Comparing Machine Learning Algorithms for Pixel/Object-Based Classifications of Semi-Arid Grassland in Northern China Using Multisource Medium Resolution Imageries
Abstract
1. Introduction
2. Materials and Methods
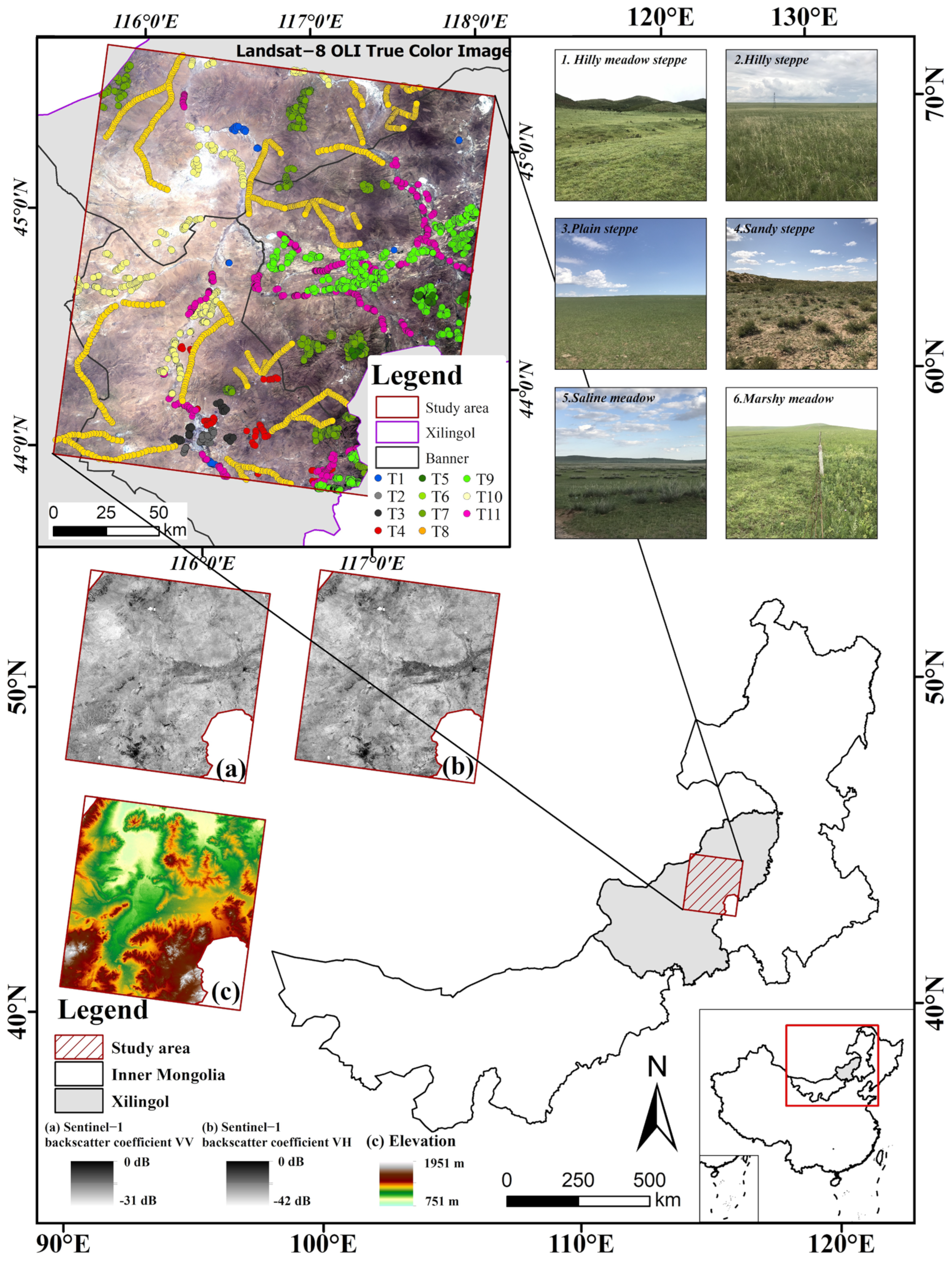
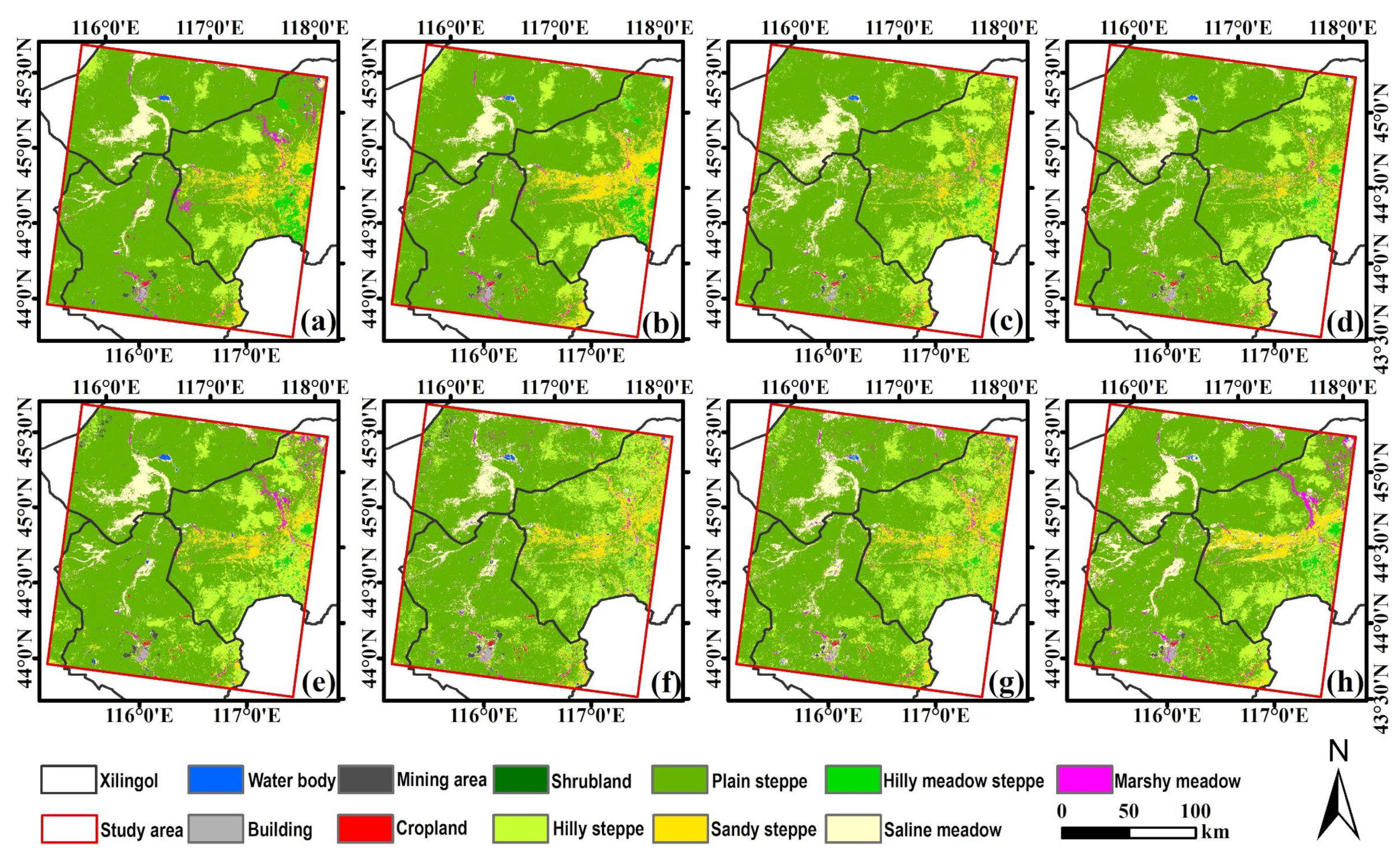
2.1. Study Area
2.2. Classification System and Sample Data
- (1)
- Hilly meadow steppe occurs mainly in the high relief sites of 900~1500 m with relatively moist and fertile soils, and the dominant species include Stipa baicalensis, Filifolium sibiricum, and Leymus chinensis.
