1. Introduction
Eutrophication is predominantly an anthropogenic process characterized by an excessive accumulation of nutrients, primarily nitrogen and phosphorus, in surface freshwater ecosystems such as lakes. This nutrient excess promotes the rapid growth of algae and aquatic plants which can increase both water turbidity and, as algae die and decompose, water oxygen depletion, leading to negative impacts on aquatic life and lake ecosystems [
1]. Human disturbances to the water cycle, such as agricultural runoff, urban development, and wastewater discharge, mostly contribute to eutrophication [
2]. Therefore, controlling and mitigating lake eutrophication is essential to protect both freshwater ecosystems and human well-being by maintaining the ecological and economic value of lakes [
3]. The need to preserve freshwater ecosystems is further enforced by their direct connection with the United Nations Sustainable Development Goal 6 (SDG 6: Ensure availability and sustainable management of water and sanitation for all) [
4]. Accordingly, effective control and mitigation actions towards freshwater ecosystem protection are imperative and require both space- and time-resolved monitoring and quantification of eutrophication levels in surface water bodies [
5].
A significant indicator of eutrophication is the concentration of Chlorophyll in water, which is a major component of algae pigments and cyanobacteria and allows for the estimation of algal biomass in water bodies [
6]. Specifically, Chlorophyll-a (Chl-a) is mostly used as a proxy for total algal biomass [
7]. Chl-a concentration is relatively easy to measure using various techniques, including imaging spectroscopy [
8], and both in-situ and remote sensing methods are often applied in the practice [
9]. In-situ monitoring generally suffers from limitations in terms of space–time coverage of measurements [
10]. Conversely, remote sensing methods, such as satellite multispectral and hyperspectral imagery, allow for the synoptic assessment of Chl-a concentration over the whole water body’s surface and provide repeated measurements over time, which are critical to capturing eutrophication space–time dynamics [
11]. Imaging spectroscopy exploits characteristic Chl-a sunlight absorption and reflection patterns at specific wavelengths including green, blue, red, and near-infrared bands [
12] to determine its concentration in water. Airborne and spaceborne imagery has been employed since the 1980s for monitoring Chl-a concentration, proving to be more successful in ocean and seawater applications rather than inland waters due mainly to the limited spatial and spectral resolution of data available at that time [
10]. Moreover, the optical complexity of inland waters, primarily caused by a high presence of suspended particles, reduces the reliability of both atmospheric corrections and estimation models initially designed for land and ocean applications [
12]. Nonetheless, the detailed and frequent retrieval of inland water biochemical parameters, including Chl-a, has become possible thanks to the latest generation of medium to high spatial resolution multispectral spaceborne sensors, such as those onboard Landsat-8/9, Sentinel-2, and Sentinel-3 satellites [
13].
Frontier applications of inland water quality remote monitoring involve hyperspectral satellite imagery, on which bio-optical algorithms demonstrated improved performances compared with multispectral imagery [
14,
15]. These applications are also favoured by new advancements in global hyperspectral remote sensing, proven by the recent or upcoming launches of hyperspectral satellites [
16]. An early example of the above is the Hyperion imager, launched by the United States (US) National Aeronautics and Space Administration (NASA) in 2000 and operational until 2017 [
17]. Relevant examples of the most recent missions that provide publicly available imagery are as follows: the German Aerospace Center Earth Sensing Imaging Spectrometer (DESIS) [
18] and the hyperspectral imager aboard the Environmental Mapping and Analysis Program (EnMAP) satellite mission [
19], the Chinese Advanced Hyperspectral Imager (AHSI) aboard the GaoFeng-5 satellite [
20], followed by the launch of the PRecursore IperSpettrale della Missione Applicative (PRISMA) sensor by the Italian Space Agency (ASI) [
21], and HyperScout instruments launched on nanosatellites by the European Space Agency (ESA) [
22]. These imaging systems offer data cubes where each pixel is composed of several spectral bands enabling space, time and spectral resolved detection of water biochemical constituents [
23].
