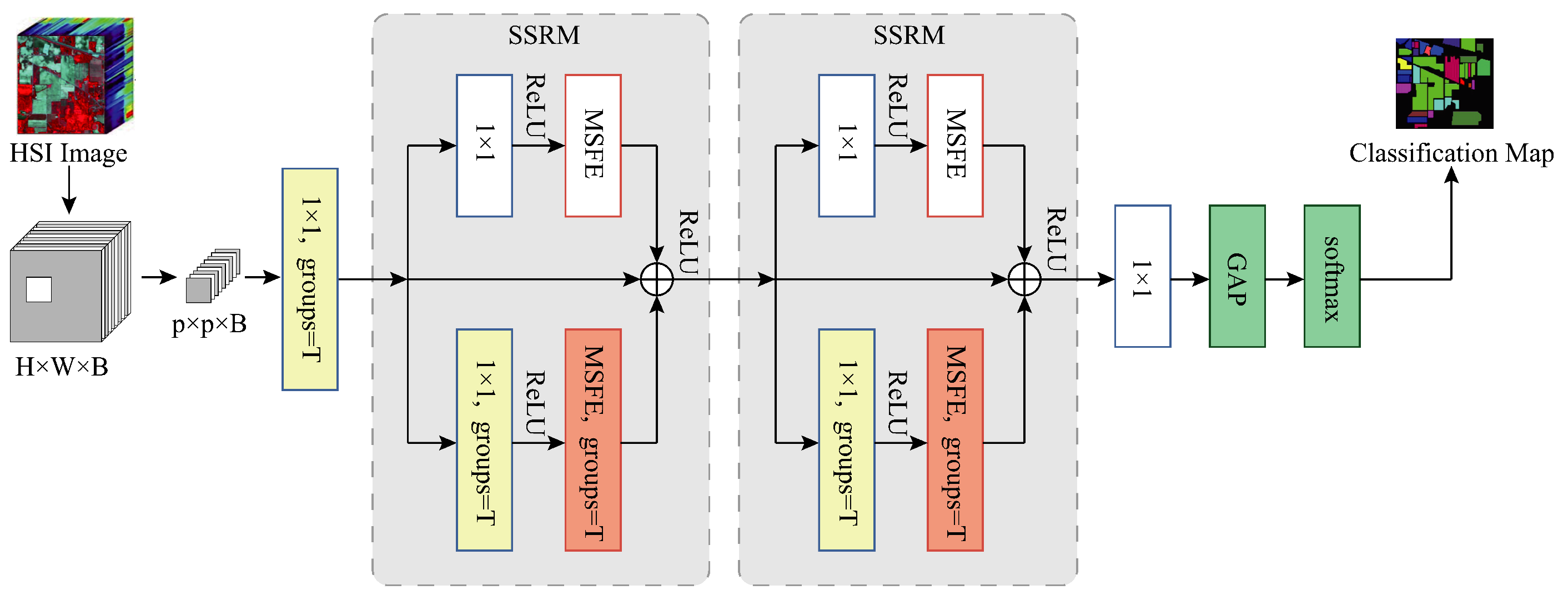
3.1. Dataset
In this section, we design experiments to assess how well our suggested model performs on three commonly used datasets: Indian Pines, Salinas, and Pavia University.
Indian Pines (IP) dataset was collected over the Indian Pines Test Site in northwest Indiana by the 224-band Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) [
39], covering 224 spectral bands between 0.4 and 2.5 um in wavelength. The image size was 145 × 145 and contained 16 vegetation classes. Twenty-four water absorption bands were removed, leaving 200 bands available for training.
Salinas (SA) dataset, which spans 224 spectral bands with wavelengths ranging from 0.4 to 2.5 um, was collected by AVIRIS over the Salinas Valley in California. The image size is 512 × 217 and contains 16 land cover classes. Twenty water absorption bands were removed from the experiment, and the actual bands used for training were 204.
Pavia University (PU) dataset was acquired over Pavia, northern Italy, by the Reflection Optical System Imaging Spectrometer (ROSIS) [
40] and covers 103 spectral bands between 0.38 and 0.86 um in wavelength. The image size is 610 × 340 and contains nine land cover classes.
We pre-processed the data by first normalizing the values to the range [0, 1], followed by a data augmentation operation that flipped the input patch vertically or horizontally and then randomly rotated it by , or .
For the IP dataset, we used 5% of the labeled samples for training, 5% for validation, and 90% for testing. On the other hand, for the SA and PU datasets, we used 0.5% of the labeled samples for training, 0.5% for validation, and 99% for testing. The number of training, validation, and testing samples for each class is displayed in
Table 2,
Table 3 and
Table 4. We selected the best model on the validation set to be evaluated on the testing set, and we divided the labeled samples 10 times at random, with all results being the average of these 10 runs.
3.2. Experimental Setup
To assess the performance of the MFERN model, we conducted experiments on an Intel(R) Core(TM) i7-9700K CPU @ 3.60 GHz and an NVIDIA GeForce RTX 2080 using the Pytorch framework. We used the Adam [
41] optimizer to update all training parameters in the framework.