- (2)
- Hilly steppe is mainly formed at elevations between 600 and 1300 m and is dominated by xerophytic or semi-xeric bunchgrass.
- (3)
- Plain steppe is the most widely distributed grassland type in this area, and occurs under a semi-arid climate with annual precipitation around 350 mm. The most common communities are dominated by Stipa grandis, Stipa krylovii, Leymus chinensis, Cleistogenes squarrosa, and Artemisia frigida.
- (4)
- Sandy steppe has distinctive zonal characteristics and is mainly found in Hunshandak sandland.
- (5)
- Saline meadow occurs mainly on salinized depression sites, broad valleys, fringes of lake basins, and river flats within steppe and desert regions. It is primarily composed of mesic perennial halophytes, such as Achnatherum splendens and Leymus chinensis.
- (6)
- Marshy meadow is primarily composed of hygrophilous herbs, such as Phragmites australis, and has transitional characteristics between a meadow and a marsh.
| Land Covers | No. of Samples | |||
|---|---|---|---|---|
| Non-vegetation | Waterbody | T1 | 47 | |
| Building | T2 | 97 | ||
| Mining area | T3 | 50 | ||
| Vegetation | Non-grassland | Cropland | T4 | 59 |
| Shrubland | T5 | 61 | ||
| Grassland (CGCS) | Hilly meadow steppe | T6 | 191 | |
| Hilly steppe | T7 | 555 | ||
| Plain steppe | T8 | 826 | ||
| Sandy steppe | T9 | 870 | ||
| Saline meadow | T10 | 340 | ||
| Marshy meadow | T11 | 524 | ||
2.3. Remote Sensing Data and Preprocessing
2.3.1. Multispectral Imagery
2.3.2. Synthetic Aperture Radar Data
2.3.3. Topographic Data
2.4. Image Segmentation
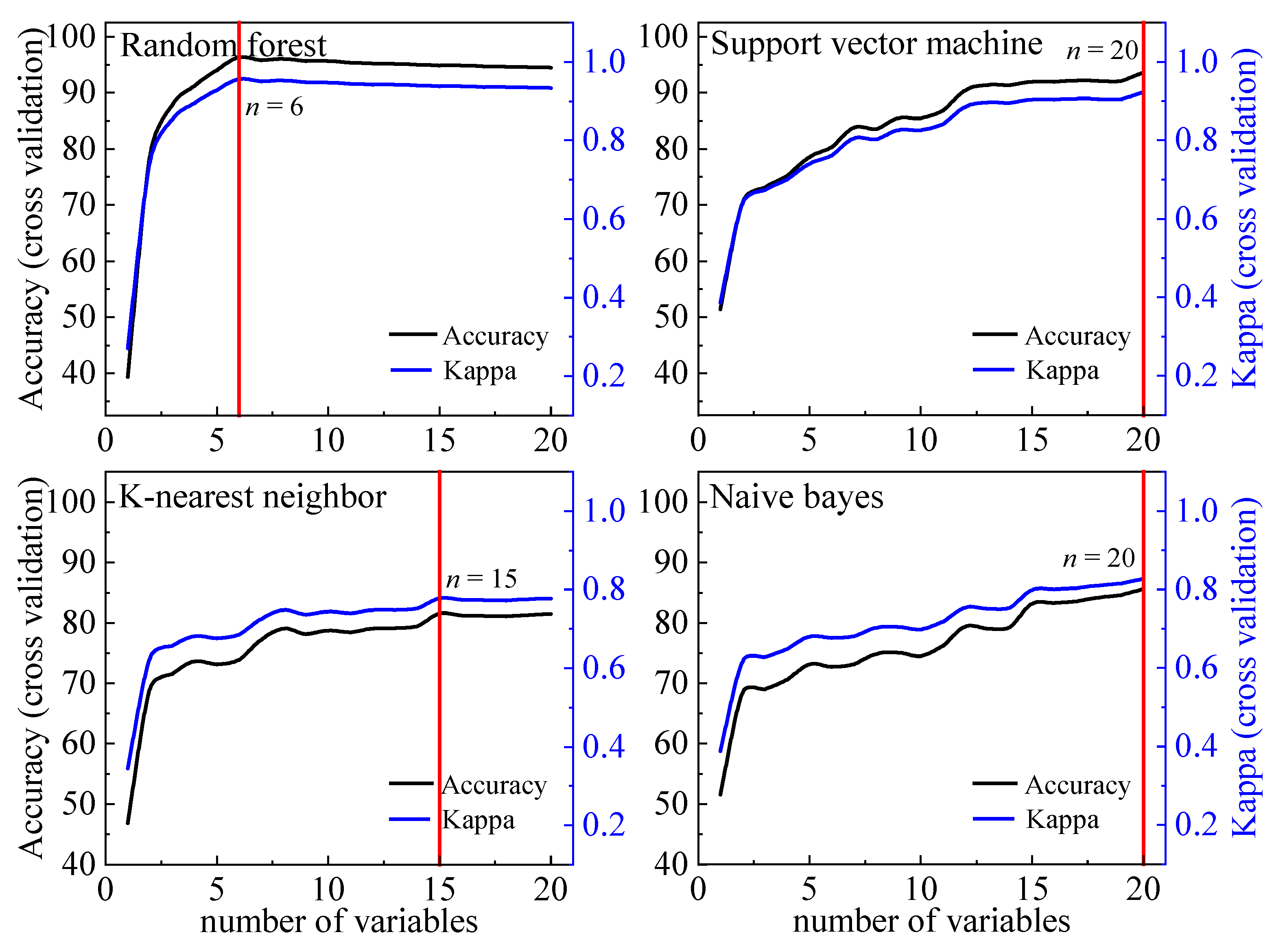
2.5. Feature Selection
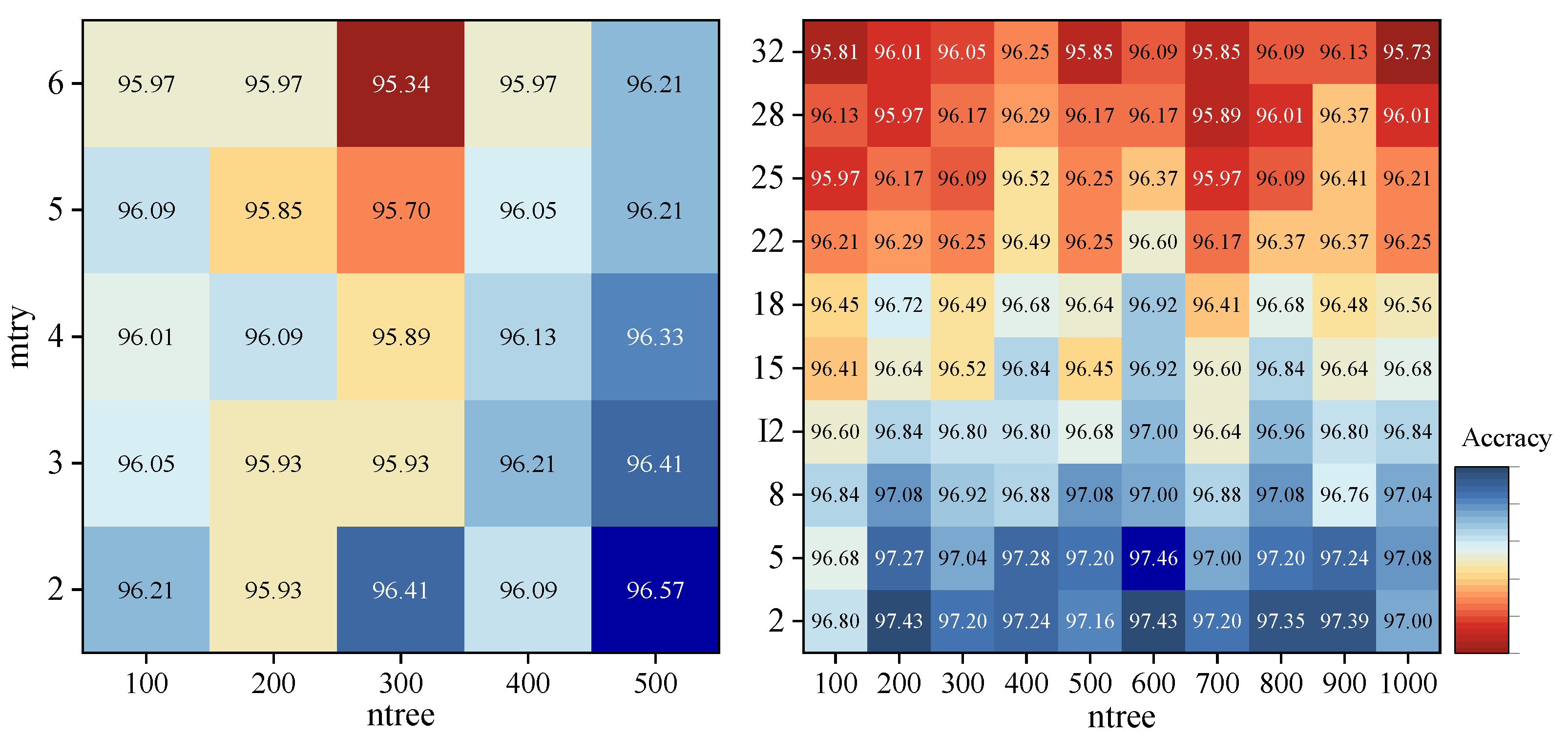
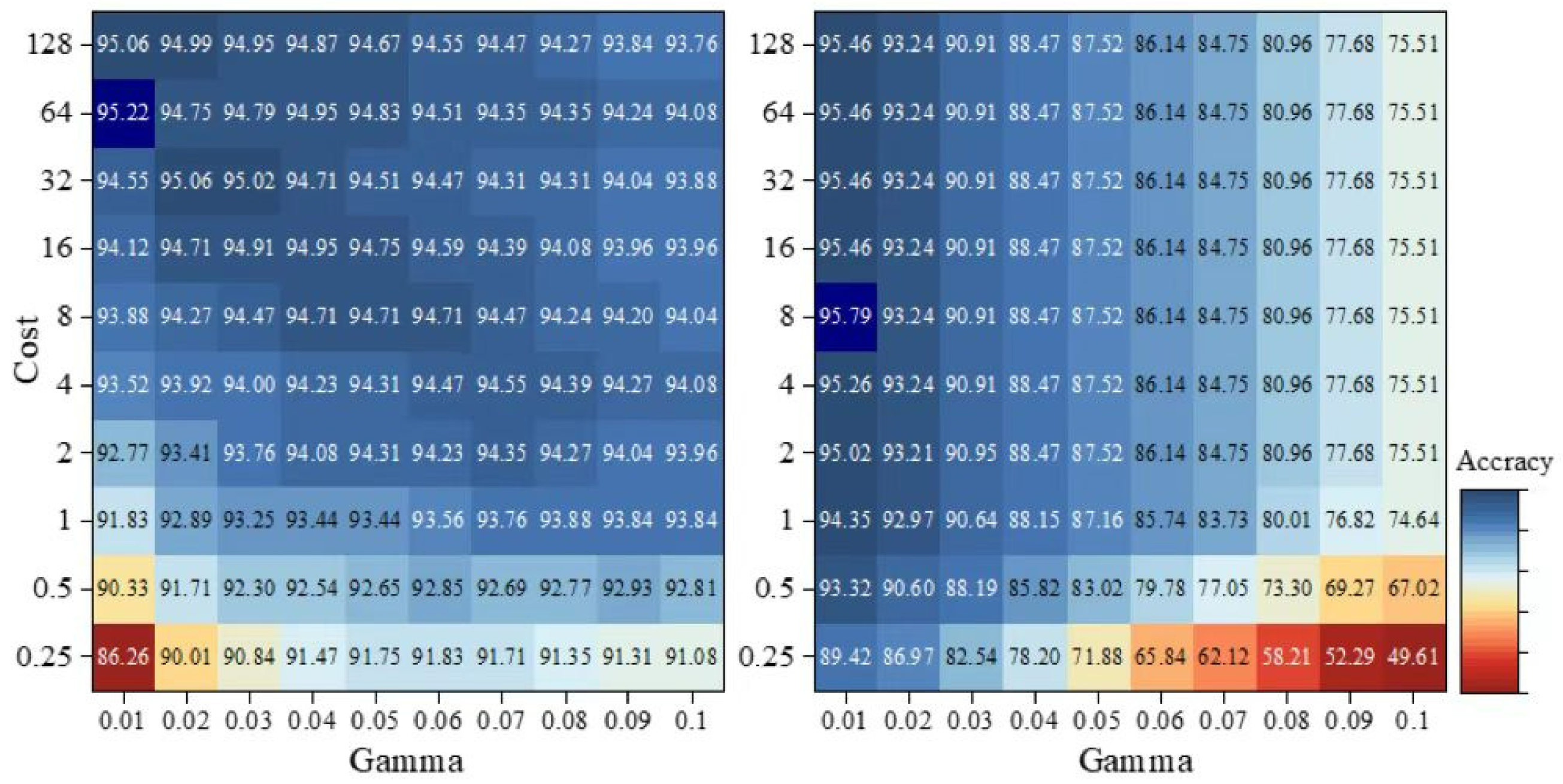
2.6. Classification Algorithms
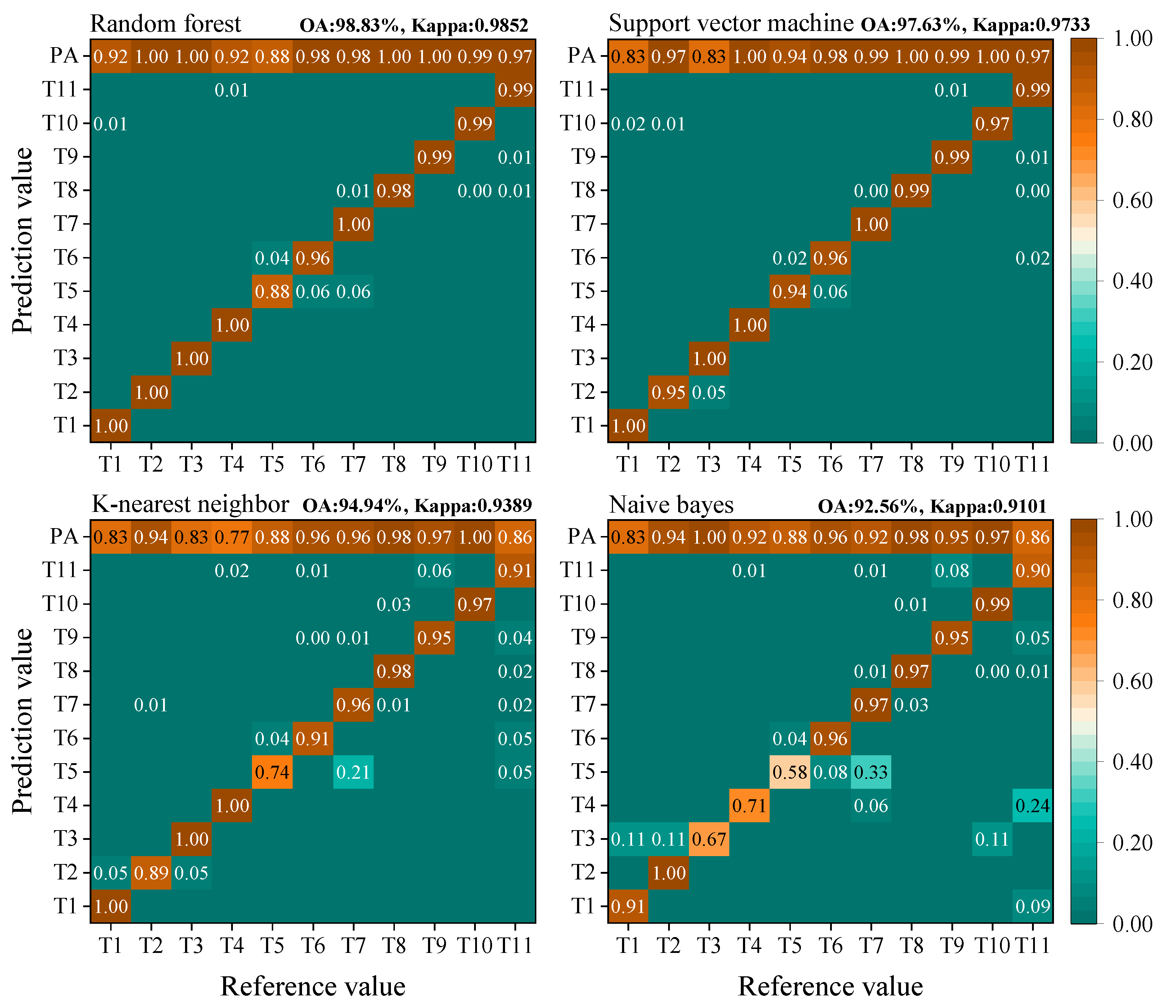
2.7. Accuracy Assessment
3. Results
3.1. Pixel-Based Classifications
3.2. Object-Based Classifications
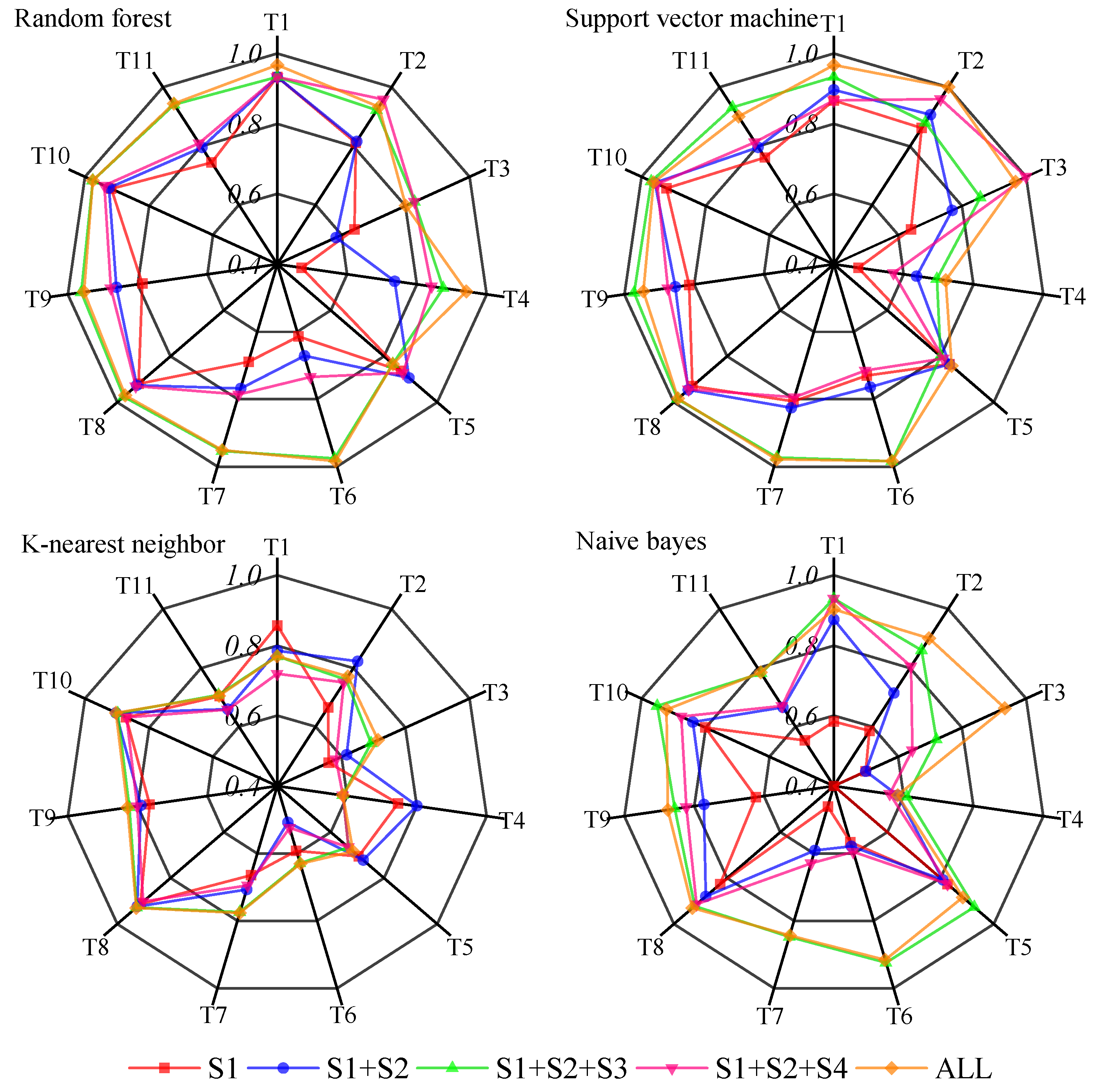
3.3. Comparison of Pixel-Based and Object-Based Classifications
4. Discussion
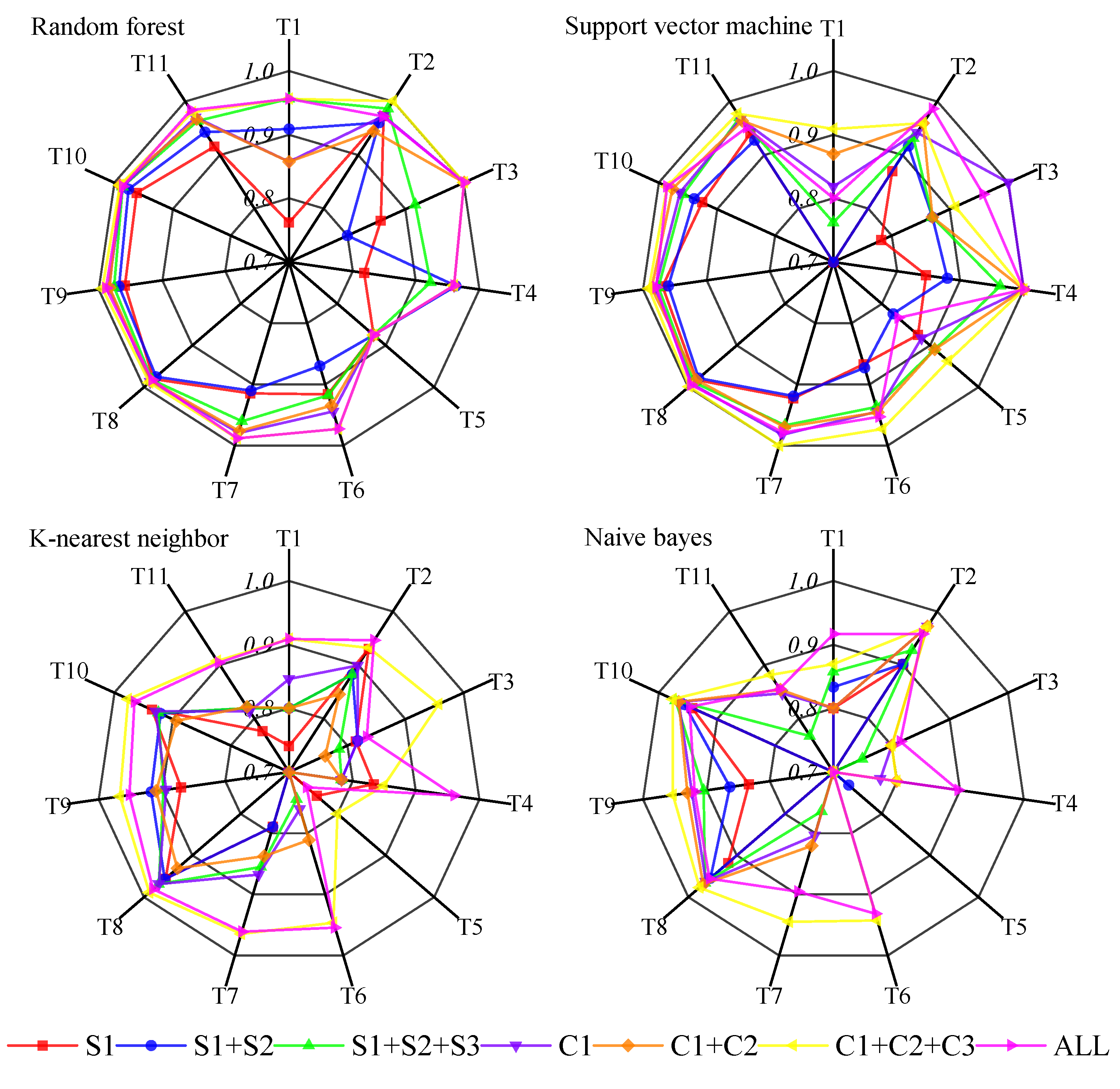
4.1. Comparison of Classification Methods for Semi-Arid Grassland