As the resolution and coverage of satellite hyperspectral images continue to improve, cutting-edge data technologies, particularly the implementation of machine and deep learning algorithms [
24], are playing a pivotal role in advancing the diffusion and enhancing the capabilities of Chl-a estimation models. Alongside traditional spectral indices and physics-based models [
25], machine and deep learning approaches have been frequently exploited in the literature within hyperspectral imaging for Chl-a and other biochemical constituents estimation in water bodies [
26]. Recent and pertinent examples are as follows. In [
27], Partial Least Squares (PLS) is utilized to determine Chl-a and Total Suspended Matter (TSM). Ref. [
28] models in-situ measurements using linear models and Support Vector Machines (SVM) to predict Chl-a concentrations in Lake Taihu (China). Ref. [
29] estimates water quality parameters, including Chl-a, for the Elbe River using ten different machine-learning regression models. Ref. [
30] evaluates Random Forest (RF), SVM, and Artificial Neural Networks (ANN) for predicting Chl-a concentrations in various inland water bodies, also exploring the inclusion of spectral derivatives as input data. Ref. [
31] developed a PLS-ANN model for Chl-a prediction in Lake Erie. Additionally, Ref. [
32] utilizes simulated hyperspectral satellite data to predict Chl-a concentrations in lakes, employing an array of models including RF, SVM, Multivariate Adaptive Regression Spline (MARS), and CNN. Ref. [
33] generates synthetic EnMAP hyperspectral imagery using EnMAP end-to-end simulator software (EeteS) [
34] for Chl-a prediction in Czech Republic water reservoirs using Principal Component Regression, PLS Regression, and RF models. Finally, Refs. [
35,
36] utilize hyperspectral data from the Hyperspectral Imager for the Coastal Ocean (HICO) [
37] and the PRISMA satellite, respectively, to predict Chl-a concentrations using Mixture Density Network (MDN) models.
Despite the availability of machine and deep learning algorithms, there are persistent challenges in implementing them for operational monitoring tasks, often due to a lack of the space–time resolved reference data necessary to train and validate such models [
25,
38]. With this in mind, the present study aims to employ a variety of machine and deep learning regression models and subsequently conduct a comparative assessment across diverse experimental setups, with the goal of predicting Chl-a concentration maps from medium-resolution hyperspectral satellite imagery through the training and evaluation of these models with reference data characterized by lower spatial resolution, heightened acquisition frequency up to 2 days, and a wide swath width. The objective of the analysis is twofold. Firstly, it aims to verify that the use of the rich spectral information of hyperspectral imagery, coupled with machine and deep learning models, is suitable for reconstructing Chl-a concentration maps using pre-existing and widely accessible reference data. Secondly, it aims to assess the potential for enhancing the spatial resolution of pre-existing Chl-a concentration maps by aligning it with the hyperspectral imagery employed as the regressor in model implementation.