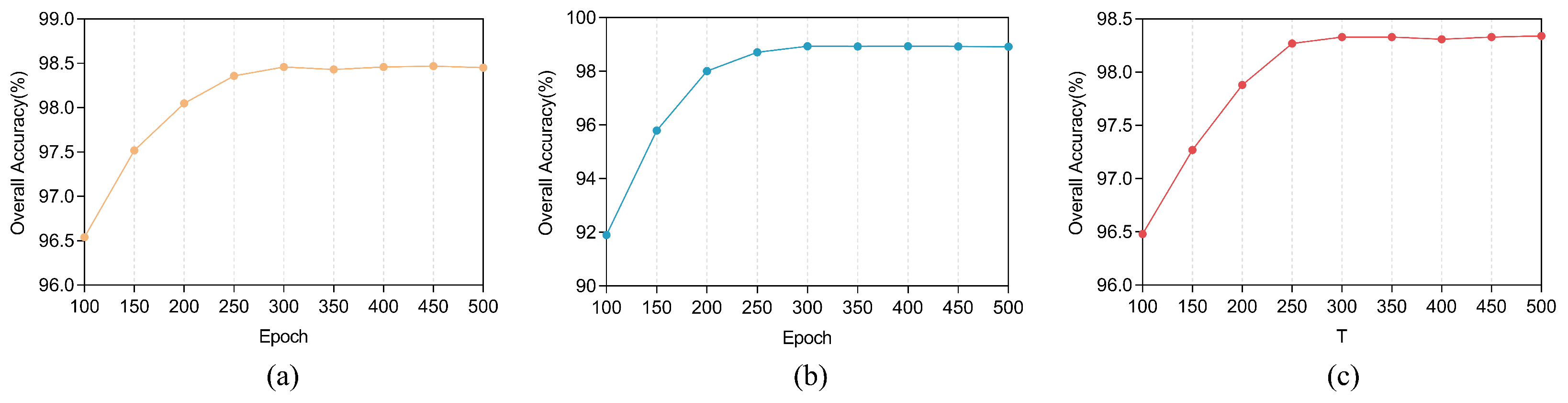
Training epochs have a direct impact on the model. Fewer training rounds may not be enough for the model to fully learn the complex patterns of the data, while more training epochs may cause the model to overfit the training data. Additionally, as the training epochs increase, the model training time also increases. Therefore, in order to find a suitable training epochs setting that avoids overfitting and underfitting while making the training time as small as possible, we conducted experimental exploration. Specifically, we initially set the maximum training epoch = 500 when training on each dataset and output the OA value of the model on the validation set when epoch
during the training process, and then finally plot a graph to observe the change of OA value with the increase in epochs, the experimental results are shown in
Figure 7, and it can be observed that on the three datasets when the epoch is less than 250, the model has poor accuracy and exhibits the characteristics of underfitting. This is because the model has not yet sufficiently learned the complex patterns of the input data to capture the underlying relationships of the data. At this point, the model’s fitting ability is weak and cannot match the training data well. As the training epochs increase, the model gradually improves its fitting ability and reduces underfitting to some extent. When epoch = 300, the model converges, and the model accuracy changes weakly when the epochs increase again after that; therefore, in order not to increase the training time of the model, all the experiments after that are trained with 300 epochs.