4.2. Feature Selection for Semi-Arid Grassland Classifications
4.3. Limitations and Uncertainties
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Bengtsson, J.; Bullock, J.M.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.J.; Smith, H.G.; Lindborg, R. Grasslands—More important for ecosystem services than you might think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- O’Mara, F.P. The role of grasslands in food security and climate change. Ann. Bot. 2012, 110, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Richter, F.; Jan, P.; El Benni, N.; Lüscher, A.; Buchmann, N.; Klaus, V.H. A guide to assess and value ecosystem services of grasslands. Ecosyst. Serv. 2021, 52, 101376. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Z.; Wu, J. Grassland ecosystem services: A systematic review of research advances and future directions. Landsc. Ecol. 2020, 35, 793–814. [Google Scholar] [CrossRef]
- Tilman, D.; Reich, P.B.; Knops, J.M. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 2006, 441, 629–632. [Google Scholar] [CrossRef]
- Nan, Z. The grassland farming system and sustainable agricultural development in China. Grassl. Sci. 2005, 51, 15–19. [Google Scholar] [CrossRef]
- Wu, N.; Liu, A.; Ye, R.; Yu, D.; Du, W.; Chaolumeng, Q.; Liu, G.; Yu, S. Quantitative analysis of relative impacts of climate change and human activities on Xilingol grassland in recent 40 years. Glob. Ecol. Conserv. 2021, 32, e01884. [Google Scholar] [CrossRef]
- Xu, D.; Chen, B.; Shen, B.; Wang, X.; Yan, Y.; Xu, L.; Xin, X. The Classification of Grassland Types Based on Object-Based Image Analysis with Multisource Data. Rangel. Ecol. Manag. 2019, 72, 318–326. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, Y.; Zhang, Y.; Shang, J. Review of Remote Sensing Applications in Grassland Monitoring. Remote Sens. 2022, 14, 2903. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Barrett, B.; Nitze, I.; Green, S.; Cawkwell, F. Assessment of multi-temporal, multi-sensor radar and ancillary spatial data for grasslands monitoring in Ireland using machine learning approaches. Remote Sens. Environ. 