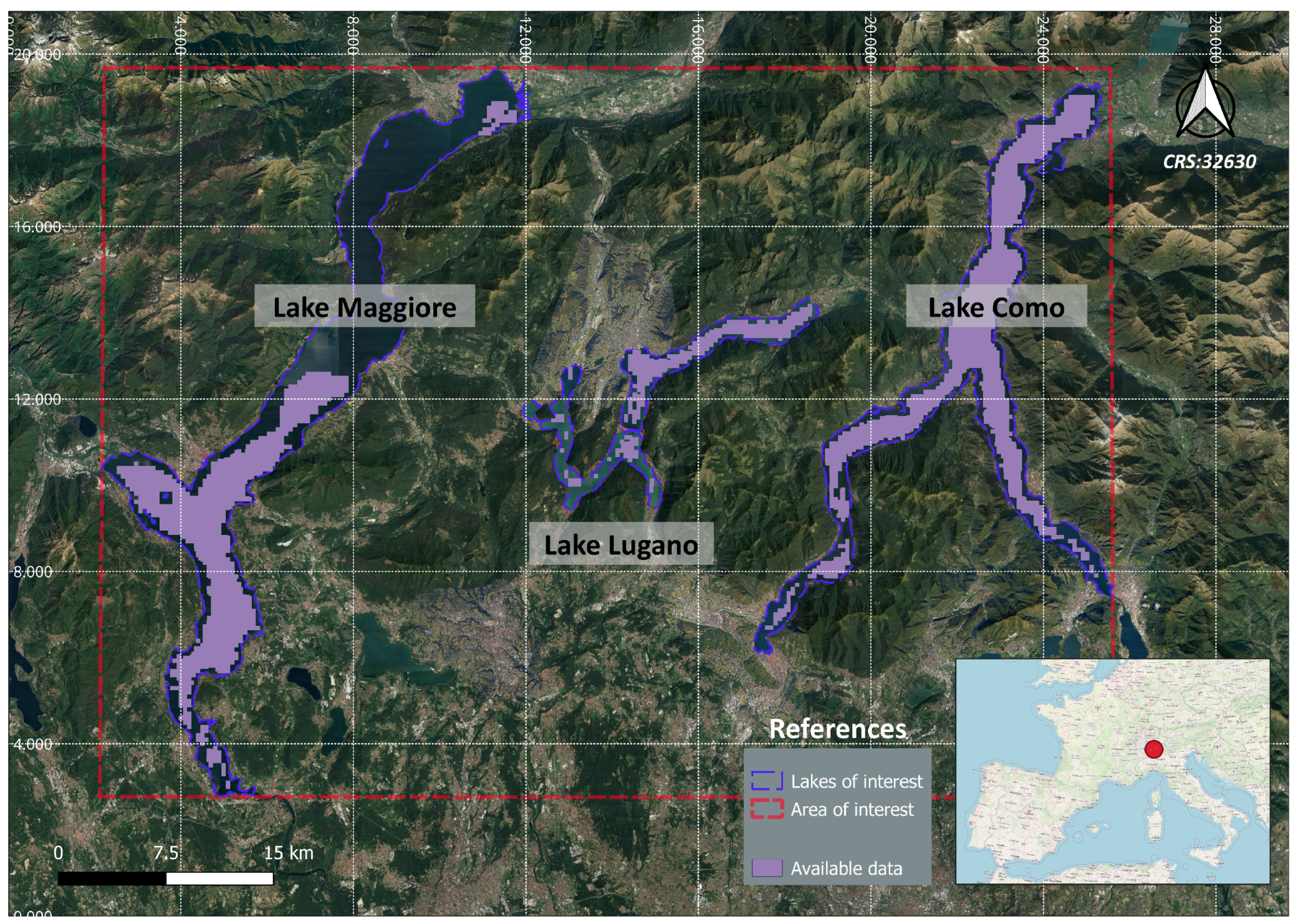
The selected study area includes three sub-alpine lakes between Northern Italy and Switzerland, specifically Lake Como, Lake Maggiore, and Lake Lugano (see
Figure 1). These lakes were chosen because they align with the selection made by the “Informative System for the Integrated Monitoring of Insubric Lakes and their Ecosystems” (SIMILE) project, within which this research is conducted. The SIMILE project is funded by the Interreg program of the European Union, which primarily focuses on enhancing coordinated management and stakeholder involvement in monitoring the water quality of sub-alpine lakes between Northern Italy and Switzerland [
39,
40]. It exploits a combination of in-situ measurements and remote sensing techniques to fulfil its objectives. Within the realm of satellite remote sensing, the project computes three key indicators for assessing lake water quality: Lake Water Surface Temperature derived from Landsat 8 imagery, Total Suspended Matter, and Chl-a concentrations [
41] derived from the Sentinel-3 A/B Ocean and Land Colour Instrument (OLCI) imagery at 300 m resolution, which provides a revisit time of less than 2 days. Each of these indicators is monitored by generating time-series of raster maps [
42].
In this study, hyperspectral images obtained from the PRISMA mission are used. PRISMA imagery features 239 bands spanning the Visible and Near-Infrared (VNIR) and Short-Wave Infrared (SWIR) regions of the electromagnetic spectrum (400–2500 nm) [
21]. PRISMA images have a spatial resolution of 30 m, a Spectral Sampling Interval (SSI) of 12 nm, and a revisit time of 29 days [
21]. For model training and testing, time-series maps of Chl-a concentration generated by the SIMILE project team from Sentinel-3 data are employed as low-resolution reference data. Quality assessment for these maps was provided by [
40] through comparisons with in-situ measurements, supporting their use as reference Chl-a concentration data in this work. The study considers both machine learning models, such as RF Regressor and SVR, as well as deep learning models such as Long Short-Term Memory (LSTM) networks [
43,
44] and Gated Recurrent Unit (GRU) networks [
45]. The choice of these models is based on empirical evidence from the literature and is primarily guided by two key characteristics, as suggested by [
24] and summarized as follows. First, when considering RF Regressor and SVR models, their effectiveness in handling non-linear dependencies within the input data is a primary factor. Moreover, when integrated with dimensionality reduction techniques, these models excel in reducing redundant spectral information. Second, LSTM and GRU models are preferred for their suitability in dealing with hyperspectral imagery. This preference is rooted in the sequential nature of the hyperspectral data, enabling them to capture both long and short-range dependencies of the contiguous bands in the spectral dimension.
