of PALSAR-2 Data. In Proceedings of the IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 7172–7175. [Google Scholar]
- Mahdianpari, M.; Mohammadimanesh, F.; McNairn, H.; Davidson, A.; Rezaee, M.; Salehi, B.; Homayouni, S. Mid-season crop classification using dual-, compact-, and full-polarization in preparation for the Radarsat Constellation Mission (RCM). Remote Sens. 2019, 11, 1582. [Google Scholar] [CrossRef]
- Skakun, S.; Kussul, N.; Shelestov, A.; Lavreniuk, M.; Kussul, O. Efficiency Assessment of Multitemporal C-Band Radarsat-2 Intensity and Landsat-8 Surface Reflectance Satellite Imagery for Crop Classification in Ukraine. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3712–3719. [Google Scholar] [CrossRef]
- Huang, X.; Liao, C.; Xing, M.; Ziniti, B.; Wang, J.; Shang, J.; Liu, J.; Dong, T.; Xie, Q.; Torbick, N. A multi-temporal binary-tree classification using polarimetric RADARSAT-2 imagery. Remote Sens. Environ. 2019, 235, 111478. [Google Scholar] [CrossRef]
- Liao, C.; Wang, J.; Xie, Q.; Baz, A.A.; Huang, X.; Shang, J.; He, Y. Synergistic Use of multi-temporal RADARSAT-2 and VENµS data for crop classification based on 1D convolutional neural network. Remote Sens. 2020, 12, 832. [Google Scholar] [CrossRef]
- Chen, S.-W.; Tao, C.-S. Multi-temporal PolSAR crops classification using polarimetric-feature-driven deep convolutional neural network. In Proceedings of the 2017 International Workshop on Remote Sensing with Intelligent Processing (RSIP), Shanghai, China, 18–21 May 2017; pp. 1–4. [Google Scholar]
- Wei, S.; Zhang, H.; Wang, C.; Wang, Y.; Xu, L. Multi-temporal SAR data large-scale crop mapping based on U-Net model. Remote Sens. 2019, 11, 68. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Zhou, H. Deep learning based multi-temporal crop classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- La Rosa, L.E.C.; Feitosa, R.Q.; Happ, P.N.; Sanches, I.D.; da Costa, G.A.O.P. Combining deep learning and prior knowledge for crop mapping in tropical regions from multitemporal SAR image sequences. Remote Sens. 2019, 11, 2029. [Google Scholar] [CrossRef]
- Teimouri, N.; Dyrmann, M.; Jørgensen, R.N. A novel spatio-temporal FCN-LSTM network for recognizing various crop types using multi-temporal radar images. Remote Sens. 2019, 11, 990. [Google Scholar] [CrossRef]
- Zhou, Y.N.; Luo, J.; Feng, L.; Zhou, X. DCN-based spatial features for improving parcel-based crop classification using high-resolution optical images and multi-temporal SAR data. Remote Sens. 2019, 11, 1619. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Z.; Jiang, H.; Jing, W.; Sun, L.; Feng, M. Evaluation of three deep learning models for early crop classification using sentinel-1A imagery time series—A case study in Zhanjiang, China. Remote Sens. 2019, 11, 2673. [Google Scholar] [CrossRef]
- de Castro Filho, H.C.; de Carvalho Júnior, O.A.; de Carvalho, O.L.F.; de Bem, P.P.; dos Santos de Moura, R.; de Albuquerque, A.O.; Silva, C.R.; Ferreira, P.H.G.; Guimarães, R.F.; Gomes, R.A.T. Rice crop detection using LSTM, Bi-LSTM, and machine learning models from sentinel-1 time series. Remote Sens. 2020, 12, 2655. [Google Scholar] [CrossRef]
- Wu, M.-C.; Alkhaleefah, M.; Chang, L.; Chang, Y.-L.; Shie, M.-H.; Liu, S.-J.; Chang, W.-Y. Recurrent Deep Learning for Rice Fields Detection from SAR Images. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 1548–1551. [Google Scholar]
- Jo, H.-W.; Lee, S.; Park, E.; Lim, C.-H.; Song, C.; Lee, H.; Ko, Y.; Cha, S.; Yoon, H.; Lee, W.-K. Deep learning applications on multitemporal SAR (Sentinel-1) image classification using confined labeled data: The case of detecting rice paddy in South Korea. IEEE Trans. Geosci. Remote Sens. 2020, 58, 7589–7601. [Google Scholar] [CrossRef]