2014, 152, 109–124. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Vanden Borre, J.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status—New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Lopatin, J.; Fassnacht, F.E.; Kattenborn, T.; Schmidtlein, S. Mapping plant species in mixed grassland communities using close range imaging spectroscopy. Remote Sens. Environ. 2017, 201, 12–23. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocińska, A.; Sławik, Ł. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef]
- Mansour, K.; Mutanga, O.; Everson, T.; Adam, E. Discriminating indicator grass species for rangeland degradation assessment using hyperspectral data resampled to AISA Eagle resolution. ISPRS J. Photogramm. Remote Sens. 2012, 70, 56–65. [Google Scholar] [CrossRef]
- Meng, B.; Yang, Z.; Yu, H.; Qin, Y.; Sun, Y.; Zhang, J.; Chen, J.; Wang, Z.; Zhang, W.; Li, M.; et al. Mapping of Kobresia pygmaea Community Based on Umanned Aerial Vehicle Technology and Gaofen Remote Sensing Data in Alpine Meadow Grassland: A Case Study in Eastern of Qinghai–Tibetan Plateau. Remote Sens. 2021, 13, 2483. [Google Scholar] [CrossRef]
- Corbane, C.; Alleaume, S.; Deshayes, M. Mapping natural habitats using remote sensing and sparse partial least square discriminant analysis. Int. J. Remote Sens. 2013, 34, 7625–7647. [Google Scholar] [CrossRef]
- Buck, O.; Millán, V.E.G.; Klink, A.; Pakzad, K. Using information layers for mapping grassland habitat distribution at local to regional scales. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 83–89. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, W.; Wei, P.; Fang, P.; Zhang, X.; Yan, N.; Liu, W.; Zhao, H.; Wu, Q. Classification of Zambian grasslands using random forest feature importance selection during the optimal phenological period. Ecol. Indic. 2022, 135, 108529. [Google Scholar] [CrossRef]
- Rapinel, S.; Mony, C.; Lecoq, L.; Clément, B.; Thomas, A.; Hubert-Moy, L. Evaluation of Sentinel-2 time-series for mapping floodplain grassland plant communities. Remote Sens. Environ. 2019, 223, 115–129. [Google Scholar] [CrossRef]