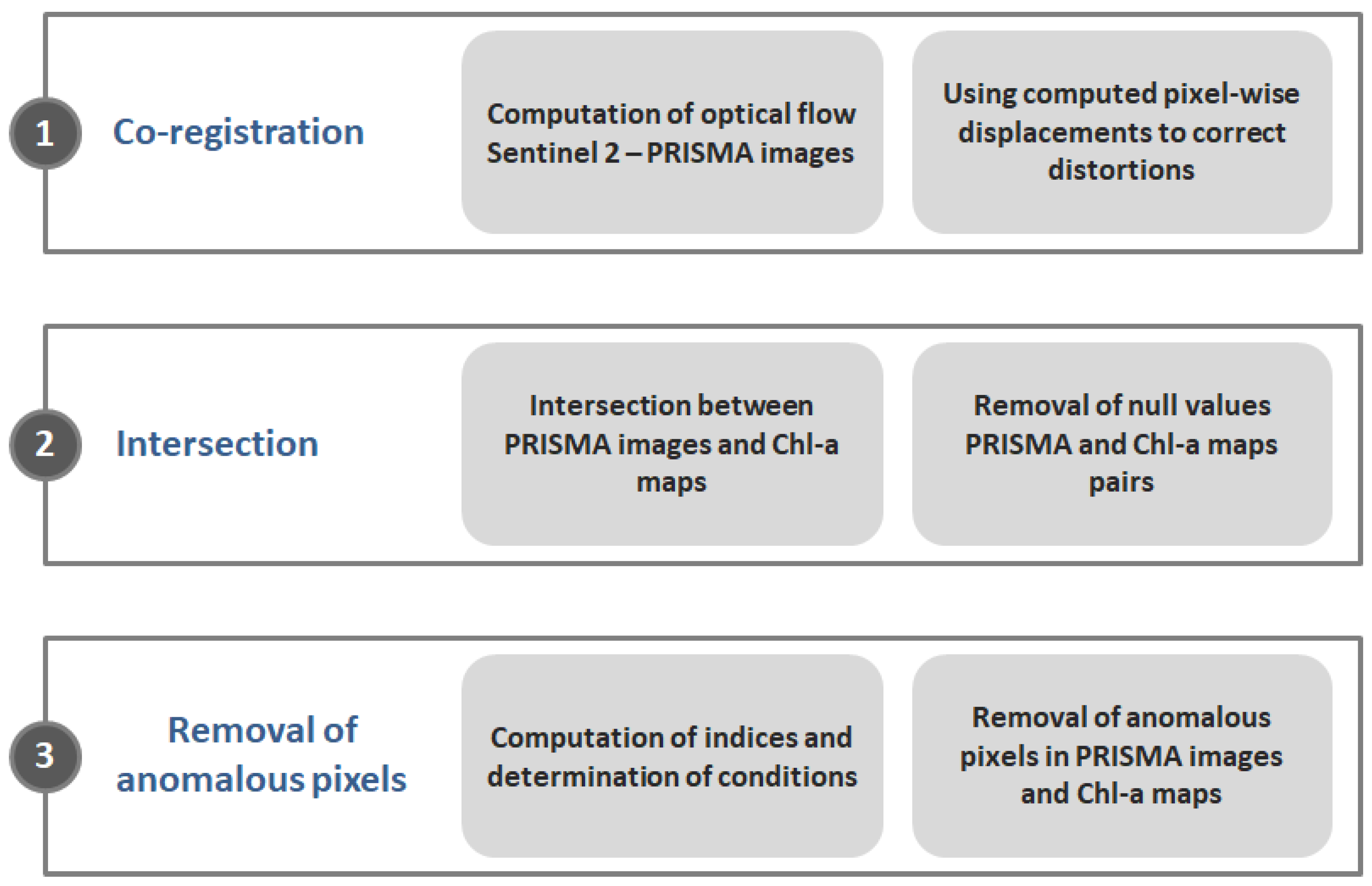
The models are employed to (i) reconstruct reference Chl-a concentrations maps computed from Sentinel-3 data, and (ii) augment the spatial resolution of such maps from 300 m to 30 m, thereby aligning them with the resolution of PRISMA imagery. The ultimate goal of such applications is to evaluate the performance of different models in reconstructing the reference Chl-a maps exploiting PRISMA images. Several experiments involving different configurations of model hyperparameters and resolutions for the training/testing datasets were conducted. The resulting accuracies were analyzed statistically to delineate and recommend the most effective models and experimental settings. The SVR model performed best for reconstructing reference Chl-a maps at 300 m spatial resolution, while the RF Regressor model proved to be the most effective for predicting Chl-a maps at 30 m spatial resolution.
The remainder of the paper is as follows.
Section 2 describes the data utilized in the study, detailing the criteria for dataset selection, outlining data preparation techniques, and introducing the models considered along with their respective hyperparameter settings.
Section 3 presents the outcomes of the modelling experiments, offering a discussion of the significant findings compared with the experimental settings adopted. Finally,
Section 4 includes conclusions and future directions of the work.
3. Results and Discussion
This section reports the results of the modelling experiments described in
Section 2.4. The predictive performances of the different models under different settings are reported and compared using well-known metrics, such as the Mean Absolute Error (MAE) and the Root Mean Square Error (RMSE). The evaluation is based on the acquisitions from the test set, specifically acquisitions 4, 23, and 24 (see
Section 2.2).
3.1. RF Regressor
A total of 12 experiments with different settings were performed using the RF Regressor model. Experimental settings and results are reported in
Table 5.
Considering the PRISMA image normalization approaches introduced in
Section 2.3, three experiments were carried out (RF-1 to RF-3) to establish the best option. The results achieved by the three experiments are identical in terms of predictive performances, suggesting a negligible effect of the normalization approach on the output.
The fourth experiment (RF-4) was used to evaluate the benefits of applying the PCA technique to reduce the spectral dimension to 30 PCs. As observed in
Table 5, this experiment yielded a worse performance in comparison with the previous three cases. This may be attributed to the ensemble nature of RF which utilizes decision trees, known to be robust against multicollinearity. Consequently, in this particular context, PCA may not yield substantial advantages, given that the algorithm inherently handles a multitude of features and their complex interactions.
