Abstract
VIS-NIR-SWIR hyperspectroscopy is a significant technique used in remote sensing for classification of prediction-based chemometrics and machine learning. Chemometrics, together with biophysical and biochemical parameters, is a laborious technique; however, researchers are very interested in this field because of the benefits in terms of optimizing crop yields. In this study, we investigated the hypothesis that VIS-NIR-SWIR could be efficiently applied for classification and prediction of leaf thickness and pigment profiling of green lettuce in terms of reflectance, transmittance, and absorbance data according to the variety. For this purpose, we used a spectroradiometer in the visible, near-infrared, and shortwave ranges (VIS-NIR-SWIR). The results showed many chemometric parameters and fingerprints in the 400–2500 nm spectral curve range. Therefore, this technique, combined with rapid data mining, machine learning algorithms, and other multivariate statistical analyses such as PCA, MCR, LDA, SVM, KNN, and PLSR, can be used as a tool to classify plants with the highest accuracy and precision. The fingerprints of the hyperspectral data indicated the presence of functional groups associated with biophysical and biochemical components in green lettuce, allowing the plants to be correctly classified with higher accuracy (99 to 100%). Biophysical parameters such as thickness could be predicted using PLSR models, which showed R2P and RMSEP values greater than >0.991 and 6.21, respectively, according to the relationship between absorbance and reflectance or transmittance spectroscopy curves. Thus, we report the methodology and confirm the ability of VIS-NIR-SWIR hyperspectroscopy to simultaneously classify and predict data with high accuracy and precision, at low cost and with rapid acquisition, based on a remote sensing tool, which can enable the successful management of crops such as green lettuce and other plants using precision agriculture systems.
1. Introduction
Lettuce (Lactuca sativa L.) is a vegetable of great importance for a healthy diet due to its nutritional composition [1,2]. Green lettuce varieties are among the most economically important and popular vegetables consumed in Brazil and around the world. Their annual production worldwide is estimated at 28 million tons, and many studies have applied different techniques and tools to correctly predict the chemometric parameters (biophysical and biochemical) of these plants (FAO, 2022).
Leaf pigments such as carotenoids and chlorophyll directly influence plant biochemical processes and therefore crop development and nutritional value, and the amount of pigments is influenced by the plant variety and environmental conditions [3,4]. In Brazil, green lettuce varieties, with a higher content of green pigments and carotenoids, as well as increased leaf thickness (which influences additional light-scattering structures) and other sensory characteristics (i.e., crispness and succulence), are among the most popular vegetables consumed [5,6]. Accordingly, predicting and classifying green lettuce varieties is of great interest for indoor farms, greenhouses, and classical agricultural production, as different varieties have different biophysical and nutritional properties, and the knowledge of these differences can enable the production of specific varieties.
VIS-NIR-SWIR spectroscopy is a significant technique for managing green lettuce crops using precision agriculture systems [7,8,9]. For example, high-throughput sensor spectroradiometer technology can be used to monitor reflectance properties based on hundreds of contiguous narrow bands [10,11,12]. Furthermore, these remote sensing tools enable rapid and nondestructive classification and monitoring of lettuce and other vegetables [8,10,13,14,15]. The principle of their operation is based on interactions between leaves and the vibrational relationship of light with molecular organic bonds, mainly –C–H, –N–H, –COOH, –NH3, and –O–H. This results in vibrational excitation at specific wavelengths (fingerprints) in the visible (VIS: 400–700 nm), near-infrared (NIR: 700–1100 nm), and shortwave infrared (SWIR: 1100–2400 nm) spectral regions. For example, according to pioneering studies [7,8,9,16], VIS–NIR–SWIR-based reflectance (R), transmittance (T), and absorbance (A) spectra are associated with certain fingerprints and promote high accuracy and precision, reduced risk of bias and noise, and high repeatability compared with other methods based on an integrating sphere for the classification and prediction of many chemometric attributes in plants [7,8,9,16]. In addition, with these remote sensing tools, there is no need to prepare reagents or exert extensive labor using high-cost equipment (e.g., UHPLC, 1H-NMR, FTIR, or DRX) for the acquisition of samples/spectra to classify plants [8,16].
An ever-expanding method is the use of data mining and machine learning to directly classify models, which are frequently used along with plant parameters in remote sensing tools [15,17,18]. In this sense, the portability, speed, accuracy, and sensitivity of spectroradiometer devices, combined with the capacity of algorithms and multivariate tools for modeling [7,8,10,14,18,19,20], allow advances in the accuracy of classification, chemometric analysis, and monitoring of plant growth and development, as well as thickness characteristics in leaves. Thus, this method can be used to classify and predict many commonly investigated biophysical plant parameters by implementing remote sensing tools [7,8,21], including biochemical and morphological attributes, facilitated by the new era of remote sensing [21,22,23,24,25]. For example, a calibration model can be used to predict the variable of interest in unknown samples based on individual and specific spectral signatures in green lettuce following the flowchart proposed in Figure 1.
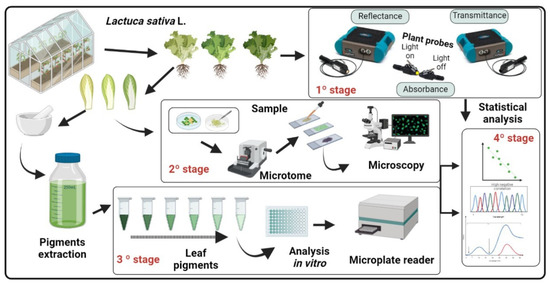
Figure 1.
Flowchart of methodology used in data mining and machine learning based on hyperspectroscopy and anatomical analysis of classification and prediction parameters in green lettuce.
The main objectives of this study were (a) to evaluate the prediction of biophysical and biochemical components of green lettuce by VIS-NIR-SWIR hyperspectral reflectance, transmittance, and absorbance combined with machine learning models, and (b) to evaluate the capacity of these biophysical and biochemical attributes to classify green lettuce varieties. For this purpose, the full spectra of the VIS-NIR-SWIR hyperspectroradiometer (400–2400 nm) were analysed in green lettuce plants.
2. Material and Methods
2.1. Plant Material, Growth Conditions, and Experimental Design
Experiments were conducted at the Department of Agronomy at the State University of Maringá, Maringá, Paraná, Brazil. Lettuce plants (Lactuca sativa L.) grown in classical hydroponic culture in a greenhouse were analysed. The experimental design was a random scheme with 3 varieties: Lisa, Crespa, and Americana. A total of 600 samples were used to collect data. On the 21st day after hydroponic cultivation, the samples were analysed (Figure 1 and Figure 2).

Figure 2.
Left to right Lisa, Crespa, and Americana varieties of lettuce; full plants on top, leaves on bottom.
2.2. Extraction of Leaf Pigments
Quantification of carotenoids (Car) and chlorophylls (a, b, and a+b) was carried out by crushing leaf segments (1 cm2) with 2 mL of methanol. The extracts were then centrifuged at 15,000 rpm for 6 min and transferred to a 1.5 mL tube. All readings and quantifications were performed exactly as described in [26]. The concentrations of chlorophylls and carotenoids (Chl a, Chl b, Chl a+b, and Car) were expressed in terms of area (mg cm−2) and mass (mg g−1) [27].
2.3. Optical Microscopy Analysis
Cross-sections of fresh leaves (hand cut) and leaf segments (1 cm2) were fixed with Karnovsky’s solution, dehydrated in a series of increasing concentrations of ethanol (50, 70, 80, 90, and 100% (3 times)), and infiltrated with methyl methacrylate (Leica Historesin®) [26]. Block sectioning was performed on a rotary microtome (Eikonal, São Paulo, SP, Brazil) and dyed with toluidine blue at pH 4.5. Images were analysed exactly as described in [26] by Fiji ImageJ v.2.9.0 software [28].