- Badreldin, N.; Prieto, B.; Fisher, R. Mapping Grasslands in Mixed Grassland Ecoregion of Saskatchewan Using Big Remote Sensing Data and Machine Learning. Remote Sens. 2021, 13, 4972. [Google Scholar] [CrossRef]
- Hong, G.; Zhang, A.; Zhou, F.; Brisco, B. Geoinformation. Integration of optical and synthetic aperture radar (SAR) images to differentiate grassland and alfalfa in Prairie area. Int. J. Appl. Earth Obs. Geoinf. 2014, 28, 12–19. [Google Scholar]
- Pulliainen, J.T.; Kurvonen, L.; Hallikainen, M.T. Multitemporal behavior of L- and C-band SAR observations of boreal forests. IEEE Trans. Geosci. Remote Sens. 1999, 37, 927–937. [Google Scholar] [CrossRef]
- Forkuor, G.; Benewinde Zoungrana, J.-B.; Dimobe, K.; Ouattara, B.; Vadrevu, K.P.; Tondoh, J.E. Above-ground biomass mapping in West African dryland forest using Sentinel-1 and 2 datasets—A case study. Remote Sens. Environ. 2020, 236, 111496. [Google Scholar] [CrossRef]
- Chang, S.; Wang, Z.; Mao, D.; Guan, K.; Jia, M.; Chen, C. Mapping the Essential Urban Land Use in Changchun by Applying Random Forest and Multi-Source Geospatial Data. Remote Sens. 2020, 12, 2488. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Pacheco, A.D.P.; Junior, J.A.d.S.; Ruiz-Armenteros, A.M.; Henriques, R.F.F. Assessment of k-Nearest Neighbor and Random Forest Classifiers for Mapping Forest Fire Areas in Central Portugal Using Landsat-8, Sentinel-2, and Terra Imagery. Remote Sens. 2021, 13, 1345. [Google Scholar] [CrossRef]
- Jamsran, B.-E.; Lin, C.; Byambakhuu, I.; Raash, J.; Akhmadi, K. Applying a support vector model to assess land cover changes in the Uvs Lake Basin ecoregion in Mongolia. Inf. Process. Agric. 2019, 6, 158–169. [Google Scholar] [CrossRef]
- Ul Din, S.; Mak, H.W.L. Retrieval of Land-Use/Land Cover Change (LUCC) Maps and Urban Expansion Dynamics of Hyderabad, Pakistan via Landsat Datasets and Support Vector Machine Framework. Remote Sens. 2021, 13, 3337. [Google Scholar] [CrossRef]