In the fifth experiment (RF-5), it was investigated whether including additional bands would benefit the performance of the model. The same normalization approach of Experiment RF-2 (standard scaling) and no dimensionality reduction were applied. For each pixel, the Mean and the Sobel x and Sobel y filters were applied. According to the result, the addition of these new features was not helpful in terms of predictive performance.
In order to determine the optimal configuration for the model’s hyperparameters, five experiments were undertaken, denoted as RF-6 through RF-11. These experiments assessed various combinations of hyperparameters to ascertain which yielded the most favourable outcomes. Experiment RF-10 emerged as the top-performing RF Regressor model configuration.
The final experiment (RF-12) aimed to assess whether utilizing input data at a 30-m spatial resolution could lead to improved performance compared to the previously identified best-performing model configuration (i.e., RF-10). To achieve this, Chl-a maps needed to be upsampled to match the spatial resolution of the PRISMA images. The Nearest Neighbour method was employed for this purpose. It is important to note that, due to computational limitations, the number of trees was reduced to 1000 in this experiment compared to RF-10. Under these specified conditions, the results of Experiment RF-12 demonstrate a higher level of error compared to RF-10.
3.2. SVR
A total of 10 experiments with different settings were performed using the SVR model. The experimental settings and results are reported in
Table 6.
The effect of PRISMA image normalization approaches was analysed through three experiments (SVR-1 to SVR-3). The standard scaling resulted in the most effective approach in terms of prediction performances. The effect of PCA application for reducing the spectral dimension of the PRISMA images was evaluated in Experiment SVR-4. In this case, prediction performance resulted to be significantly improved with the use of 30 PCs instead of the original PRISMA bands. This result may be explained by the fact that the SVR model is based on an RBF kernel, which incorporates feature distances and may be sensitive to multicollinearity, thereby possibly resulting in over-fitting. Experiment SVR-5 investigated the advantages of employing data augmentation on the input data. This did not lead to an improvement in the model’s performance, achieving a less favourable outcome compared to SVR-4. Different setups for the SVR model hyperparameters, C and gamma, were assessed in experiments SVR-6 to SVR-9. Despite the evaluations, it was found that Experiment SVR-4 consistently yielded the most favourable outcomes. Therefore, SVR-4 was identified as the best-performing model configuration. The final experiment (SVR-10) assessed the impact of employing a 30-m spatial resolution for the input data, which included the original PRISMA images and the Chl-a maps upsampled via the Nearest Neighbor technique. Unfortunately, the results from SVR-10 were not satisfactory, with both MAE and RMSE metrics surpassing those attained by the previously identified top-performing model configuration, SVR-4.
3.3. LSTM Network
A total of 14 experiments with different settings were performed using the LSTM network. Experimental settings and results are reported in
Table 7.
The first three experiments (LSTM-1 to LSTM-3) explored the best normalization approach. The best performance was achieved with the experiment LSTM-2 which corresponds to the standard scaling method. Experiment LSTM-4 explored the spectral dimensionality reduction, and determined that this technique was useful for improving the performance as it achieved a lower level of error. From Experiment LSTM-5 to Experiment LSTM-12, all the hyperparameters of this model architecture were systematically adjusted. Among these experiments, the best-performing configuration was the Experiment LSTM-10. Furthermore, Experiment LSTM-13 used the same hyperparameter settings as LSTM-10 but incorporated bidirectional flow. Notably, this modification improved the performance compared to LSTM-10. The utilization of 30-m resolution inputs was examined in Experiment LSTM-14. The model hyperparameters and input normalization were kept identical to those in Experiment LSTM-13. However, the outcome did not show any improvement over the performance metrics of the best-performing model configuration, LSTM-13.
3.4. GRU Network
A total of 17 experiments with different settings were performed using the GRU network. Experimental settings and results are reported in
Table 8.
The first three experiments (GRU-1 to GRU-3) focused on investigating the effect of normalization approaches. The standard scaling method (GRU-2) emerged once again as the most effective approach. The results obtained from Experiment GRU-4 indicate that employing the PCA method for this model did not yield any significant benefit. Hyperparameter tuning was carried out by experiments GRU-5 to GRU-15, with Experiment GRU-8 resulting as the best-performing model configuration. Experiment GRU-16 was used to determine whether configuring the GRU-8 network with bidirectional flow would enhance its performance. The results indicate a decrease in performance. Finally, Experiment GRU-17 maintained an identical configuration to Experiment GRU-8, except for the utilization of 30-m input data. However, this adjustment did not yield any improvements in the model’s performance. As a result, the best-performing model configuration was identified as that of Experiment GRU-8.
3.5. Summary of Best Models and Inference on 30 m
Drawing from the aforementioned experiments, it is evident that the best performances were attained when training and assessing the models with 300-m input datasets. The details of the best-performing configurations for each model are consolidated in
Table 9, with SVR (Experiment SVR-4) emerging as the top-performing model overall.