- de Bem, P.P.; de Carvalho Júnior, O.A.; de Carvalho, O.L.F.; Gomes, R.A.T.; Guimarāes, R.F.; Pimentel, C.M.M. Irrigated rice crop identification in Southern Brazil using convolutional neural networks and Sentinel-1 time series. Remote Sens. Appl. Soc. Environ. 2021, 24, 100627. [Google Scholar] [CrossRef]
- Wei, P.; Chai, D.; Lin, T.; Tang, C.; Du, M.; Huang, J. Large-scale rice mapping under different years based on time-series Sentinel-1 images using deep semantic segmentation model. ISPRS J. Photogramm. Remote Sens. 2021, 174, 198–214. [Google Scholar] [CrossRef]
- Pang, J.; Zhang, R.; Yu, B.; Liao, M.; Lv, J.; Xie, L.; Li, S.; Zhan, J. Pixel-level rice planting information monitoring in Fujin City based on time-series SAR imagery. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102551. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, W.; Chen, S.; Ye, T. Mapping Crop Rotation by Using Deeply Synergistic Optical and SAR Time Series. Remote Sens. 2021, 13, 4160. [Google Scholar] [CrossRef]
- Thorp, K.; Drajat, D. Deep machine learning with Sentinel satellite data to map paddy rice production stages across West Java, Indonesia. Remote Sens. Environ. 2021, 265, 112679. [Google Scholar] [CrossRef]
- Lin, Z.; Zhong, R.; Xiong, X.; Guo, C.; Xu, J.; Zhu, Y.; Xu, J.; Ying, Y.; Ting, K.; Huang, J. Large-Scale Rice Mapping Using Multi-Task Spatiotemporal Deep Learning and Sentinel-1 SAR Time Series. Remote Sens. 2022, 14, 699. [Google Scholar] [CrossRef]
- Chang, Y.-L.; Tan, T.-H.; Chen, T.-H.; Chuah, J.H.; Chang, L.; Wu, M.-C.; Tatini, N.B.; Ma, S.-C.; Alkhaleefah, M. Spatial-Temporal Neural Network for Rice Field Classification from SAR Images. Remote Sens. 2022, 14, 1929. [Google Scholar] [CrossRef]
- Chakraborty, S.; Tomsett, R.; Raghavendra, R.; Harborne, D.; Alzantot, M.; Cerutti, F.; Srivastava, M.; Preece, A.; Julier, S.; Rao, R.M. Interpretability of deep learning models: A survey of results. In Proceedings of the 2017 IEEE Smartworld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computed, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (Smartworld/SCALCOM/UIC/ATC/CBDcom/IOP/SCI), San Francisco, CA, USA, 4–8 August 2017; pp. 1–6. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.-R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4765–4774. [Google Scholar]
- Ancona, M.; Oztireli, C.; Gross, M. Explaining deep neural networks with a polynomial time algorithm for shapley value approximation. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 272–281. [Google Scholar]
- Arras, L.; Montavon, G.; Müller, K.-R.; Samek, W. Explaining recurrent neural network predictions in sentiment analysis. arXiv 2017. [Google Scholar] [CrossRef]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 3319–3328. [Google Scholar]
- Hendricks, L.A.; Akata, Z.; Rohrbach, M.; Donahue, J.; Schiele, B.; Darrell, T. Generating visual explanations. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 3–19. [Google Scholar]
- Lei, T.; Barzilay, R.; Jaakkola, T. Rationalizing neural predictions. arXiv 2016. [Google Scholar] [CrossRef]
- Kim, J.; Rohrbach, A.; Darrell, T.; Canny, J.; Akata, Z. Textual explanations for self-driving vehicles. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 563–578. [Google Scholar]
- Park, D.H.; Hendricks, L.A.; Akata, Z.; Rohrbach, A.; Schiele, B.; Darrell, T.; Rohrbach, M. Multimodal explanations: Justifying decisions and pointing to the evidence. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8779–8788. [Google Scholar]
- Yoon, J.; Jordon, J.; van der Schaar, M. INVASE: Instance-wise variable selection using neural networks. In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Rajani, N.F.; McCann, B.; Xiong, C.; Socher, R. Explain yourself! leveraging language models for commonsense reasoning. arXiv 2019. [Google Scholar] [CrossRef]
- Chang, S.; Zhang, Y.; Yu, M.; Jaakkola, T. A game theoretic approach to class-wise selective rationalization. Adv. Neural Inf. Process. Syst. 2019, 32, 10055–10065. [Google Scholar] [CrossRef]