- Dusseux, P.; Vertès, F.; Corpetti, T.; Corgne, S.; Hubert-Moy, L. Agricultural practices in grasslands detected by spatial remote sensing. Environ. Monit. Assess. 2014, 186, 8249–8265. [Google Scholar] [CrossRef] [PubMed]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Liu, D.; Xia, F. Assessing object-based classification: Advantages and limitations. Remote Sens. Lett. 2010, 1, 187–194. [Google Scholar] [CrossRef]
- Flanders, D.; Hall-Bayer, M.; Pereverzoff, J. Preliminary evaluation of eCognition object-based software for cut block delineation and feature extraction. Can. J. Remote Sens. 2003, 29, 441–452. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Wang, L.; Hu, L.; Gong, P. Comparison of Classification Algorithms and Training Sample Sizes in Urban Land Classification with Landsat Thematic Mapper Imagery. Remote Sens. 2014, 6, 964–983. [Google Scholar] [CrossRef]
- Autrey, B.; Reif, M. Segmentation and object-oriented classification of wetlands in a karst Florida landscape using multi-season Landsat7 ETM+ imagery. Int. J. Remote Sens. 2011, 32, 1471–1489. [Google Scholar] [CrossRef]
- Zerrouki, N.; Bouchaffra, D. Pixel-Based or Object-Based: Which Approach is More Appropriate for Remote Sensing Image Classification? In Proceedings of the International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2014.
- Oruc, M.; Marangoz, A.; Buyuksalih, G. Comparison of Pixel-Based and Object-Oriented Classification Approaches Using Landsat-7 ETM Spectral Bands. In Proceedings of the 20th ISPRS Congress on Technical Commission VII, Istanbul, Turkey, 12–23 July 2004; pp. 1118–1123. [Google Scholar]
- Dingle Robertson, L.; King, D. Comparison of pixel- and object-based classification in land cover change mapping. Int. J. Remote Sens. 2011, 32, 1505–1529. [Google Scholar] [CrossRef]
- Li, F.Y.; Jäschke, Y.; Guo, K.; Wesche, K. Grasslands of China. In Encyclopedia of the World’s Biomes; Goldstein, M.I., DellaSala, D.A., Eds.; Elsevier: Oxford, UK, 2020; pp. 773–784. [Google Scholar]
- Ministry of Agriculture of the People’s Republic of China. Rangeland Resources of China; China Science and Technology Press: Beijing, China, 1996.