2.4. Optical Reflectance, Transmittance, and Absorbance Properties of Leaves
Reflectance (R) and transmittance (T) were measured using 2 plant probes coupled with spectroradiometers (FieldSpec 3, ASD Inc., Boulder, CO, USA). An ASD spectroradiometer light beam plant probe was coupled (and calibrated with a standard Spectralon® dish as 100% reflectance) to another probe at the opposite leaf surface with the light off to simultaneously measure leaf reflectance and transmittance (350–2500 nm), as described in [7,8,16]. Light absorption (A) was estimated as A = 1 − (R + T) [8,16], and the measurement and statistical analysis scheme are shown in Figure 1.
2.5. Statistical Analyses
All statistical analyses were performed using XLSTAT (Addinsoft, New York, NY, USA), Excel 2021® (Microsoft Office Inc., Torrance, CA, USA), The Unscrambler X 10.4® (CAMO Software, Oslo, Norway), Statistica® 12.0 software (Statsoft Inc., Tulsa, OK, USA), SigmaPlot® 12.0 (Systat, Santa Clara, CA, USA), and the R package (R Core Team, 2020; https://www.R-project.org; accessed on 5 October 2022) [29].
2.5.1. Descriptive and Univariate Statistical Analyses
Mean and standard error, maximum, minimum, and coefficient of variation (CV, %) were calculated, following [30]. CV was classified exactly as described in [8]. The data (pigments and anatomy) were submitted to one-way ANOVA for mean comparisons. Duncan’s post hoc test was considered significant at p < 0.01 [31].
2.5.2. Analysis of Leaf Spectral Fingerprints
The hyperspectral curves, parameters derived from the hyperspectral data, data mining, and machine learning algorithms were used for decision analysis. The effect of the green lettuce variety (Lisa, Crespa, or Americana) on the leaf traits was analysed by one-way analysis of variance (ANOVA). The effects of the variety on the reflectance (R), transmittance (T), and absorbance (A) profiles were assessed using PERMANOVA exactly as described in [8,29].
2.5.3. Principal Component Analysis (PCA)
Principal component analysis (PCA) (p < 0.05) was performed using The Unscrambler X software, version 10.4 (CAMO Software, Oslo, Norway). To avoid underfitting and overfitting the ideal number of PCs, we assumed the number of PCs corresponding to the first maximum value of the overall accuracy to classify the base PCA and other statistical methods based on data mining and machine learning as described in [32].
2.5.4. Multivariate Curve Resolution (MCR)
Multivariate curve resolution (MCR) was used as a group of techniques, also known as blind source separation or self-modelling mixture analysis. The MCR components were used to an unresolved extent using a minimal number of assumptions about the biophysical (thickness and interaction of light with matter) and biochemical (pigments and other molecules) properties of the samples [10]. In addition, variable importance in projection (VIP) was selected after the analysis based on hyperspectral curves [8].
2.5.5. Linear Discriminant Analysis (LDA)
LDA was carried out to obtain models to classify each reflectance, transmittance, and absorbance spectrum of green lettuce. Before obtaining the discriminant models, the PCA–LDA procedure (3 components) was performed, allowing the selection of wavelengths that would best explain the differences between varieties [33]. Linear, quadratic, and Mahalanobis models of analysis were applied to classify green lettuce using machine learning algorithms [14,34].
2.5.6. Support Vector Machine (SVM)
SVM analysis is a supervised, kernel-based nonlinear learning method that uses nuclear techniques to manage complex nonlinear problems with good performance [32]. The function can take many forms, thus providing the ability to handle nonlinear regression cases, as described in [8]. The final decision function of the SVM is determined by only a few support vectors (machine learning). The complexity of the calculation depends on the number of support vectors [35].
2.5.7. K-Nearest Neighbour (KNN)
KNN is a supervised learning algorithm that can describe nonlinear relationships between collected samples and hyperspectral data, and it is widely used to solve classification problems. KNN works as follows: (1) given test samples, a specific distance evaluation method is used to determine the k samples closest to them, and (2) prediction classification is performed based on these k samples. The KNN results strongly depend on the choice of k-based prediction model. The data were processed by k-linear, k-sigmoid, k-log, and k-weighted models based on the hyperspectral data [36]. KNN algorithms were applied to test different functions for each classifier to build the prediction model and different numbers of VIPs and PCs were used to perform the sample classification with higher accuracy.
2.5.8. Partial Least Squares Regression (PLSR) by Analysis of Spectroscopy Data
Hyperspectral data were centred on the mean and subjected to PLSR. To obtain prediction models of thickness based on biophysical and biochemical compounds, the spectral data of the different parameters of 600 samples were divided into two groups: 75% (450) of the samples in the first group, with the aim of creating the model (training), and the remaining 25% (150) in the second group, with the aim of testing (prediction) to adjust the model, as described in [7,8]. This proportion (75:25) was selected to assure estimates that were (1) valid, in the sense that they did not overestimate the accuracy (i.e., did not underestimate the approximation error), and (2) the most accurate among all valid estimates (i.e., their overestimation of the approximation error was the smallest possible), based on a previous analysis described in [30,31,37,38,39,40].
Calibration (Cal) and cross-validation (Cva) were used to predict the quality attributes based on thickness (biophysical and biochemical compounds in leaves) with respect to data mining and machine learning algorithms. In addition, the predictive ability of the calibration model was evaluated by calculating metrics such as coefficient of determination (R2), offset, root mean square error (RMSE), and ratio of performance to deviation (RPD), and bias was used to assess the quality, precision, and accuracy of the model, as described in [33,41].
3. Results
3.1. Descriptive Analysis-Based Biochemical and Biophysical Attributes of Lettuce
The coefficient of variation (CV%) of the leaf pigment base area, mass, and thickness parameters of the 3 lettuce varieties, Lisa, Crespa, and Americana, are given in Figure 3 and Table 1. The CV values were between 3.7 and 25.8% (Table 1). For example, of the 11 parameters analysed and reported, 9 show CV values classified as low to high, with 2 low-medium (Chl a/Chl b, Car/Chl a+b), 4 medium (Chl a (mass), Chl b (mass), Chl a+b (mass), thickness), and 5 high (Chl a (area), Chl b (area), Chl a+b (area), Car (area), Car (mass)). In addition, significant differences (p < 0.01) for many parameters based on pigmented area and mass were noted, except for the Car/Chl a+b ratio (p > 0.05), as shown in Table 1.
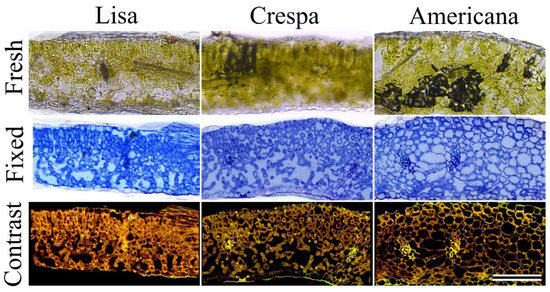
Figure 3.
Microscopy of fresh leaf cross-section (top), historesin-fixed (middle), and phase contrast analysis (bottom) of representative leaves from Lisa, Crespa, Americana lettuce varieties. Scale bar = 150 µm.

Table 1.
Descriptive analysis of lettuce plants. Photosynthetic pigment content expressed by leaf area (mg m−2), mass (mg g−1), and thickness (µm). Different letters in rows show significant differences between Lisa, Crespa, and Americana lettuce varieties (Duncan’s test; p < 0.01) (n = 600 ± SE).