- Chen, J.; Song, L.; Wainwright, M.J.; Jordan, M.I. L-shapley and c-shapley: Efficient model interpretation for structured data. arXiv 2018. [Google Scholar] [CrossRef]
- Shendryk, Y.; Davy, R.; Thorburn, P. Integrating satellite imagery and environmental data to predict field-level cane and sugar yields in Australia using machine learning. Field Crops Res. 2021, 260, 107984. [Google Scholar] [CrossRef]
- Al-Najjar, H.A.; Pradhan, B.; Beydoun, G.; Sarkar, R.; Park, H.-J.; Alamri, A. A novel method using explainable artificial intelligence (XAI)-based Shapley Additive Explanations for spatial landslide prediction using Time-Series SAR dataset. Gondwana Res. 2022; in press. [Google Scholar] [CrossRef]
- Panati, C.; Wagner, S.; Brüggenwirth, S. Feature Relevance Evaluation Using Grad-CAM, LIME and SHAP for Deep Learning SAR Data Classification. In Proceedings of the 2022 23rd International Radar Symposium (IRS), Gdansk, Poland, 12–14 September 2022; pp. 457–462. [Google Scholar]
- Amri, E.; Dardouillet, P.; Benoit, A.; Courteille, H.; Bolon, P.; Dubucq, D.; Credoz, A. Offshore oil slick detection: From photo-interpreter to explainable multi-modal deep learning models using SAR images and contextual data. Remote Sens. 2022, 14, 3565. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Inouye, D.I. Shapley explanation networks. arXiv 2021. [Google Scholar] [CrossRef]
- Zanaga, D.; Van De Kerchove, R.; Daems, D.; De Keersmaecker, W.; Brockmann, C.; Kirches, G.; Wevers, J.; Cartus, O.; Santoro, M.; Fritz, S. ESA WorldCover 10 m 2021 v200; The European Space Agency: Paris, France, 2022.
- Cloude, S.; Zebker, H. Polarisation: Applications in Remote Sensing. Phys. Today 2010, 63, 53. [Google Scholar] [CrossRef]
- Freeman, A.; Durden, S.L. A three-component scattering model for polarimetric SAR data. IEEE Trans. Geosci. Remote Sens. 1998, 36, 963–973. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Moriyama, T.; Ishido, M.; Yamada, H. Four-component scattering model for polarimetric SAR image decomposition. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1699–1706. [Google Scholar] [CrossRef]
- Jong-Sen, L.; Pottier, E. Polarimetric Radar Imaging: From Basics to Applications; Taylor & Francis: Oxfordshire, UK; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar] [CrossRef]
- Huynen, J.R. Phenomenological Theory of Radar Targets. Ph.D. Thesis, Technical University, Delft, The Netherlands, 1970. [Google Scholar]
- Cloude, S.R.; Pottier, E. A review of target decomposition theorems in radar polarimetry. IEEE Trans. Geosci. Remote Sens. 1996, 34, 498–518. [Google Scholar] [CrossRef]
- Riedel, T.; Liebeskind, P.; Schmullius, C. Seasonal and diurnal changes of polarimetric parameters from crops derived by the Cloude decomposition theorem at L-band. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Toronto, ON, Canada, 24–28 June 2002; pp. 2714–2716. [Google Scholar]
- Jiao, X.; Kovacs, J.M.; Shang, J.; McNairn, H.; Walters, D.; Ma, B.; Geng, X. Object-oriented crop mapping and monitoring using multi-temporal polarimetric RADARSAT-2 data. ISPRS J. Photogramm. Remote Sens. 2014, 96, 38–46. [Google Scholar] [CrossRef]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 3145–3153. [Google Scholar]
- Peng, W.; Li, S.; He, Z.; Ning, S.; Liu, Y.; Su, Z. Random forest classification of rice planting area using multi-temporal polarimetric Radarsat-2 data. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 2411–2414. [Google Scholar]
- Mansaray, L.R.; Wang, F.; Huang, J.; Yang, L.; Kanu, A.S. Accuracies of support vector machine and random forest in rice mapping with Sentinel-1A, Landsat-8 and Sentinel-2A datasets. Geocarto Int. 2020, 35, 1088–1108. [Google Scholar] [CrossRef]
- Xie, Q.; Dou, Q.; Peng, X.; Wang, J.; Lopez-Sanchez, J.M.; Shang, J.; Fu, H.; Zhu, J. Crop Classification Based on the Physically Constrained General Model-Based Decomposition Using Multi-Temporal RADARSAT-2 Data. Remote Sens. 2022, 14, 2668. [Google Scholar] [CrossRef]