The batch size is set to 128, the initial learning rate (lr) is set to 0.001, and the network was trained from scratch without using predefined weights. At 100 epochs, the lr is multiplied by a factor of 0.1, changing to 0.0001, and at 250 epochs, the lr continues to be multiplied by a factor of 0.1, changing to 0.00001. The precision (OA), average precision (AA), and Kappa coefficient act as evaluation metrics to assess the effectiveness of the suggested approach.
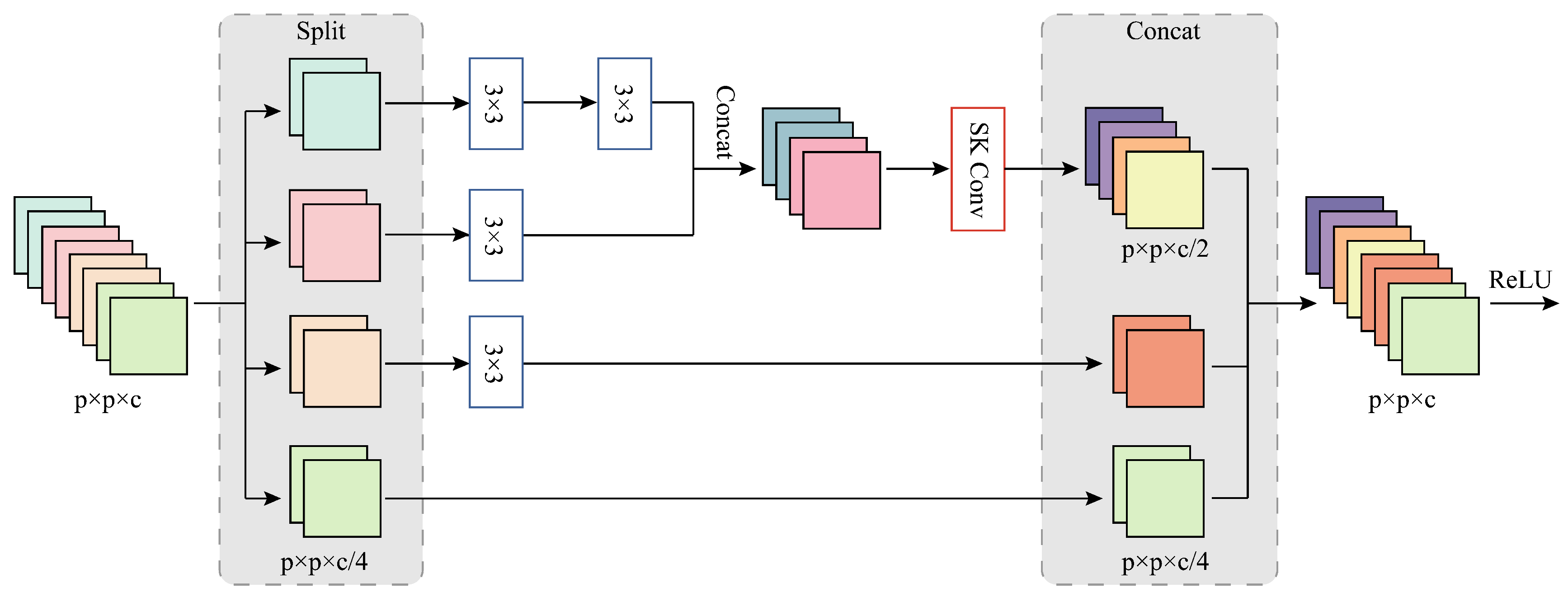
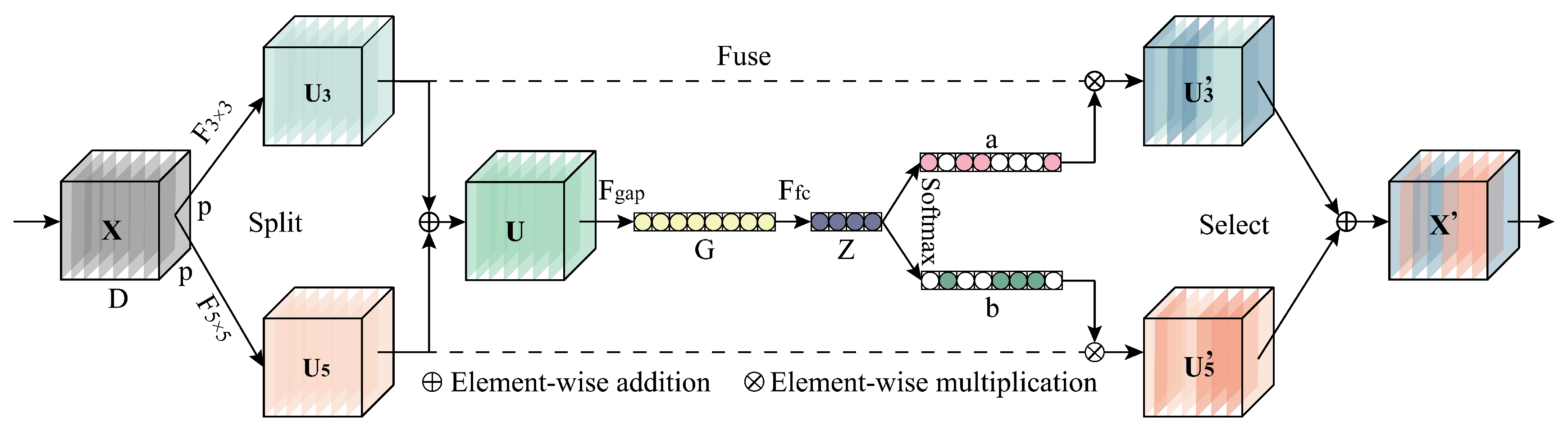
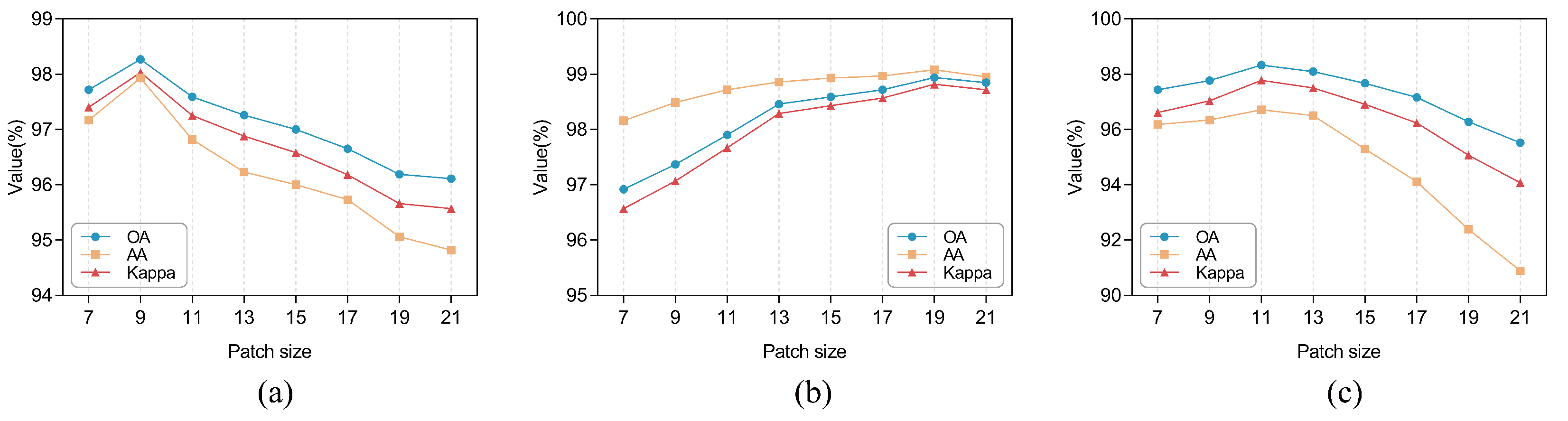
3.4. Effect of the Number of MSFE Divisions
For the MSFE module, the number of divisions
s determines the maximum RF size that can be used to extract spatial features. The larger the
s, the more 3 × 3 convolutions are stacked on the branch, and the larger the RF of the convolutional layer to which the branch is applied. However, for different datasets, the applicable RF size varies, so in order to obtain the optimal setting of the number of MSFE divisions, we conducted experiments by taking the number of MSFE divisions s as
and observing the variation of OA, AA, and Kappa values of the model on the three datasets, respectively. The results of the experiment are displayed in
Figure 9. It can be seen that the model performs best for the SA dataset and PU dataset when
, while for the IP dataset,
provides the best classification performance. This is because the input patches of SA and PU are larger, and their spatial information is more complex, so it is better to use a larger RF for classification, while the input patches of the IP dataset are smaller and suitable for using a relatively small RF. However, for all three datasets, the model performance tends to decrease when