Biophysical (anatomical) analysis revealed different characteristics between Lisa and Americana varieties, such as increased heterogeneity, anisodiametric cells, and lacuna in tissues from homogeneous, isodiametric, and compact parenchyma tissues in leaves (Figure 3). Anatomical characteristics, such as the distribution and format of cells in the mesophyll and leaf thickness (palisade and spongy) of parenchyma cells, showed responses to higher concentrations of chlorophylls (pigment vs. thickness-based area and mass) in green lettuce (Figure 3). The data show that leaf thickness increased 41.2 and 44.5% compared to thickness and Chl a+b in Lisa and Americana (Table 1, Figure 3).
3.2. Hyperspectral Analysis of Leaves
Leaf hyperspectral reflectance (R), transmittance (T), and absorbance (A) data for the three varieties of lettuce used for cultivation (600 samples; average of hyperspectroscopy data) are shown in Figure 4. PERMANOVA discriminated significance among wavelengths from the spectra (F: 4.83, 3.62, 4.28; p < 0.001) (Figure 4A–C). There were significant and slight variations in R, T, and A hyperspectral parameters, particularly in the visible (VIS) region (400–700 nm), owing to leaf pigments such as carotenoids and chlorophyll, and in the near-infrared (NIR) region (700–1100 nm), due to structural differences in the leaf mesophyll. The majority of functional groups (molecular vibrational) distinguished in the shortwave infrared (SWIR) region (1150–2400 nm) showed differences between varieties. Lisa and Crespa showed increased reflectance and transmittance in the green, NIR, and SWIR spectral regions, while the values were lower for the absorbance spectra of increased thickness and pigment in leaf tissue (Figure 4).
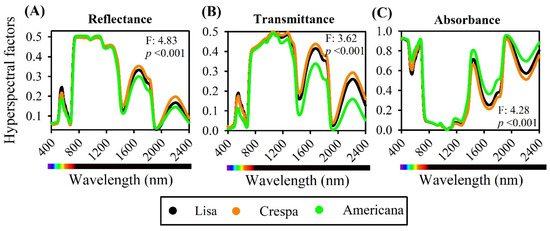
Figure 4.
Average foliar hyperspectral factor profiles of fully expanded leaves of Lisa, Crespa, and Americana varieties of green lettuce. (A) Reflectance, (B) transmittance, and (C) absorbance of adaxial light source. F- and p-values from permutation analysis of variance (PERMANOVA) of full range (400–2400 nm) hyperspectral reflectance, transmittance, and absorbance of lettuce leaves reported in top-right or bottom-right corner of panel (n = 600).
The near-infrared region (700–1300 nm) showed reflectance of up to 47% of incident light and absorbance, with a maximum of 15% at 710 nm, progressively decreasing to 1058 nm and reaching values close to zero (Figure 4). At approximately 1300–2500 nm, the shortwave infrared region (1300–2500 nm), absorbance increased and reflectance decreased (Figure 4A,B), particularly for the Americana variety (Figure 4C).
3.3. Principal Component Analysis (PCA)
The first, second, and third principal components (PCs) represent 99.8, 100, and 100% of reflectance, transmittance, and absorbance, respectively, of the total variance that can be explained (Figure 5).
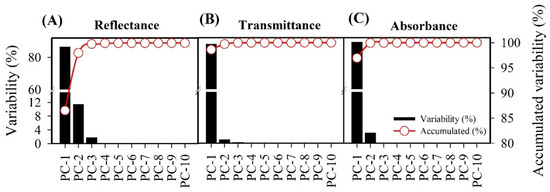
Figure 5.
Principal component analysis (PCA) and PC scores obtained from hyperspectral analysis of lettuce plants: (A) reflectance, (B) transmittance, (C) absorbance. Black bars indicate variability of PC, red circles indicate accumulated variability.
PCA at 400–2400 nm indicated the formation of distinct clusters between varieties through the analysis of reflectance, transmittance, and absorbance data (Figure 6). PC1 to PC3 score plots show a dense clustering formation with clear boundaries between the three lettuce varieties, with large dispersion along with the first three PCs. Wavelengths also contribute to pigments, structural components, and water based on vibrational bands.
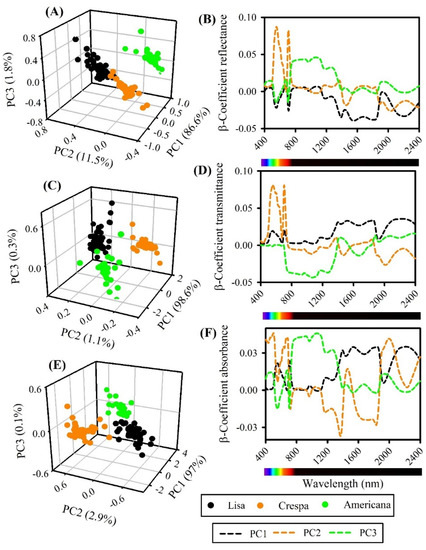
Figure 6.
(A) Contribution of principal component analysis (PCA) and β-coefficients for hyperspectral data (400–2400 nm) of lettuce plants: (A,B) reflectance, (C,D) transmittance, (E,F) absorbance. (A,C,E) PC score 3D plots. (B,D,F) β-coefficients for reflectance, transmittance, and absorbance, respectively (n = 600).
The reflectance of the β-coefficients showed that the main variance occurred for the green (535–580 nm) and NIR-SWIR (1300–1800 nm) bands, but with contributions from almost all wavelengths in the composition by green and red bands (535–580 and 660–680 nm) in PC1, SWIR (1900–2400 nm) in PC2, and green (535–580 nm) and NIR (710–1400 nm) in PC3 (Table 2, Figure 6A,B).

Table 2.
Most responsive variable importance for projection (VIP) values by wavelength selected according to the first three regression coefficients for principal component (PC) and multivariate curve resolution (MCR) components by spectroscopy from reflectance, transmittance, and absorbance of leaves.
A similar pattern was observed in transmittance β-coefficient spectra; the sample variety groups formed a cluster with few dispersions in the first three PCs (Table 2, Figure 6C,D) and a dense cluster with clear boundaries between green lettuce samples. The loading plot showed that the main variance occurred for VIS and SWIR (500–600 and 1900–2200 nm) in PC1, higher loading in green and red bands (540–595 and 654–690 nm) in PC2, and higher correspondence to NIR (702–1500 nm) (Figure 6C,D). In addition, the inverse of NIR-SWIR was observed between reflectance and transmittance β-coefficients, even if similar VIP wavenumbers were observed (Table 2, Figure 6A–D).
The absorbance spectrum range (400–2400 nm) is displayed in Figure 6E,F. The clustering formation between the Lisa, Crespa, and Americana varieties remained the same as before, showing good dispersion with a clear boundary separation. β-coefficients showed the full contribution of each wavelength to classification and distinct cluster formation in PC1, PC2, and PC3 by VIS-NIR-SWIR bands.
3.4. Multivariate Curve Resolution (MCR)
Multivariate curve resolution (400–2400 nm) indicated the formation of distinct MCR components between green lettuce varieties. The first three MCR components (MCR1, MCR2, MCR3) were identified through the analysis of reflectance, transmittance, and absorbance data (Figure 7). MCR1-3 did not show any overlap and explained over 98% of the total data variance obtained for reflectance, transmittance, and absorbance at all wavelengths analysed (Figure 7).
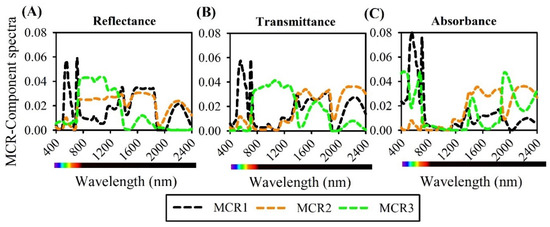
Figure 7.