- Xilingol League Grassland Station. Xilingol Grassland Resources; Inner Mongolia Daily Youth Press: Hohhot, China, 1988. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Zha, Y.; Gao, J.; Ni, S. Use of normalized difference built-up index in automatically mapping urban areas from TM imagery. Int. J. Remote Sens. 2003, 24, 583–594. [Google Scholar] [CrossRef]
- McFeeters, S.K. The use of the Normalized Difference Water Index (NDWI) in the delineation of open water features. Int. J. Remote Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Soudani, K.; Delpierre, N.; Berveiller, D.; Hmimina, G.; Vincent, G.; Morfin, A.; Dufrêne, É. Potential of C-band Synthetic Aperture Radar Sentinel-1 time-series for the monitoring of phenological cycles in a deciduous forest. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102505. [Google Scholar] [CrossRef]
- Mullissa, A.; Vollrath, A.; Odongo-Braun, C.; Slager, B.; Balling, J.; Gou, Y.; Gorelick, N.; Reiche, J. Sentinel-1 SAR Backscatter Analysis Ready Data Preparation in Google Earth Engine. Remote Sens. 2021, 13, 1954. [Google Scholar] [CrossRef]
- Abrams, M.; Crippen, R.; Fujisada, H. ASTER Global Digital Elevation Model (GDEM) and ASTER Global Water Body Dataset (ASTWBD). Remote Sens. 2020, 12, 1156. [Google Scholar] [CrossRef]
- Tachikawa, T.; Kaku, M.; Iwasaki, A.; Gesch, D.; Oimoen, M.; Zhang, Z.; Danielson, J.; Krieger, T.; Curtis, B.; Haase, J. ASTER Global Digital Elevation Model Version 2—Summary of Validation Results. NASA: Washington, DC, USA, 2011. [Google Scholar]
- Nussbaum, S.; Menz, G. eCognition Image Analysis Software; Springer: Berlin/Heidelberg, Germany, 2008; pp. 29–39. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Probst, P.; Bischl, B.; Boulesteix, A.-L. Tunability: Importance of hyperparameters of machine learning algorithms. J. Mach. Learn. Res. 2018, 20, 1934–1965. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support Vector Regression. In Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers; Awad, M., Khanna, R., Eds.; Apress: Berkeley, CA, USA, 2015; pp. 67–80. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inform. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Landau, S.; Leese, M.; Stahl, D.; Everitt, B.S. Cluster Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Russell, S.J. Artificial Intelligence: A Modern Approach, 3rd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2010. [Google Scholar]
- Townsend, J.T. Theoretical analysis of an alphabetic confusion matrix. Percept. Psychophys. 1971, 9, 40–50. [Google Scholar] [CrossRef]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
- Whiteside, T.G.; Boggs, G.S.; Maier, S.W. Comparing object-based and pixel-based classifications for mapping savannas. Int. J. Appl. Earth Observ. Geoinform. 2011, 13, 884–893. [Google Scholar] [CrossRef]
- Fu, B.; Wang, Y.; Campbell, A.; Li, Y.; Zhang, B.; Yin, S.; Xing, Z.; Jin, X. Comparison of object-based and pixel-based Random Forest algorithm for wetland vegetation mapping using high spatial resolution GF-1 and SAR data. Ecol. Indic. 2017, 73, 105–117. [Google Scholar] [CrossRef]
- Du, B.; Mao, D.; Wang, Z.; Qiu, Z.; Yan, H.; Feng, K.; Zhang, Z. Mapping Wetland Plant Communities Using Unmanned Aerial Vehicle Hyperspectral Imagery by Comparing Object/Pixel-Based Classifications Combining Multiple Machine-Learning Algorithms. IEEE J. Select. Top. Appl. Earth Observ. Remote Sens. 2021, 14, 8249–8258. [Google Scholar] [CrossRef]
- Qu, L.a.; Chen, Z.; Li, M.; Zhi, J.; Wang, H. Accuracy Improvements to Pixel-Based and Object-Based LULC Classification with Auxiliary Datasets from Google Earth Engine. Remote Sens. 2021, 13, 453. [Google Scholar] [CrossRef]
- Tassi, A.; Gigante, D.; Modica, G.; Di Martino, L.; Vizzari, M. Pixel- vs. Object-Based Landsat 8 Data Classification in Google Earth Engine Using Random Forest: The Case Study of Maiella National Park. Remote Sens. 2021, 13, 2299. [Google Scholar] [CrossRef]
- Qian, Y.; Zhou, W.; Yan, J.; Li, W.; Han, L. Comparing Machine Learning Classifiers for Object-Based Land Cover Classification Using Very High Resolution Imagery. Remote Sens. 2015, 7, 153–168. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support Vector Machine Versus Random Forest for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Abdi, A.M. Land cover and land use classification performance of machine learning algorithms in a boreal landscape using Sentinel-2 data. GIScience Remote Sens. 2020, 57, 1–20. [Google Scholar] [CrossRef]
- Parikh, H.; Patel, S.; Patel, V. Classification of SAR and PolSAR images using deep learning: A review. Int. J. Image Data Fusion 2020, 11, 1–32. [Google Scholar] [CrossRef]
- Joshi, N.; Baumann, M.; Ehammer, A.; Fensholt, R.; Grogan, K.; Hostert, P.; Jepsen, M.R.; Kuemmerle, T.; Meyfroidt, P.; Mitchard, E.T.A.; et al. A Review of the Application of Optical and Radar Remote Sensing Data Fusion to Land Use Mapping and Monitoring. Remote Sens. 2016, 8, 70. [Google Scholar] [CrossRef]
- Flores-Anderson, A.I.; Herndon, K.E.; Thapa, R.B.; Cherrington, E. The SAR Handbook: Comprehensive Methodologies for Forest Monitoring and Biomass Estimation; NASA: Washington, DC, USA, 2019.
- Kouw, W.M.; Loog, M. A Review of Domain Adaptation without Target Labels. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 766–785. [Google Scholar] [CrossRef] [PubMed]
- Farahani, A.; Voghoei, S.; Rasheed, K.; Arabnia, H.R. A Brief Review of Domain Adaptation. Proc. Adv. Data Sci. Inf. Eng. 2021, 2021, 877–894. [Google Scholar]
- Zadrozny, B. Learning and evaluating classifiers under sample selection bias. In Proceedings of the Twenty-First International Conference on Machine Learning ICML, Banff, AB, Canada, 4–8 July 2004; Volume 2004. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2016; pp. 770–778. [Google Scholar]



| Feature Types | Image Layers | No. of Features | |
|---|---|---|---|
| S1 | Multispectral band | Blue, green, red, NIR, SWIR1, SWIR2 | 6 |
| S2 | Spectral indices | NDVI-4, NDVI-7, NDVI-9, NDWI, NDBI | 5 |
| S3 | Geography | Aspect, elevation, slope, X, Y | 5 |
| S4 | SAR | VH, VV, VH/VV, VV/VH | 4 |
| Feature Types | Object Features | No. of Features | ||