and 6 because when the RF is too large, the model may incorrectly extract many useless features, confusing the classifier and possibly overfitting the model. Additionally, it can be noted that when
, the MSFE module becomes a normal residual block and can only obtain information about the 3 × 3 size RF when the OA, AA, and Kappa values are lower on all three datasets, so this verifies that our proposed MSFE module can indeed exploit the multi-scale potential at a finer level of granularity and extract spatial information from the feature maps.
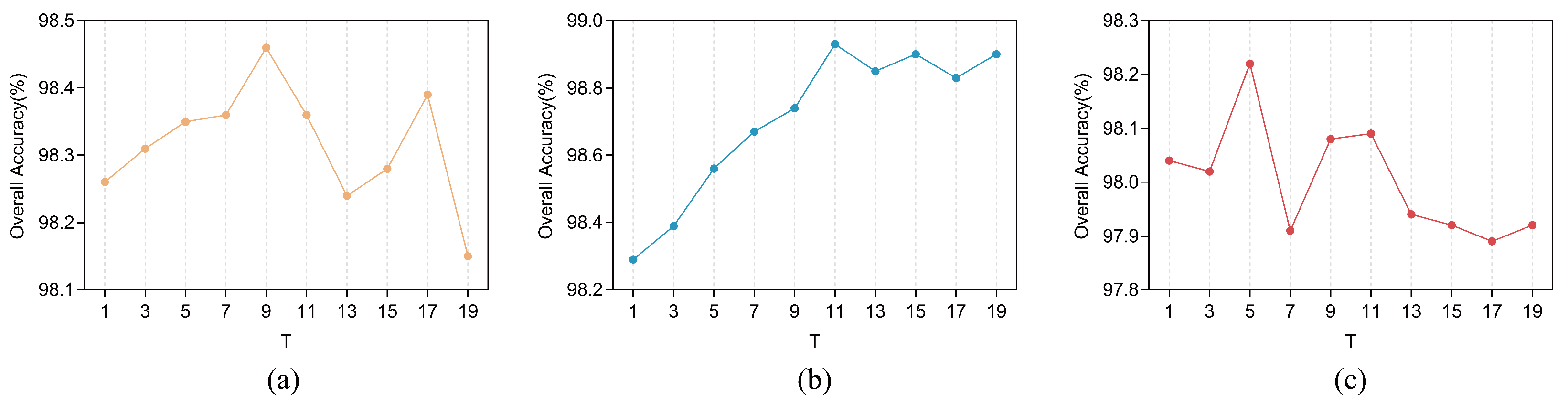
3.5. Number of Groups in SSRM
The amount of groupings in SSRM determines the number of spectral bands divided and the number of parallel MSFEs to be used for feature extraction and is, therefore, a parameter to be considered. According to the experimental findings in
Section 3.4, we set the number of MSFE divisions to be
on the IP dataset, and
on the SA and PU datasets. On this basis, we explored the OA values on the three datasets when the number of groupings
T of the SSRM takes odd values between 1 and 23, respectively. The results of the experiment are displayed in
Figure 10, where it is evident that the optimal number of groups
T varies for different datasets, with the optimal number of groups
T being 9, 11, and 5 for the IP, SA, and PU datasets, respectively. It can be noted that when
T takes the value of 1, the group convolution degenerates to a normal convolution, which is equivalent to having two global branches without local branches, and since the OA value at
is not high for all three datasets, therefore, this validates the validity of the improved grouped convolution in our presented SSRM, i.e., the validity of the local branches. Validation of the validity of global branching will be explored in a subsequent subsection on ablation studies.