Major contribution of multivariate curve resolution (MCR) component of hyperspectral (A) reflectance, (B) transmittance, and (C) absorbance (400–2400 nm) of lettuce plants (n = 600).
The MCR curves are not inversely proportional to the reflectance and transmittance curves in the VIS bands (400–700 nm). For example, the regions formed by violet (401–439 nm) and blue (440–485 nm) bands had higher and more significant contributions (p < 0.01) by classification between green lettuce varieties and showed many VIP wavelengths to distinguish these varieties (Figure 2 and Figure 7).
The MCR components also showed the highest values in reflectance and transmittance of up to 355% of the incident light in the near-infrared region (700–1300 nm) and many VIP wavelengths in relation to the absorbance spectra data (Figure 7A,C). Furthermore, the absorbance hyperspectral data showed decreases up to 750 nm, reaching values close to zero for MCR1-3 (Table 2, Figure 7C). On the other hand, in the SWIR region (1500–1600 nm range, peak 1555 nm; 1900–2300 nm, peak 2110 nm), increased reflectance and transmittance (Figure 7A,B) and increased absorbance are noted (Figure 7C). MCR analysis produced three spectral signatures that enabled classification of each spectral band. Thus, we provide additional evidence that the data mining and machine learning results are based on increased component values in VIS and SWIR data (Table 2, Figure 7).
3.5. Regression Coefficient (RC) and Variable Importance in Projection (VIP)
The RC and VIP values for the projection chemometrics of the PCA, LDA, and PLSR models are shown in Table 2. Valleys and peaks of the regions where VIP and RC values appear demonstrate a great influence, showing that machine learning and algorithms for classification and construction of the prediction model were, in general, well dispersed among all VIS-NIR-SWIR bands.
The VIP and RC values used for the machine learning algorithm models differed from 6–12 wavelengths (valleys and peaks), and there were higher RCs in regions near 410 (violet), 445 (blue), 555 (green), 672 (red), 699–750 (red edge), 1330 (NIR), 1450 (SWIR), 1945 (SWIR), and 2215 (SWIR) nm (Table 2, Figure 6 and Figure 7). Although the classification used VIP and RC values, they demonstrated high accuracy in classifying and distinguishing the varieties and contributed to the performance based on data mining (Table 2). In this sense, LDA, SVM, KNN, and RPD were good examples, displaying excellent prediction performance together with VIP values selected by wavelengths (Table 2).
3.6. Model Evaluation of Chemometric Parameters
Machine learning algorithms were applied to sample group classification using hyperspectral raw data and PCA data of full spectra (400–2400 nm). A general accuracy of 99% in the 400–2400 nm range was obtained by the LDA-PLS method using the first three PCs, carrying 99.9% of data variance. The confusion matrix shows >99.9% accuracy for LDA-linear, contributing to lower misclassification between training and test data. In addition, reflectance, transmittance, and absorbance showed higher accuracy in correctly classifying green lettuce, while other machine learning algorithms did not classify with similar precision. Linear SVM was obtained by using three PCs, carrying 99.5 and 99% reflectance and absorbance accuracy, to classify lettuce varieties (Figure 8).
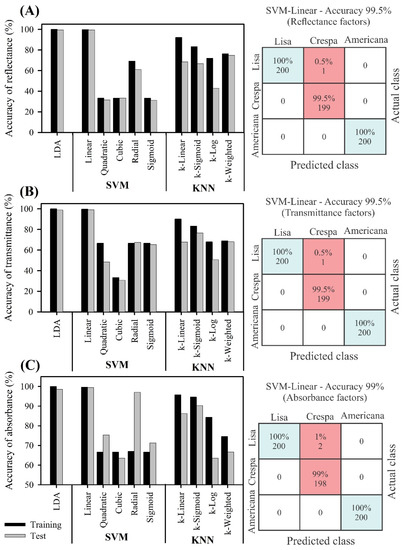
Figure 8.
Overall accuracy of machine learning algorithms (linear discriminant analysis (LDA), support vector machine (SVM), k-nearest neighbour (KNN)) based on hyperspectral (400–2400 nm) analysis. First three PCA datasets were selected from training and testing to create a confusion matrix for machine learning algorithm with higher overall accuracy. (A) Reflectance, (B) transmittance, (C) absorbance. Blue box indicates higher accuracy/precision (accept), and red box indicates lowest accuracy/precision (error). Total of 450 training samples and 150 test samples (n = 600).
LDA-PLS based on validation data was performed with the second dataset (27% of the spectral curve reflectance, transmittance, and absorbance data), and the discrimination oscillated between 94.8 and 100%. The highest successful classification was observed for Americana vs. Lisa, with 100% accuracy (Figure 8A,C).
The SVM-based linear machine can be considered the most promising algorithm, since it reached >99% accuracy with the smallest number of PCs using a smaller data matrix than the VIS-NIR-SWIR bands (Figure 8). Overall accuracy was obtained for SVM-linear and k-linear (KNN-linear) data for reflectance, transmittance, and absorbance. In addition, the reflectance (Figure 8A) and transmittance (Figure 8B) showed better results between training and test values in comparison to the absorbance data. However, higher classification accuracy was obtained with the SVM algorithms. Conversely, with the KNN algorithm, if k is small, the prediction will be more sensitive to nearby examples, which will lead to noise and prediction errors. The lowest k was determined using the fivefold cross-validation method (training vs. test sample data). Based on Figure 8, when k-models were set to three PCs, a low result was produced, and the accuracy, sensitivity, and specificity were 35 and 28% for SVM cubic of reflectance and transmittance, respectively, but up to 58% for absorbance (Figure 8). Similarly, the KNN-based (k-log) algorithm showed lower accuracy in discriminating green lettuces based on machine learning algorithms.
3.7. Estimation of Thickness-Based Data Mining and Machine Learning by PLSR Method
PLSR results for reflectance and light-absorption spectral curves (400–2400 nm) for thickness were considered excellent, calculated by the RMSE values, R2, and RPD (Table 3). In general, the models presented good capability for pigment content and thickness prediction, despite the high spectral interference of one variety with another under diverse anatomical effects on spectral curves. For example, internal validation using the leave-in-out method (evaluated with the RMSEC) and compared with the root mean square error of cross-validation (RMSECV) presented extremely low values (excellent results) associated with R2x. The predicted absorbance (RMSEC = 3.86, RMSEP = 6.21), transmittance (RMSEC = 4.43, RMSEP = 7.75), and reflectance (RMSEC = 4.95, RMSEP = 5.60) are shown in Table 3. In the model validation estimated using the root mean square error of prediction (RMSEP) obtained by external analysis, the data confirm the quality by fitting RMSEC (Table 3), i.e., the data present excellent predictive ability as obtained by hyperspectral sensors, using both reflectance and absorbance curves (Table 3).

Table 3.
PLSR model for thickness in calibration, cross-validation, and predicted reflectance, transmittance, and absorbance measurements. Model goodness-of-fit (R2), offset, root mean square error (RMSE), ratio of performance to deviation (RPD), and bias parameters from hyperspectroscopy of green lettuce leaves. Bold indicates statistically significant regression (R2).
In calculating the capacity of the PLSR models, the calculated RPD and R2P values were classified as excellent except for the underlined RPDs (Table 3). The R2P values were considered excellent (0.991) for absorbance; the reflectance and transmittance models had excellent prediction capability but were somewhat low (0.986 and 0.973, respectively). Given the RPDP parameter, the models were considered excellent (>6.09) in similar analysis to R2P (Table 3).