| No. | Date | Incidence Angle | Flight Direction | No. | Date | Incidence Angle | Flight Direction |
|---|---|---|---|---|---|---|---|
| 1 | 30 April 2017 | 35.29°~37.10° | Ascending | 13 | 13 March 2019 | 27.25°~29.84° | Descending |
| 2 | 29 May 2017 | 35.29°~37.10° | Ascending | 14 | 27 May 2019 | 29.70°~31.89° | Descending |
| 3 | 29 May 2017 | 35.29°~37.11° | Ascending | 15 | 27 May 2019 | 29.69°~31.88° | Descending |
| 4 | 24 August 2017 | 35.33°~37.12° | Ascending | 16 | 19 July 2019 | 24.18°~26.81° | Descending |
| 5 | 24 August 2017 | 35.29°~37.11° | Ascending | 17 | 12 February 2020 | 35.62°~37.43° | Descending |
| 6 | 24 August 2017 | 35.29°~37.11° | Ascending | 18 | 12 February 2020 | 35.69°~37.49° | Descending |
| 7 | 10 June 2018 | 29.34°~31.39° | Ascending | 19 | 12 February 2020 | 35.63°~37.44° | Descending |
| 8 | 10 June 2018 | 29.36°~31.42° | Ascending | 20 | 12 June 2020 | 47.07°~48.25° | Descending |
| 9 | 10 August 2018 | 36.84°~38.31° | Descending | 21 | 25 May 2021 | 48.24°~49.26° | Ascending |
| 10 | 10 August 2018 | 36.84°~38.31° | Descending | 22 | 25 May 2021 | 48.21°~49.26° | Ascending |
| 11 | 10 August 2018 | 36.85°~38.32° | Descending | 23 | 25 May 2021 | 48.21°~49.27° | Ascending |
| 12 | 2 January 2019 | 35.29°~37.12° | Descending | 24 | 14 September 2021 | 31.28°~33.41° | Descending |
| The Characteristic Parameter | Physical Significance |
|---|---|
| α | The size of the average scattering angle α is closely related to the scattering type. α = 0° indicates surface scattering. As α increases, the surface becomes anisotropy. An α-value of 45° represents a dipole. If α reaches 90° the scattering process is characterized by dihedral scattering interactions. |
| H | Scattering entropy (H) is an indicator for the number of effective scattering mechanisms, whereby H = 0 belongs to deterministic scattering and H = 1 to totally random scattering. |
| A | Anisotropy (A) only yields additional information for medium values of H. High A signifies that besides the first scattering mechanism only one secondary process contributes to the radar signal. For low A both secondary scattering processes play an important role. |
| Freeman_Odd | Surface scattering of Freeman–Durden decomposition |
| Freeman_Dbl | Dihedral scattering of Freeman–Durden decomposition |
| Freeman_Vol | Volume scattering of Freeman–Durden decomposition |
| Yamaguchi_Odd | Single-bounce of Yamaguchi decomposition |
| Yamaguchi_Dbl | Dihedral scattering of Yamaguchi decomposition |
| Yamaguchi_Vol | Volume scattering of Yamaguchi decomposition |
| Yamaguchi_Hlx | Helix scattering of Yamaguchi decomposition |
| Model | Overall Accuracy | Precision | Recall | F1 | Kappa |
|---|---|---|---|---|---|
| RF | 89.76 | 89.69 | 91.41 | 90.54 | 0.7939 |
| Deeplabv3+ | 93.01 | 90.31 | 97.45 | 93.74 | 0.8586 |
| SGEM | 95.73 | 97.15 | 94.82 | 95.97 | 0.9143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, J.; Zhang, H.; Xu, L.; Sun, C.; Duan, H.; Guo, Z.; Wang, C. A Physically Interpretable Rice Field Extraction Model for PolSAR Imagery. Remote Sens. 2023, 15, 974. https://doi.org/10.3390/rs15040974
Ge J, Zhang H, Xu L, Sun C, Duan H, Guo Z, Wang C. A Physically Interpretable Rice Field Extraction Model for PolSAR Imagery. Remote Sensing. 2023; 15(4):974. https://doi.org/10.3390/rs15040974
Chicago/Turabian StyleGe, Ji, Hong Zhang, Lu Xu, Chunling Sun, Haoxuan Duan, Zihuan Guo, and Chao Wang. 2023. "A Physically Interpretable Rice Field Extraction Model for PolSAR Imagery" Remote Sensing 15, no. 4: 974. https://doi.org/10.3390/rs15040974
APA StyleGe, J., Zhang, H., Xu, L., Sun, C., Duan, H., Guo, Z., & Wang, C. (2023). A Physically Interpretable Rice Field Extraction Model for PolSAR Imagery. Remote Sensing, 15(4), 974. https://doi.org/10.3390/rs15040974