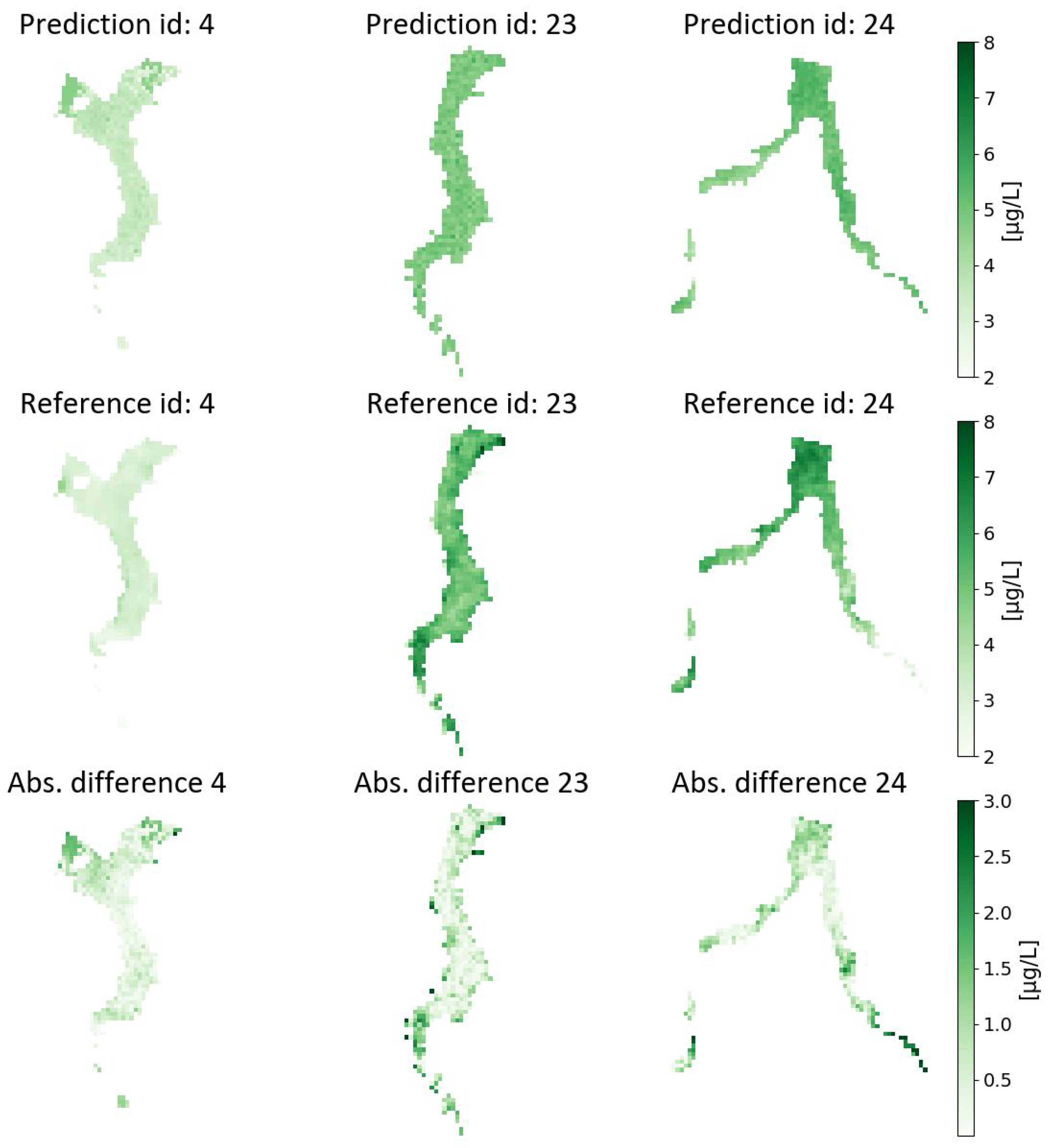
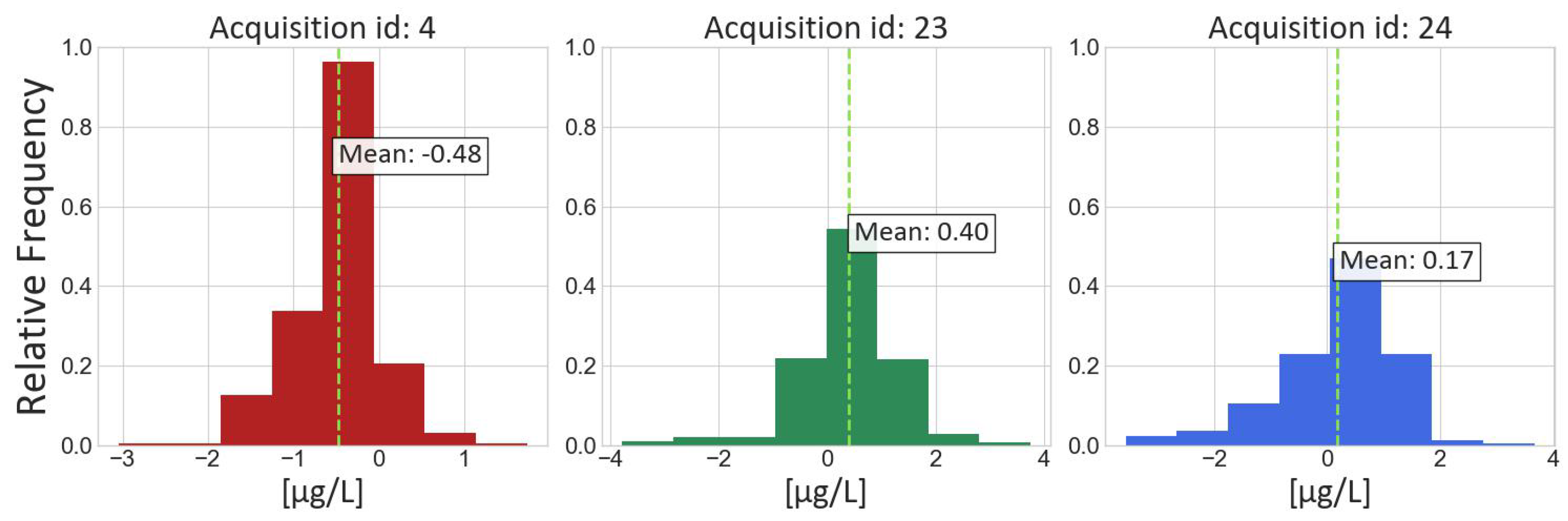
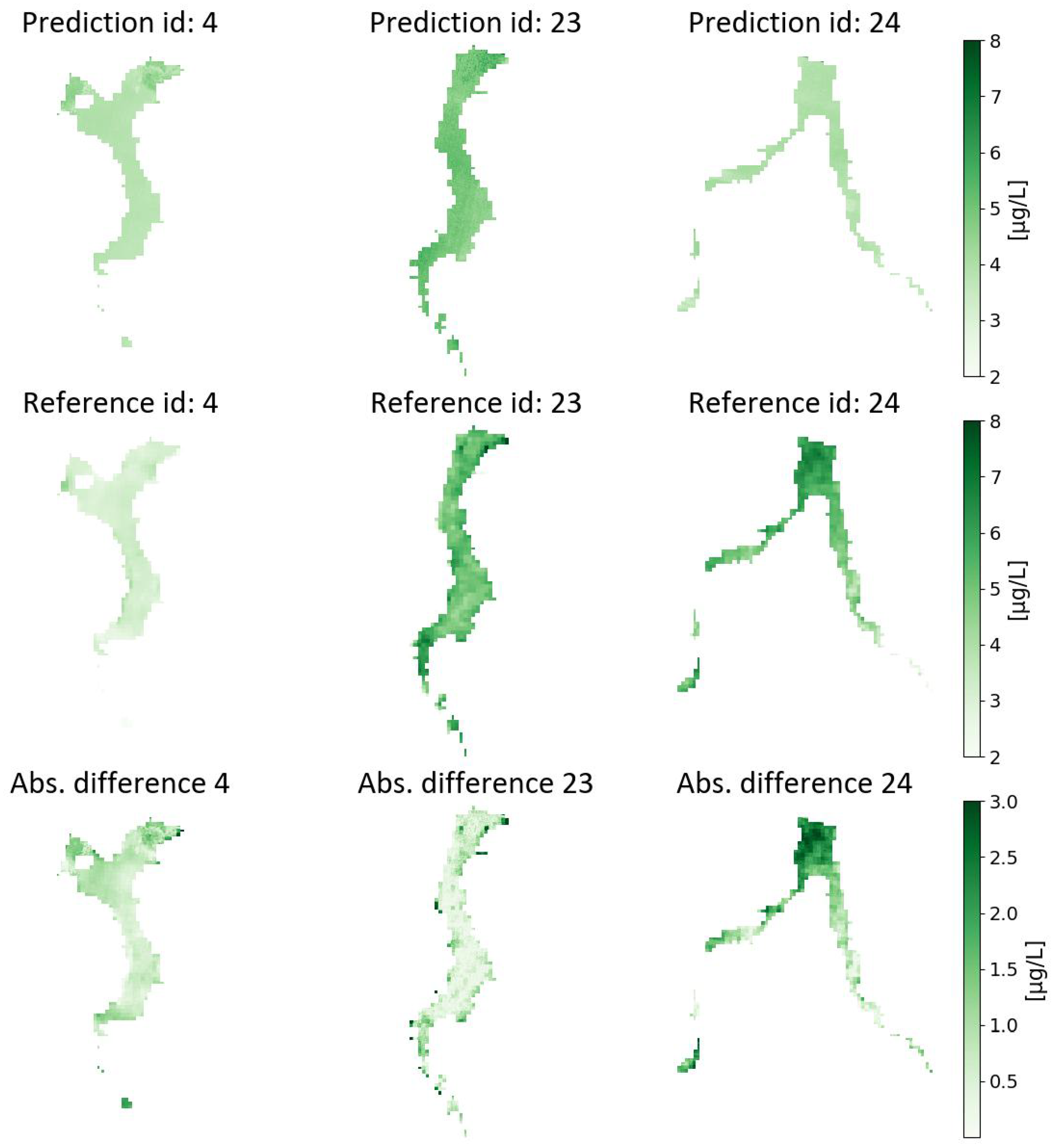
Figure 5 presents the visual results for Experiment SVR-4 with its model setting applied to the test acquisitions (ID 4, 23 and 24; see
Table 1) and
Figure 6 shows the distribution of the errors for each of the acquisitions in the test set.