4. Discussion
4.1. Descriptive Analysis
The lowest variability in green lettuce plants demonstrated efficient estimation and classification of parameters such as thickness based on the VIS-NIR-SWIR bands (Figure 3). The trained and tested algorithms demonstrated higher accuracy. Statistical-based learning methods of analysis of the lettuce by spectroscopy combined with data mining and machine learning demonstrated the great value of deep learning methods for the screening of green lettuce [36,42]. Higher chlorophyll content and leaf thickness maximize the likelihood that algorithms will classify correctly based on larger differences in biophysical and biochemical parameters, thus they are considered as having higher prediction and classification accuracy (based on high linear SVM, KNN, and RPD values) (Table 2).
All methods reported here (Figure 1) allow the estimation and classification of physiological and remote sensing parameters and the monitoring of plant attributes. In general, remote sensing associated with data mining techniques should play a significant part in the development of fast, accurate, simple, and efficient prediction of crop phenotyping of lettuce plants in response to growth and development under environmental conditions [17,18,43]. Based on the main objective of this research, the use of VIS-NIR-SWIR spectroscopy associated with machine and deep learning methods may be able to correctly provide classification and prediction in green hydroponic indoor farming (Figure 1 and Figure 2).
4.2. Analysis of Hyperspectral Curves
The clustering between VIS-NIR-SWIR bands is marked on the all-hyperspectral curve. VIS showed variations arising from the absorbance of pigments, such as chloroplastic pigments (Chls and Cars). In this sense, machine learning algorithms can be applied to classify alterations of these biochemical and biophysical compounds more efficiently in leaves.
Furthermore, the NIR region showed higher reflectance values and larger differences in anatomical and physiological traits in plants [44,45]. Thus, the distinct green varieties are related to radiation scattering within the thickness of mesophyll cells [7,10,14,23]. Particularly in green lettuce plants, which are quite plastic regarding the structure and thickness of their mesophyll as well as biochemical properties, compounds, and accumulated calorific energy of their leaves, there are different reflectance spectra, mainly in the NIR region. Thus, it is an important band for classification, and data mining and machine learning algorithms can be used to quantify and monitor the status of the dynamics of the data, since they have improved computational processing and less misclassification [46]. In this sense, agriculture 4.0 combined with remote sensing represents an important type of monitoring in crop sciences.
The SWIR spectrum should contribute to obtaining fingerprints, especially at 1400 and 1950 nm, which are significant in the classification of water bonds and compounds associated with plant cell compounds, such as lignin, cellulose, structural carbohydrates, and other molecules [11,43,47]. Many of these compounds are linked to a higher energetic status and construction cost of the tissues in leaves, especially in green lettuce. In this sense, remote sensing tools and chemometric parameters are related to characteristics of the distinct biophysical and biochemical chemometric phenotypes of lettuce plants [48,49].
4.3. Machine Learning-Based PCA Classification
Many algorithms can be applied to machine learning for sample group classification using hyperspectral raw data and PCA data. In this study, the overall accuracy was slightly improved by using PCA data; for the full spectrum, larger differences in the accuracy of algorithms were observed for both datasets. The first three PCAs improved the computational processing because they contained a reduced dataset. Overall accuracy of 99% for linear SVM was obtained by using three PCs with ≈97% data variance. Similarly, LDA-linear obtained 99.9% of data variance but with higher accuracy in reflectance, transmittance, and absorbance spectroscopy. Following [32], data mining and machine learning can be enhanced to classify with higher precision in log, cubic, or quadratic SVM or KNN, and these can be considered the most promising algorithms since they showed precision with the smallest number of principal components. However, that is not what we observed in this machine learning experiment and algorithm test based on green lettuce groups (Figure 3, Figure 4, Figure 5 and Figure 6).
Combined PCAs have been used in research to extract the most useful hyperspectral information, which may be a better method to classify and discriminate the most responsive wavelengths. This method has shown good results in situations with fewer impurities [14,50]. However, there will be many impurities that affect the results of compounds in leaves by using hyperspectral analysis, producing undesirable effects. Choosing appropriate bands from all available bands to obtain useful information requires expert knowledge and rich practical experience, and it is not easy to achieve automated and rapid detection. SVM and KNN are often used for quantitative analysis of spectra, but SVM easily learns too many features when processing hyperspectral data, which leads to overfitting [21,32,36]. KNN is computationally expensive, and when the sample size is small, the classification accuracy is not very high. The use of decision algorithms along with data enhancement technology, deep learning, and hyperspectral data can yield good results in predicting the classification of pigments by concentration and thickness, which is a good alternative that exceeds the standard [32,36,51]. Combined with data enhancement, SVM and KNN achieve excellent performance on classification [10].
4.4. Partial Least Squares Regression (PLSR) of Predicted Chemometrics
The R2CV, R2P, RPDCV, RPDP and RMSECV, and RMSEP values regarding the full VIS-NIR-SWIR bands were applied according to the evaluated parameters (Table 2 and Table 3). For example, the approach used (Figure 1) produced very good results [7,8]. Furthermore, multivariate statistical analysis achieved similar predictive results by VIP, RC, STEPwise, and other methods or processes with decreased noise and better efficiency [14,52,53].
The results of the cross-validation statistical phase were slightly higher than those of the prediction phase, as expected, since the number of samples used to obtain the model was smaller in the calibration phase [23]. Similarly, when using NIR-SWIR bands to predict lettuce chemometrics, an intensification in RMSE in the estimate of each stage was found in [10,23].
4.5. Data Mining and Machine Learning-Based Modelling by Hyperspectroscopy Data
There was a clear advantage of using the whole-spectrum-based approaches (PLSR, LDA, and SVR) over VIS to correctly classify and predict leaf properties (e.g., minimum differences in mesophyll; Table 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 and Figure 8). For example, thickness, for which excellent predictions were shown, was obtained by reflectance, transmittance, and hyperspectral absorbance (Table 3). Some studies have reported that when monitoring and classifying pigments, vegetation indices (VIs) or VIS could predict satisfactorily, and models developed by KNN, LDA, PLSR, and SVR performed even better. For example, in research on the other leaf properties in which VIS predicted only fairly or poorly, PLSR and SVR still yielded moderately satisfactory predictions, possibly based on full spectral data to the detriment of the spectral range [17,54]. According to [55], remote sensing-based hyperspectral data, such as narrow-band VIS, are computationally simple [55]. However, by selecting only a few (usually two) bands from hundreds or thousands of hyperspectral bands, a large amount of useful information is discarded [7,16]. In addition, some authors have reported that vegetation indices (VIs) could provide better classification than VIS bands [10,14].
Plant leaves are a complex mixture of many chemical compounds (such as water, pigments, N-containing proteins, structural carbohydrates, etc.), which all contribute to the overall shape of leaf spectra [10,19,56]. Thus, the biophysical state of the leaf (such as thickness and surface roughness) not only affects its reflectance spectra [55], but also its transmittance and absorbance spectra (Table 3). Using PLSR and linear SVR, which employ the entire spectra, makes the models more flexible in accentuating the spectral features that are correlated with the target property while suppressing the bands whose variation is sensitive to other confounding factors. An example matrix vector is shown in Figure 8. With the rapid advancement of computing, PLSR and SVR modelling can be performed very efficiently, following the suggestion of [55]. Moreover, other machine learning approaches, such as ridge/lasso regression, artificial neural networks, and random forest, can also be considered, giving researchers a wide range of choices for their data [10,55]. In addition, to enable the use of PLS-DA on resolved MCR fingerprints, a strategy of MCR resampling is needed so that a sufficient number of spectral signatures for each population to be compared is available [20,57,58]. Some of these approaches might work particularly well under certain conditions. We therefore suggest that whole-spectrum-based modelling should be used for the phenotyping of plant leaf physiological and biochemical traits using VIS-NIR-SWIR, as there are practically no computational barriers, as compared with earlier studies [8,19,45,55].