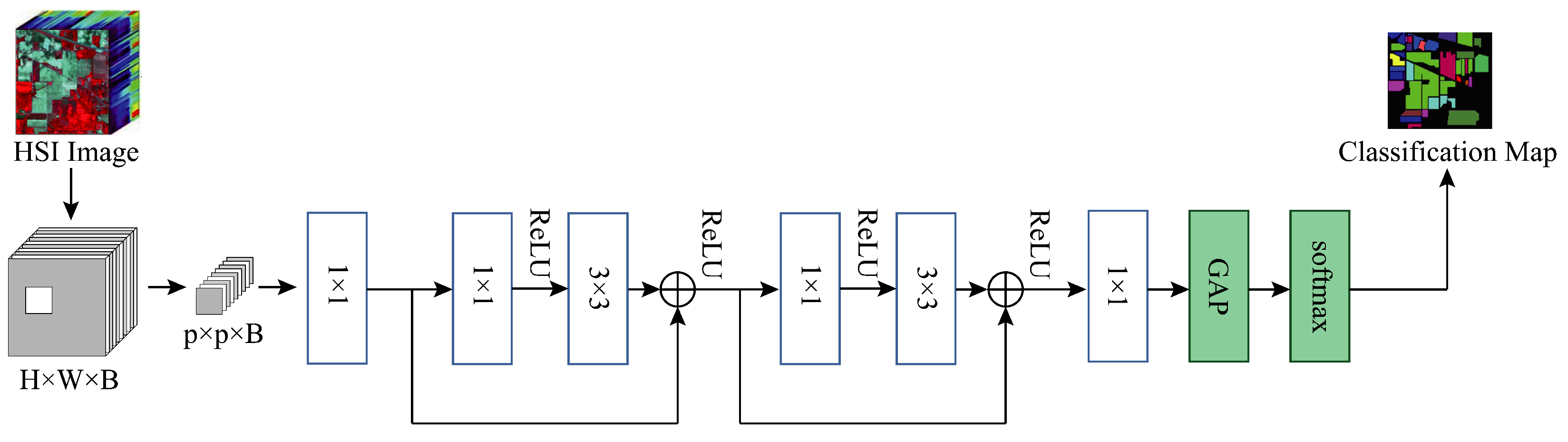
3.6. Ablation Study
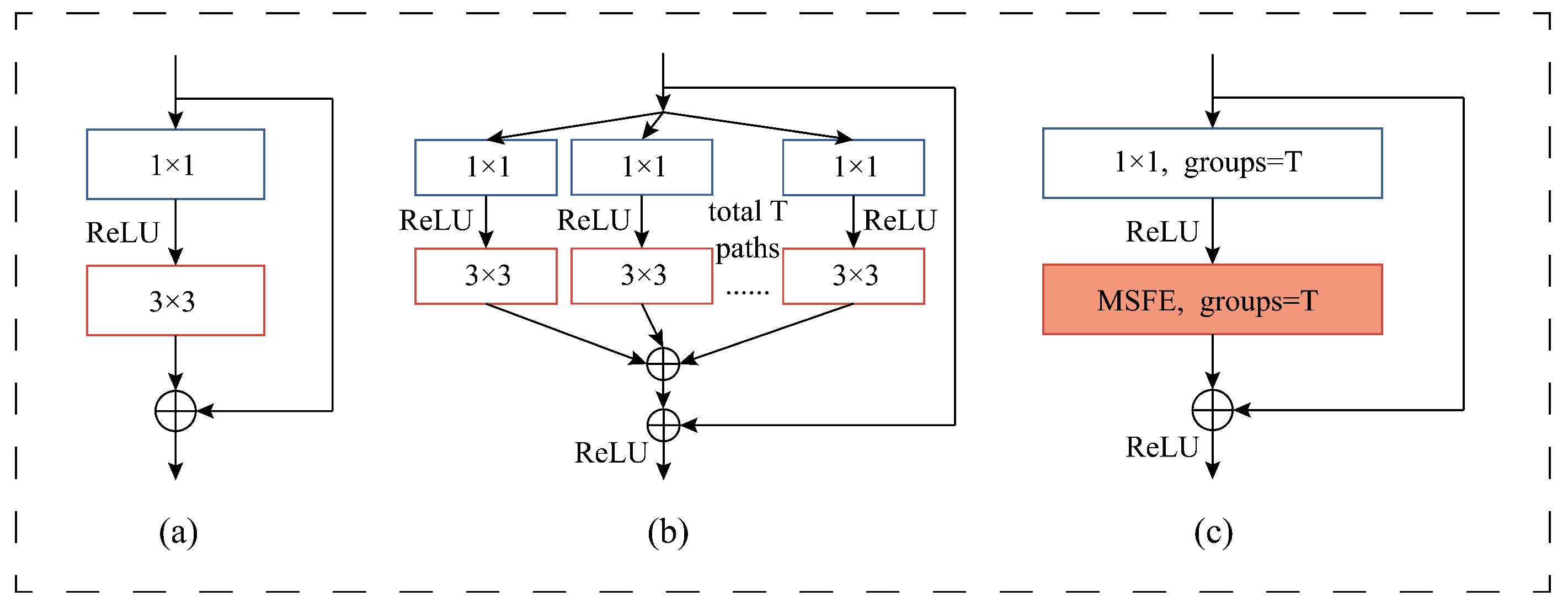
To demonstrate the contribution of the MSFE and SSRM modules of our proposed method to the final classification results of the model, we conducted an ablation study on three datasets. Specifically, we kept the other experimental settings unchanged while replacing our SSRM module with the basic module designed in DenseNet in
Figure 4a, i.e., at this point the model does not use the MSFE module, the convolutions are all normal convolutions rather than grouped convolutions, and there is no global branching. We use this as the baseline model, which we name Baseline, and
Figure 11 depicts the model’s construction. We add the modules we have designed to this model in turn to verify the validity of the different designs and modules.