Until this moment, the output spatial resolution of 300 m was overlooked, and only the resulting performance was analyzed. However, recognizing that output with a finer spatial resolution of 30 m could yield more valuable results, efforts were directed towards determining the optimal approach to achieve predictions at this higher resolution.
For this purpose, two alternative approaches were identified. The first, which has already been investigated, consisted of using 30-m data for model training, validation, and evaluation (testing). The second approach involved the use of the best-performing configurations of each considered model, which were trained with 300 m data (a summary of the results is included in
Table 9), and to perform an inference on 30-m data for their evaluation.
Figure 7 provides a schematic of these two alternative approaches.
After evaluating both approaches with the best-performing configurations of the four model typologies, the RF Regressor (Experiment RF-12) both trained and evaluated with 30-m data, emerged as the best alternative for achieving a prediction with 30 m of spatial resolution. The summary of these results is reported in
Table 10.
Figure 8 presents the visual results of Experiment RF-12, trained and evaluated at 30-m spatial resolution data and applied to each of the acquisitions in the test set. The associated errors’ distributions are shown in
Figure 9.
A final observation drawn from the presented results is the tendency of the considered machine learning models to underestimate Chl-a values in high local concentration spots. Further considerations on the above are reported in the following section.
4. Conclusions and Outlook
This study addressed the implementation aspects related to the generation of Chl-a concentration maps utilizing PRISMA hyperspectral imagery, with low-resolution training data derived from Sentinel-3 imagery. The complete workflow for preparing input data for a range of machine and deep learning models was outlined. Performances of the models under various hyperparameter configurations were compared to offer empirical insights into the best-performing solutions for estimating multi-resolution Chl-a concentrations in lakes using hyperspectral imagery and pre-existing Chl-a concentration maps at lower spatial but higher temporal resolution.
By conducting several modelling experiments, the optimal configurations for each of the four analyzed models were determined. Specifically, the most favourable performances were attained when employing 300 m spatial resolution inputs for all experiments. The best results were achieved with the SVR model. Supplementary experiments were conducted to evaluate model performances in enhancing the spatial resolution of Chl-a concentration predictions from the original 300 m reference data (i.e., Sentinel-3 derived Chl-a concentration maps) to 30 m resolution such as the one of PRISMA hyperspectral imagery. The RF Regressor proved to deliver the best performance for this last objective.
While the obtained performances are relevant for all model typologies, it is worth noting that these results could be potentially improved with the availability of additional PRISMA acquisitions. As discernible from the presented results, the selected machine learning models demonstrated a tendency to underestimate regions characterized by high Chl-a concentrations. The inclusion of supplementary PRISMA acquisitions linked to high Chl-a concentration spots in the input dataset (which were limited in the dataset used for this study) is expected to mitigate this discrepancy and represents a critical improvement for future developments of this work.
Given the limitations to the accessibility of ground truth data for training and evaluating machine and deep learning models, the approach outlined in this study is promising for preliminary large-scale estimates of Chl-a concentrations in freshwater bodies. This is because it suggests strategies for the use of low-resolution and widely accessible training and testing datasets by leading to a final product with a significantly higher spatial resolution than the reference data while maintaining an acceptable margin of error. This enhancement is achieved by leveraging both spectral and spatial characteristics of the emerging hyperspectral satellite imagery. It is worth remarking that operations such as resampling low-resolution reference data for model evaluation on 30 m resolution outputs were tested for purely experimental purposes. The use of high-resolution reference data is envisaged to improve both the quality and reliability of the proposed procedure, especially of local gradients of Chl-a concentrations in each single water body.
The outcomes of the suggested method have the potential to function as supportive resources for the monitoring and administration of the lakes under investigation. The use of global coverage and freely available data, coupled with open modelling tools, additionally strengthens this groundwork for enhancements and replications in different geographic regions.