4.6. Regression Coefficient (RC) and Variable Importance in Projection (VIP)
VIP and RC are important to avoid mistaken correlations, as reported in [17]. Some researchers have explored the potential of remote sensing as a technique for classifying lettuce plants at indoor farms, mainly considering that the VIS-NIR or just the NIR-SWIR region [23,43,52] can estimate low-accuracy outputs or biased parameters. For example, high RC values in the SWIR region highlight the importance of predicting attributes (on average, 68% of VIP values), according to the premise in our research. It was reported in [59] that differences in leaf biochemical and biophysical structure are associated with SWIR spectroscopy curves. According to [10,23], reflectance data in the SWIR bands offer important information for the potential classification of lettuce plants. It is important to note that VIS-NIR-SWIR does not measure leaf physiological or chemical properties directly. High-resolution sensors based on VIS-NIR-SWIR spectroscopy can generate enough accurate data to allow for classification between substances that absorb in the same spectral region through RC and VIP analysis. It is noteworthy that absorbed light data are, in a way, “unbiased” from the effects induced by the epidermis and anatomical and ultrastructural aspects, as well as leaf thickness, in relation to reflectance or transmittance data [7,16,60]. Thus, VIS-NIR-SWIR spectroscopy-based reflectance, transmittance, and absorbance, together with artificial intelligence algorithms, could be a promising method for the classification of other lettuce varieties as well as for use in other remote sensing applications to monitor and manage crops as well as food quality and safety.
5. Conclusions
This study explored the hypothesis that, by using hyperspectroscopy, it would be possible to classify varieties with higher accuracy with VIS-NIR-SWIR spectroscopy together with machine learning and data-mining algorithms. Here, green lettuce varieties were found to have unique spectral signatures, as well as typical inflections of -COOH and -NH stretching from aromatic rings linked to many compounds.
Algorithms were shown to have higher accuracy and discrimination of >99.9, 100. and 100% for base reflectance, transmittance, and absorbance hyperspectral data. In addition, it was possible to adjust the PLSR model in the prediction (test) phase with R2P values of 0.973, 0.986, and 0.991 and RPDP values between 6.08 and 10.54 for reflectance, transmittance, and absorbance for many variables predicted by using the full spectrum (400–2400 nm) with the hyperspectral technique. Therefore, this study offers a promising alternative, as the technique provides advantages such as being rapid and not requiring previous sample preparation (chemical reagents), as well as providing a new way to classify lettuce and possibly other crops. In addition, high repeatability of VIS-NIR-SWIR was found compared with other methods based on an integrating sphere. Thus, this study reports a good technique for classification and prediction of thickness parameters based on data mining and machine learning that shows efficiency using VIS-NIR-SWIR hyperspectroscopy, which can improve the management of crops as well as food quality and safety using precision agriculture systems.
Author Contributions
Conceptualization, R.F.; Data curation, R.F.; Formal analysis, R.F., J.V.F.G., K.M.d.O., W.C.A. and M.R.N.; Funding acquisition, R.F., W.C.A. and M.R.N.; Investigation, R.F. and W.C.A.; Methodology, R.F., J.V.F.G., K.M.d.O. and M.R.N.; Project administration, R.F.; Resources, R.F.; Software, R.F. and M.R.N.; Supervision, R.F., W.C.A. and M.R.N.; Validation, R.F. and M.R.N.; Visualization, R.F., J.V.F.G., K.M.d.O., W.C.A. and M.R.N.; Writing—original draft, R.F., J.V.F.G., K.M.d.O., W.C.A. and M.R.N.; Writing—review and editing, R.F., J.V.F.G., K.M.d.O. and M.R.N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001.
Data Availability Statement
Not applicable.
Acknowledgments
Thanks are due to Programa de Pós-Graduação em Agronomia (PGA-UEM) at the State University of Maringá for encouragement and supporting communication.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Buturi, C.V.; Sabatino, L.; Mauro, R.P.; Navarro-León, E.; Blasco, B.; Leonardi, C.; Giuffrida, F. Iron Biofortification of Greenhouse Soilless Lettuce: An Effective Agronomic Tool to Improve the Dietary Mineral Intake. Agronomy 2022, 12, 1793. [Google Scholar] [CrossRef]
- Hong, J.; Xu, F.; Chen, G.; Huang, X.; Wang, S.; Du, L.; Ding, G. Evaluation of the Effects of Nitrogen, Phosphorus, and Potassium Applications on the Growth, Yield, and Quality of Lettuce (Lactuca Sativa L.). Agronomy 2022, 12, 2477. [Google Scholar] [CrossRef]
- Muneer, S.; Kim, E.; Park, J.; Lee, J. Influence of Green, Red and Blue Light Emitting Diodes on Multiprotein Complex Proteins and Photosynthetic Activity under Different Light Intensities in Lettuce Leaves (Lactuca Sativa L.). Int. J. Mol. Sci. 2014, 15, 4657–4670. [Google Scholar] [CrossRef] [PubMed]
- Horf, M.; Vogel, S.; Drücker, H.; Gebbers, R.; Olfs, H.-W. Optical Spectrometry to Determine Nutrient Concentrations and Other Physicochemical Parameters in Liquid Organic Manures: A Review. Agronomy 2022, 12, 514. [Google Scholar] [CrossRef]
- Giordano, M.; El-Nakhel, C.; Carillo, P.; Colla, G.; Graziani, G.; Di Mola, I.; Mori, M.; Kyriacou, M.C.; Rouphael, Y.; Soteriou, G.A.; et al. Plant-Derived Biostimulants Differentially Modulate Primary and Secondary Metabolites and Improve the Yield Potential of Red and Green Lettuce Cultivars. Agronomy 2022, 12, 1361. [Google Scholar] [CrossRef]
- Matysiak, B.; Ropelewska, E.; Wrzodak, A.; Kowalski, A.; Kaniszewski, S. Yield and Quality of Romaine Lettuce at Different Daily Light Integral in an Indoor Controlled Environment. Agronomy 2022, 12, 1026. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Pattaro, M.; Herrig Furlanetto, R.; Nanni, M.R.; Camargos Antunes, W. High Resolution Leaf Spectral Signature as a Tool for Foliar Pigment Estimation Displaying Potential for Species Differentiation. J. Plant Physiol. 2020, 249, 153161. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Antunes, W.C.; Nanni, M.R. Rapid Quantification Method for Yield, Calorimetric Energy and Chlorophyll a Fluorescence Parameters in Nicotiana Tabacum L. Using Vis-NIR-SWIR Hyperspectroscopy. Plants 2022, 11, 2406. [Google Scholar] [CrossRef]
- Moriwaki, T.; Falcioni, R.; Tanaka, F.A.O.; Cardoso, K.A.K.; Souza, L.A.; Benedito, E.; Nanni, M.R.; Bonato, C.M.; Antunes, W.C. Nitrogen-Improved Photosynthesis Quantum Yield Is Driven by Increased Thylakoid Density, Enhancing Green Light Absorption. Plant Sci. 2019, 278, 1–11. [Google Scholar] [CrossRef]