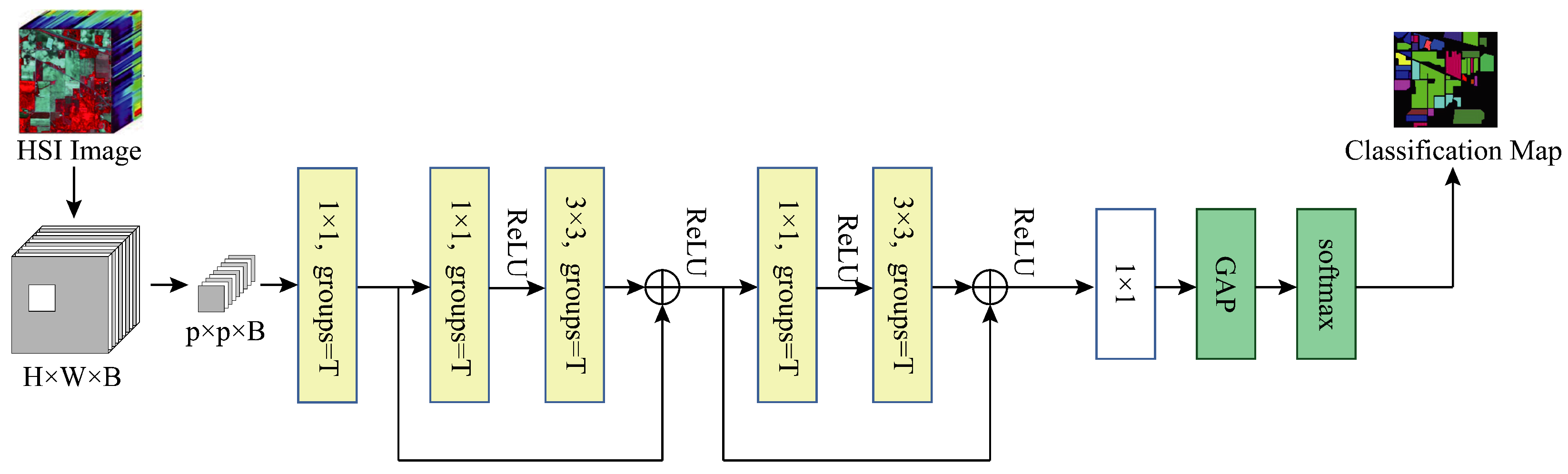
Base+GC: We replace all the convolution layers except the last one with grouped convolution on top of the Baseline to verify the effectiveness of using grouped convolution, at which point the model structure is shown in
Figure 12.
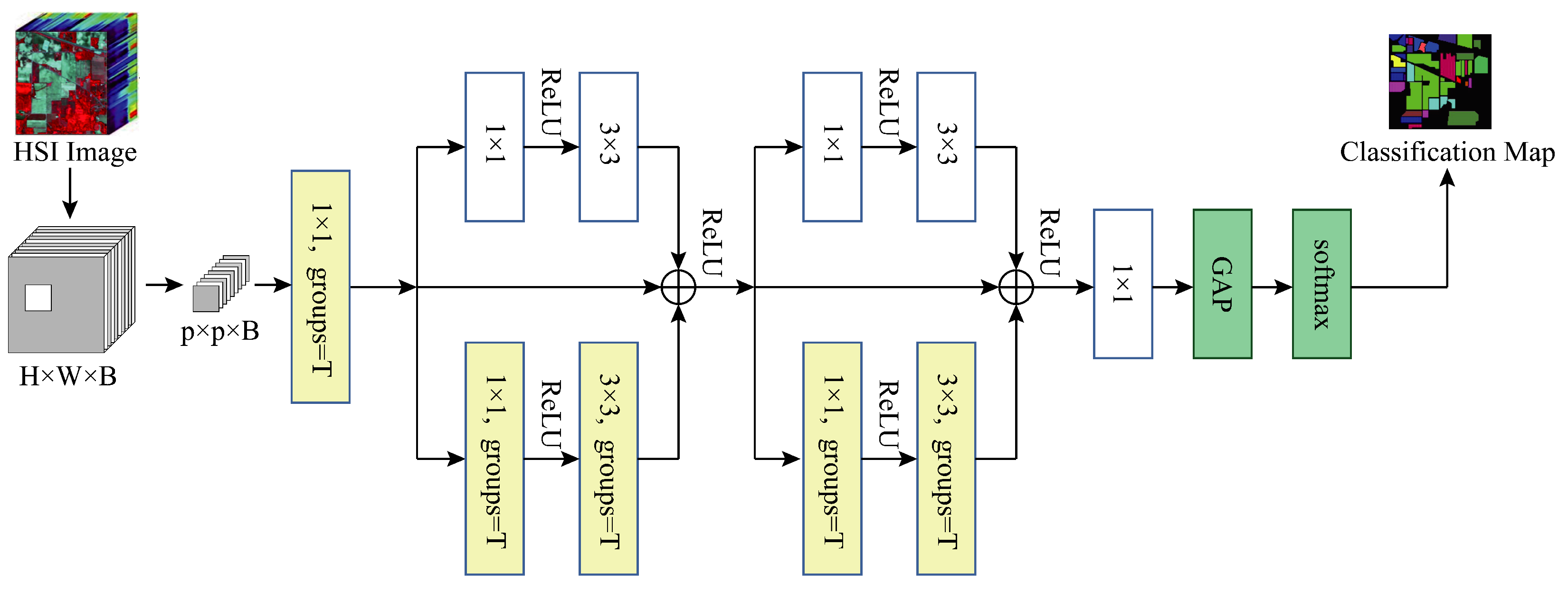
Base+GC+GB: We verify the validity of our proposed global branching by adding global branches to the two residual blocks in the model based on the use of grouped convolution, with only normal convolutional layers on the global branches, specifically a 1 × 1 kernel size convolutional layer and a 3 × 3 kernel size convolutional layer.