|---|---|---|---|---|
| C1 | S1 | Blue, green, red, NIR, SWIR1, SWIR2 | Mean, standard deviation | 12 |
| S2 | NDVI-4, NDVI-7, NDVI-9, NDWI, NDBI | 10 | ||
| S3 | Aspect, elevation, slope | 6 | ||
| S4 | VH, VV, VH/VV, VV/VH | 8 | ||
| C2 | Area, width, length, length/width, asymmetry, density, compactness, roundness | Geometry | 8 | |
| C3 | X max, X min, X center, Y max, Y min, Y center | Position | 6 | |
| C4 | Blue, green, red, NIR, SWIR1, SWIR2, NDVI-4, NDVI-7, NDVI-9, NDWI, NDBI, aspect, elevation, slope, VH, VV, VH/VV, VV/VH | Mean, homogeneity, dissimilarity, entropy, contrast, correlation | 108 | |
| RF | SVM | KNN | NB | |||||
|---|---|---|---|---|---|---|---|---|
| OA (%) | Kappa | OA (%) | Kappa | OA (%) | Kappa | OA (%) | Kappa | |
| S1 | 79.76 | 0.7553 | 82.98 | 0.7945 | 77.09 | 0.723 | 64.77 | 0.5813 |
| S1 + S2 | 84.27 | 0.8096 | 85.92 | 0.8304 | 78.01 | 0.7338 | 74.15 | 0.6918 |
| S1 + S2 + S3 | 95.58 | 0.9467 | 95.31 | 0.9435 | 80.40 | 0.7632 | 86.84 | 0.8416 |
| S1 + S2 + S4 | 86.46 | 0.8365 | 86.48 | 0.837 | 76.63 | 0.7171 | 77.55 | 0.7323 |
| S1 + S2 + S3 + S4 | 95.40 | 0.9445 | 95.77 | 0.949 | 79.21 | 0.7487 | 87.49 | 0.8495 |
| RF | SVM | KNN | NB | |||||
|---|---|---|---|---|---|---|---|---|
| OA (%) | Kappa | OA (%) | Kappa | OA (%) | Kappa | OA (%) | Kappa | |
| S1 | 94.30 | 0.9311 | 94.03 | 0.9279 | 85.48 | 0.8247 | 77.21 | 0.7269 |
| S1 + S2 | 94.85 | 0.9379 | 93.84 | 0.9257 | 87.13 | 0.8445 | 81.71 | 0.7804 |
| S1 + S2 + S3 | 96.88 | 0.9623 | 96.69 | 0.9601 | 88.33 | 0.8591 | 86.21 | 0.8344 |
| C1 | 97.70 | 0.9722 | 97.43 | 0.9689 | 88.60 | 0.8625 | 88.79 | 0.8653 |
| C1 + C2 | 97.43 | 0.9689 | 97.24 | 0.9667 | 87.50 | 0.8488 | 89.43 | 0.873 |
| C1 + C2 + C3 | 98.62 | 0.9833 | 97.59 | 0.9754 | 94.68 | 0.9378 | 92.12 | 0.9091 |
| C1 + C2 + C3 + C4 | 98.25 | 0.9789 | 97.43 | 0.9689 | 94.94 | 0.9389 | 91.36 | 0.8958 |
| Object-Based Algorithms | Selected Variables | No. of Variables |
|---|---|---|
| RF | Mean (S1, S2, elevation, VV, VH), standard deviation (blue, red, NDVI-7, NDWI, elevation), C3, homogeneity (blue, green, elevation), entropy (blue, elevation, slope), correlation (elevation) | 32 |
| SVM | Mean (S1, S2, S3, VV, VH), standard deviation (S1, S2, elevation), width, length, length/width, C3, GLCM mean (S1, S2), homogeneity (red, green, S2, S3), dissimilarity (S2), entropy (S1, elevation, S3, VV, VH), contrast (blue, green, NIR, S2, S3), correlation (red, S2, S3) | 95 |
| KNN | All | 158 |
| NB | Mean (red, NIR, SWIR1, SWIR2, S2, aspect, VV, VH), standard deviation (SWIR1, SWIR2, NDVI-4, NDVI-7, Aspect), Xmin, Xmax, Xcenter, GLCM mean (blue, green, red, NDVI-7, NDWI), homogeneity (red, SWIR1, SWIR2, NDVI-7, NDVI-9, NDBI, aspect, slope), entropy (SWIR1, SWIR2, aspect, slope, VV), dissimilarity (NDVI-7, NDVI-9) | 40 |
| All | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | T11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RF vs. SVM | 0.000 | 0.500 | 0.001 | 0.000 | 1.000 | 0.250 | 0.039 | 0.021 | 0.001 | 0.092 | 0.302 | 0.000 |
| RF vs. KNN | 0.000 | 0.500 | 0.000 | 0.000 | 0.000 | 0.002 | 0.125 | 0.375 | 0.000 | 0.000 | 0.000 | 0.000 |
| RF vs. NB | 0.000 | 0.500 | 0.000 | 0.000 | 0.006 | 0.063 | 0.219 | 0.375 | 0.000 | 0.000 | 0.000 | 0.000 |
| SVM vs. KNN | 0.000 | 1.000 | 0.038 | 0.000 | 0.000 | 0.016 | 0.625 | 0.001 | 0.035 | 0.000 | 0.000 | 0.000 |
| SVM vs. NB | 0.000 | 1.000 | 0.038 | 0.000 | 0.006 | 0.500 | 0.250 | 0.001 | 0.008 | 0.000 | 0.000 | 0.000 |
| KNN vs. NB | 0.002 | 1.000 | 1.000 | 0.286 | 0.000 | 0.063 | 1.000 | 1.000 | 0.375 | 1.000 | 0.388 | 0.222 |
| All | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | T11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RF vs. SVM | 0.618 | 1.000 | 0.000 | 0.000 | 0.016 | 0.063 | 0.125 | 0.004 | 0.089 | 0.519 | 0.000 | 0.066 |
| RF vs. KNN | 0.000 | 1.000 | 0.000 | 0.038 | 0.031 | 0.250 | 1.000 | 0.008 | 0.238 | 0.001 | 0.000 | 0.000 |
| RF vs. NB | 0.004 | 0.500 | 0.000 | 0.000 | 0.500 | 1.000 | 0.500 | 0.063 | 0.388 | 0.222 | 0.710 | 0.085 |
| SVM vs. KNN | 0.000 | 1.000 | 1.000 | 0.004 | 1.000 | 0.500 | 0.250 | 1.000 | 0.000 | 0.000 | 0.016 | 0.007 |
| SVM vs. NB | 0.002 | 1.000 | 0.125 | 0.675 | 0.125 | 0.125 | 0.625 | 0.125 | 0.011 | 0.101 | 0.000 | 0.000 |
| KNN vs. NB | 0.000 | 0.500 | 0.219 | 0.029 | 0.219 | 0.500 | 1.000 | 0.250 | 0.804 | 0.000 | 0.000 | 0.000 |
| All | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | T11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RF vs. RF | 0.000 | 1.000 | 0.000 | 0.000 | 1.000 | 1.000 | 1.000 | 0.125 | 0.000 | 0.000 | 0.000 | 0.000 |
| SVM vs. SVM | 0.318 | 1.000 | 0.000 | 0.403 | 0.070 | 0.727 | 0.289 | 0.629 | 0.054 | 0.007 | 0.424 | 1.000 |
| KNN vs. KNN | 0.000 | 0.500 | 0.000 | 0.000 | 0.000 | 0.065 | 0.063 | 0.039 | 0.007 | 0.000 | 0.000 | 0.000 |
| Bayes vs. NB | 0.000 | 1.000 | 0.000 | 0.000 | 0.012 | 0.219 | 0.250 | 0.219 | 0.003 | 0.000 | 0.000 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, N.; Crusiol, L.G.T.; Liu, G.; Wuyun, D.; Han, G. Comparing Machine Learning Algorithms for Pixel/Object-Based Classifications of Semi-Arid Grassland in Northern China Using Multisource Medium Resolution Imageries. Remote Sens. 2023, 15, 750. https://doi.org/10.3390/rs15030750
Wu N, Crusiol LGT, Liu G, Wuyun D, Han G. Comparing Machine Learning Algorithms for Pixel/Object-Based Classifications of Semi-Arid Grassland in Northern China Using Multisource Medium Resolution Imageries. Remote Sensing. 2023; 15(3):750. https://doi.org/10.3390/rs15030750
Chicago/Turabian StyleWu, Nitu, Luís Guilherme Teixeira Crusiol, Guixiang Liu, Deji Wuyun, and Guodong Han. 2023. "Comparing Machine Learning Algorithms for Pixel/Object-Based Classifications of Semi-Arid Grassland in Northern China Using Multisource Medium Resolution Imageries" Remote Sensing 15, no. 3: 750. https://doi.org/10.3390/rs15030750
APA StyleWu, N., Crusiol, L. G. T., Liu, G., Wuyun, D., & Han, G. (2023). Comparing Machine Learning Algorithms for Pixel/Object-Based Classifications of Semi-Arid Grassland in Northern China Using Multisource Medium Resolution Imageries. Remote Sensing, 15(3), 750. https://doi.org/10.3390/rs15030750