- Rodrigues, M.; Berti de Oliveira, R.; Leboso Alemparte Abrantes dos Santos, G.; Mayara de Oliveira, K.; Silveira Reis, A.; Herrig Furlanetto, R.; Antônio Yanes Bernardo Júnior, L.; Silva Coelho, F.; Rafael Nanni, M. Rapid Quantification of Alkaloids, Sugar and Yield of Tobacco (Nicotiana Tabacum L.) Varieties by Using Vis–NIR–SWIR Spectroradiometry. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 274, 121082. [Google Scholar] [CrossRef]
- Sobejano-Paz, V.; Mikkelsen, T.N.; Baum, A.; Mo, X.; Liu, S.; Köppl, C.J.; Johnson, M.S.; Gulyas, L.; García, M. Hyperspectral and Thermal Sensing of Stomatal Conductance, Transpiration, and Photosynthesis for Soybean and Maize under Drought. Remote Sens. 2020, 12, 3182. [Google Scholar] [CrossRef]
- Tahir, H.E.; Xiaobo, Z.; Zhihua, L.; Jiyong, S.; Zhai, X.; Wang, S.; Mariod, A.A. Rapid Prediction of Phenolic Compounds and Antioxidant Activity of Sudanese Honey Using Raman and Fourier Transform Infrared (FT-IR) Spectroscopy. Food Chem. 2017, 226, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Chicati, M.S.; Nanni, M.R.; Chicati, M.L.; Furlanetto, R.H.; Cezar, E.; De Oliveira, R.B. Hyperspectral Remote Detection as an Alternative to Correlate Data of Soil Constituents. Remote Sens. Appl. Soc. Environ. 2019, 16, 100270. [Google Scholar] [CrossRef]
- Furlanetto, R.H.; Moriwaki, T.; Falcioni, R.; Pattaro, M.; Vollmann, A.; Sturion Junior, A.C.; Antunes, W.C.; Nanni, M.R. Hyperspectral Reflectance Imaging to Classify Lettuce Varieties by Optimum Selected Wavelengths and Linear Discriminant Analysis. Remote Sens. Appl. Soc. Environ. 2020, 20, 100400. [Google Scholar] [CrossRef]
- da Silva Junior, C.A.; Nanni, M.R.; Shakir, M.; Teodoro, P.E.; de Oliveira-Júnior, J.F.; Cezar, E.; de Gois, G.; Lima, M.; Wojciechowski, J.C.; Shiratsuchi, L.S. Soybean Varieties Discrimination Using Non-Imaging Hyperspectral Sensor. Infrared Phys. Technol. 2018, 89, 338–350. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Bonato, C.M.; de Souza, L.A.; Nanni, M.R.; Antunes, W.C. Distinct Growth Light and Gibberellin Regimes Alter Leaf Anatomy and Reveal Their Influence on Leaf Optical Properties. Environ. Exp. Bot. 2017, 140, 86–95. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pellegrini, E.; Saponaro, V.; Hoshika, Y.; Arab, L.; Rennenberg, H.; Paoletti, E. Hyperspectral Reflectance of Light-Adapted Leaves Can Predict Both Dark- and Light-Adapted Chl Fluorescence Parameters, and the Effects of Chronic Ozone Exposure on Date Palm (Phoenix Dactylifera). Int. J. Mol. Sci. 2020, 21, 6441. [Google Scholar] [CrossRef]
- El-Hendawy, S.; Al-Suhaibani, N.; Mubushar, M.; Tahir, M.U.; Marey, S.; Refay, Y.; Tola, E. Combining Hyperspectral Reflectance and Multivariate Regression Models to Estimate Plant Biomass of Advanced Spring Wheat Lines in Diverse Phenological Stages under Salinity Conditions. Appl. Sci. 2022, 12, 1983. [Google Scholar] [CrossRef]
- Braga, P.; Crusiol, L.G.T.; Nanni, M.R.; Caranhato, A.L.H.; Fuhrmann, M.B.; Nepomuceno, A.L.; Neumaier, N.; Farias, J.R.B.; Koltun, A.; Gonçalves, L.S.A.; et al. Vegetation Indices and NIR-SWIR Spectral Bands as a Phenotyping Tool for Water Status Determination in Soybean. Precis. Agric. 2021, 22, 249–266. [Google Scholar] [CrossRef]
- Ralbovsky, N.M.; Smith, J.P. Multivariate Curve Resolution for Analysis of Raman Hyperspectral Imaging Data Sets for Enzyme Immobilization. Chem. Data Collect. 2022, 38, 100835. [Google Scholar] [CrossRef]
- Féret, J.-B.; le Maire, G.; Jay, S.; Berveiller, D.; Bendoula, R.; Hmimina, G.; Cheraiet, A.; Oliveira, J.C.; Ponzoni, F.J.; Solanki, T.; et al. Estimating Leaf Mass per Area and Equivalent Water Thickness Based on Leaf Optical Properties: Potential and Limitations of Physical Modeling and Machine Learning. Remote Sens. Environ. 2019, 231, 110959. [Google Scholar] [CrossRef]
- Jin, J.; Arief Pratama, B.; Wang, Q. Tracing Leaf Photosynthetic Parameters Using Hyperspectral Indices in an Alpine Deciduous Forest. Remote Sens. 2020, 12, 1124. [Google Scholar] [CrossRef]
- Sexton, T.; Sankaran, S.; Cousins, A.B. Predicting Photosynthetic Capacity in Tobacco Using Shortwave Infrared Spectral Reflectance. J. Exp. Bot. 2021, 72, 4373–4383. [Google Scholar] [CrossRef] [PubMed]
- Shorten, P.R.; Leath, S.R.; Schmidt, J.; Ghamkhar, K. Predicting the Quality of Ryegrass Using Hyperspectral Imaging. Plant Methods 2019, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, G.F.; Santos, R.O.; Teixeira, I.D.; de Cassia de Souza Schneider, R.; Helfer, G.A.; Costa, A. Ben Rapid Quantification of Constituents in Tobacco by NIR Fiber-optic Probe. J. Chemom. 2020, 34, e3303. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Furlanetto, R.H.; Nanni, M.R.; Antunes, W.C. Simple, Fast and Efficient Methods for Analysing the Structural, Ultrastructural and Cellular Components of the Cell Wall. Plants 2022, 11, 995. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of Total Carotenoids and Chlorophylls a and b of Leaf Extracts in Different Solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magalhaes, P.J.; Ram, P.J. Image Processing with ImageJ. Biophotonics Int. 2012, 11, 36–42. [Google Scholar]
- R-Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2020. Available online: https://www.R-project.org (accessed on 5 October 2022).
- Pimentel-Gomes, F. Statistics Course Experimental, 1st ed.; FEALQ: Piracicaba, Brazil, 2009. [Google Scholar]
- Zar, J.H. Biostatistical Analysis, 5th ed.; Pearson Education: Upper Saddle River, NJ, USA, 2010; ISBN 0-13-100846-3. [Google Scholar]
- Franca, T.; Goncalves, D.; Cena, C. ATR-FTIR Spectroscopy Combined with Machine Learning for Classification of PVA/PVP Blends in Low Concentration. Vib. Spectrosc. 2022, 120, 103378. [Google Scholar] [CrossRef]
- Nanni, M.R.; Cezar, E.; da Silva Junior, C.A.; Silva, G.F.C.; da Silva Gualberto, A.A. Partial Least Squares Regression (PLSR) Associated with Spectral Response to Predict Soil Attributes in Transitional Lithologies. Arch. Agron. Soil Sci. 2018, 64, 682–695. [Google Scholar] [CrossRef]
- Cozzolino, D.; Vadell, A.; Ballesteros, F.; Galietta, G.; Barlocco, N. Combining Visible and Near-Infrared Spectroscopy with Chemometrics to Trace Muscles from an Autochthonous Breed of Pig Produced in Uruguay: A Feasibility Study. Anal. Bioanal. Chem. 2006, 385, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Pierna, J.A.F.; Baeten, V.; Renier, A.M.; Cogdill, R.P.; Dardenne, P. Combination of Support Vector Machines (SVM) and near-Infrared (NIR) Imaging Spectroscopy for the Detection of Meat and Bone Meal (MBM) in Compound Feeds. J. Chemom. 2004, 18, 341–349. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, Z.; Zhang, G.; Chen, S.; Zhao, Y.; Lu, J. Analysis and Classification of Kidney Stones Based on Raman Spectroscopy. Biomed. Opt. Express 2018, 9, 4175. [Google Scholar] [CrossRef] [PubMed]
- Gholamy, A.; Kreinovich, V.; Kosheleva, O. Why 70/30 or 80/20 Relation between Training and Testing Sets: A Pedagogical Explanation. El Paso, TX, USA. 2018. Available online: https://www.cs.utep.edu/vladik/2018/tr18-09.pdf (accessed on 5 October 2022).