Figure 13 illustrates the model’s current structure.
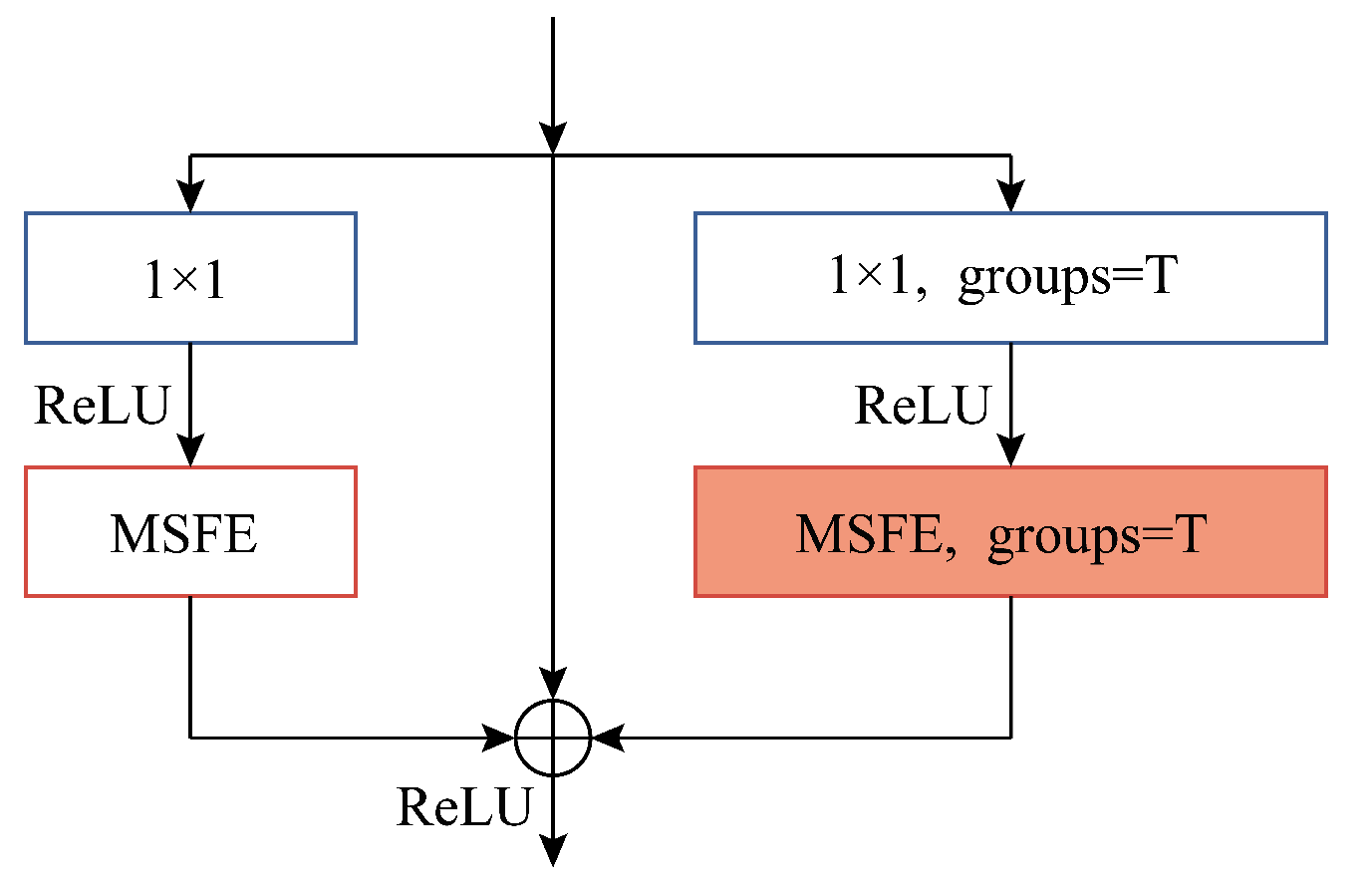
Base+GC+GB+MSFE(MFERN): We replace all the convolutional layers of kernel size 3 × 3 in the model with MSFE modules based on the use of grouped convolution and the addition of global branches, at which point the residual blocks in the model are our proposed SSRM modules and the model structure is the final structure of our presented MFERN method, as displayed in
Figure 6. We can verify the effectiveness of the suggested MSFE and SSRM modules through this experiment.