- Izenman, A.J. Modern Multivariate Statistical Techniques, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Jolliffe, I.T. (Ed.) Principal Component Analysis, 2nd ed.; Springer USA: New York, NY, USA, 2002; Volume 30. [Google Scholar]
- Sheskin, D.J. Handbook of Parametric and Nonparametric Statistical Procedures, 1st ed.; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Minasny, B.; McBratney, A.B.; Malone, B.P.; Wheeler, I. Digital Mapping of Soil Carbon. Adv. Agron. 2013, 3, 1–47. [Google Scholar]
- de Oliveira Moura, L.; de Carvalho Lopes, D.; Steidle Neto, A.J.; de Castro Louback Ferraz, L.; de Almeida Carlos, L.; Martins, L.M. Evaluation of Techniques for Automatic Classification of Lettuce Based on Spectral Reflectance. Food Anal. Methods 2016, 9, 1799–1806. [Google Scholar] [CrossRef]
- Zheng, W.; Lu, X.; Li, Y.; Li, S.; Zhang, Y. Hyperspectral Identification of Chlorophyll Fluorescence Parameters of Suaeda Salsa in Coastal Wetlands. Remote Sens. 2021, 13, 2066. [Google Scholar] [CrossRef]
- Calviño-Cancela, M.; Martín-Herrero, J. Spectral Discrimination of Vegetation Classes in Ice-Free Areas of Antarctica. Remote Sens. 2016, 8, 856. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Perez-Llorca, M.; Munné-Bosch, S.; Gibin, M.S.; Sato, F.; Pelozo, A.; Pattaro, M.C.; Giacomelli, M.E.; Rüggeberg, M.; et al. Cell Wall Structure and Composition Is Affected by Light Quality in Tomato Seedlings. J. Photochem. Photobiol. B Biol. 2020, 203, 111745. [Google Scholar] [CrossRef]
- Silva, C.A.; Nanni, M.R.; Teodoro, P.E.; Silva, G.F.C. Vegetation Indices for Discrimination of Soybean Areas: A New Approach. Agron. J. 2017, 109, 1331–1343. [Google Scholar] [CrossRef]
- Baranović, G.; Šegota, S. Infrared Spectroscopy of Flavones and Flavonols. Reexamination of the Hydroxyl and Carbonyl Vibrations in Relation to the Interactions of Flavonoids with Membrane Lipids. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 192, 473–486. [Google Scholar] [CrossRef]
- Llorach, R.; Martínez-Sánchez, A.; Tomás-Barberán, F.A.; Gil, M.I.; Ferreres, F. Characterisation of Polyphenols and Antioxidant Properties of Five Lettuce Varieties and Escarole. Food Chem. 2008, 108, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Kitazaki, K.; Fukushima, A.; Nakabayashi, R.; Okazaki, Y.; Kobayashi, M.; Mori, T.; Nishizawa, T.; Reyes-Chin-Wo, S.; Michelmore, R.W.; Saito, K.; et al. Metabolic Reprogramming in Leaf Lettuce Grown Under Different Light Quality and Intensity Conditions Using Narrow-Band LEDs. Sci. Rep. 2018, 8, 7914. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Marino, G.; LLusia, J.; Morfopoulos, C.; Farré-Armengol, G.; Filella, I. Photochemical Reflectance Index as an Indirect Estimator of Foliar Isoprenoid Emissions at the Ecosystem Level. Nat. Commun. 2013, 4, 2604. [Google Scholar] [CrossRef] [PubMed]
- Galvão, L.S.; Formaggio, A.R.; Tisot, D.A. Discrimination of Sugarcane Varieties in Southeastern Brazil with EO-1 Hyperion Data. Remote Sens. Environ. 2005, 94, 523–534. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Fortini, E.A.; de Müller, L.A.C.; Batista, D.S.; Vieira, L.M.; Silva, P.O.; do Amaral, C.H.; Poethig, R.S.; Otoni, W.C. Leaf Development Stages and Ontogenetic Changes in Passionfruit (Passiflora Edulis Sims.) Are Detected by Narrowband Spectral Signal. J. Photochem. Photobiol. B Biol. 2020, 209, 111931. [Google Scholar] [CrossRef]
- Lysenko, V.; Guo, Y.; Kosolapov, A.; Usova, E.; Varduny, T.; Krasnov, V. Polychromatic Fourier-PAM Fluorometry and Hyperspectral Analysis of Chlorophyll Fluorescence from Phaseolus Vulgaris Leaves: Effects of Green Light. Inf. Process. Agric. 2020, 7, 204–211. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Q. Selection of Informative Spectral Bands for PLS Models to Estimate Foliar Chlorophyll Content Using Hyperspectral Reflectance. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3064–3072. [Google Scholar] [CrossRef]
- Ge, Y.; Atefi, A.; Zhang, H.; Miao, C.; Ramamurthy, R.K.; Sigmon, B.; Yang, J.; Schnable, J.C. High-Throughput Analysis of Leaf Physiological and Chemical Traits with VIS–NIR–SWIR Spectroscopy: A Case Study with a Maize Diversity Panel. Plant Methods 2019, 15, 66. [Google Scholar] [CrossRef]
- Straková, P.; Larmola, T.; Andrés, J.; Ilola, N.; Launiainen, P.; Edwards, K.; Minkkinen, K.; Laiho, R. Quantification of Plant Root Species Composition in Peatlands Using FTIR Spectroscopy. Front. Plant Sci. 2020, 11, 597. [Google Scholar] [CrossRef]
- Juan, A. Multivariate Curve Resolution for Hyperspectral Image Analysis. In Hyperspectral Imaging; Amigo, J.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 115–150. [Google Scholar]
- Olmos, V.; Marro, M.; Loza-Alvarez, P.; Raldúa, D.; Prats, E.; Padrós, F.; Piña, B.; Tauler, R.; de Juan, A. Combining Hyperspectral Imaging and Chemometrics to Assess and Interpret the Effects of Environmental Stressors on Zebrafish Eye Images at Tissue Level. J. Biophotonics 2018, 11, e201700089. [Google Scholar] [CrossRef]
- Boshkovski, B.; Doupis, G.; Zapolska, A.; Kalaitzidis, C.; Koubouris, G. Hyperspectral Imagery Detects Water Deficit and Salinity Effects on Photosynthesis and Antioxidant Enzyme Activity of Three Greek Olive Varieties. Sustainability 2022, 14, 1432. [Google Scholar] [CrossRef]
- Gitelson, A.; Solovchenko, A. Non-Invasive Quantification of Foliar Pigments: Possibilities and Limitations of Reflectance- and Absorbance-Based Approaches. J. Photochem. Photobiol. B Biol. 2018, 178, 537–544. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).