Table 5 displays the findings of the ablation study using the three datasets, and it can be observed that, compared to Baseline, the addition of grouped convolution (Base+GC) to Baseline improved OA by 0.09%, AA by 0.05% and Kappa by 0.10% on the IP dataset and on the SA and PU datasets, OA, AA, and Kappa improved by 0.29%, 0.27%, 0.33%, and 0.06%, 0.09%, 0.07%, respectively. Thus, using grouped convolution instead of normal convolution is effective, especially for the SA dataset. Later, adding global branches (Base+GC+GB) to the use of grouped convolution, it can be seen that compared to Baseline, OA improves by 0.15%, AA improves by 0.29%, and Kappa improves by 0.17% on the IP dataset, OA, AA, and Kappa improve by 0.42%, 0.42%, and 0.46%, respectively, on the SA and PU datasets, 0.46% and 0.25%, 0.20% and 0.33% on the SA and PU datasets, respectively. Therefore, our proposed global branch is valid, and it is able to extract global feature information for the whole spectral band. Finally replacing all 3 × 3 kernel size convolutional layers with the MSFE module (MFERN) on top of the previous one, the OA, AA and Kappa values on all three datasets are significantly improved compared to Baseline, Specifically, for the IP dataset, OA improved by 0.30%, AA by 0.70% and Kappa by 0.34%, and for the SA and PU datasets, OA, AA, and Kappa improved by 0.52%, 0.47%, 0.58% and 0.41%, 0.50%, and 0.55%, respectively. In summary, the baseline model has the worst performance, while the performance of the model continues to improve with the addition of our suggested modules, with the best results when all of our suggested modules are used, so that the MFERN model we proposed achieves the most advanced performance.
3.7. Comparison with Other Methods
In this section, we contrast our MFERN model with other deep learning-based HSIC methods. Specifically, we compare our approach with ResNet [
27], DFFN [
42], SSRN [
43], PResNet [
29],
-ResNet [
44], HybridSN [
25], RSSAN [
45], SSTN [
46], and DCRN [
47]. Among them, ResNet, DFFN, and PResNet use 2D-CNN, SSRN,
-ResNet, and RSSAN is based on 3D-CNN, HybridSN and DCRN use a hybrid CNN of 2D-CNN and 3D-CNN, and SSTN is based on Transformer. The experimental setup for this method was set up as described in
Section 3.2, and the parameters were set to the values paired with the optimal experimental results in
Section 3.3,
Section 3.4 and
Section 3.5, as follows: the IP dataset patch size of 9 × 9, the number of divisions
, and the number of groupings
; the SA dataset patch size of 19 × 19, the number of divisions
, the number of groupings
; the PU dataset patch size of 11 × 11, the number of divisions
, the number of groupings
. The detailed architecture of MFERN on the three datasets is shown in
Table 1. The IP dataset is used as an example. All grouped convolutional layers have 288 filters in 9 groups. In other words, we divide the spectrum into 9 groups and extract features using 9 CNNs, respectively, each of which has a bandwidth of 32. The normal convolutional layer in SSRM, on the other hand, has 288 filters, and the final 1 × 1 convolutional layer has 128 filters. All the convolutional layers have a step size of 1. We chose this number of convolutional kernels in order to keep the network parameters around 10 MB on the IP and PU datasets and 50 MB on the SA dataset because of the larger input patches and higher number of subgroups in the SA dataset. For a fair comparison, the Pytorch framework was used for all compared methods; we trained the model using fewer samples, and the samples from the training, validation, and testing sets were chosen at the scale described in
Section 3.1, for the IP dataset, we used 5% of the labeled samples for training, 5% for validation, and 90% for testing; for the SA and PU datasets, we used 0.5% of the labeled samples for training, 0.5% for validation, and 99% for testing. The other hyperparameters were set as described in the original paper of the model.
Table 6,
Table 7 and
Table 8 display the outcomes of the experiment.
Overall, our proposed MFERN outperforms the other methods on all three datasets. Specifically, MFERN’s OA, AA, and Kappa values on the IP dataset were 98.46 ± 0.25, 98.13 ± 0.82 and 98.24 ± 0.29, respectively. On the SA and PU datasets, the OA, AA, and Kappa values were 98.94 ± 0.39, 99.08 ± 0.23, 98.82 ± 0.43 and 98.33 ± 0.47, 97.71 ± 0.28, and 97.78 ± 0.63. Compared to ResNet, DFFN, and PResNet, which are also based on 2D-CNN, MFERN has improved OA values by 2.40–9.57%, 1.70–8.01% and 3.20–10.26% over the IP, SA and PU datasets, and the above three methods are also based on residual networks, suggesting that our MSFE module helps to improve the model performance. Compared with the 3D-CNN-based SSRN, A2S2K-ResNet, MFERN’s method is closer to, but slightly better than, these two methods. This is because although we use a 2D-CNN, designing global branches in SSRM enables us to obtain spectral features for the whole band so that no spectral information is lost and obtain higher performance. Our method also performs better than the Transformer-based SSTN method; this is because we combine MSFE with group convolution in SSRM to reduce the input dimensionality of each MSFE module performing feature extraction while better extracting multi-scale spatial features, after which the features extracted from local and global branches are combined to obtain, for the full band, both spectral and multi-scale spatial features. It can be seen that the performance of RSSAN based on 3D-CNN and HybridSN based on hybrid CNN of 2D-CNN and 3D-CNN is not very good. The reason may be that the HybridSN network is more complex, which will increase the time and resource consumption for training and inference and is prone to overfitting problems, leading to unsatisfactory classification accuracy, while the RSSAN network is too simple, which may not be able to capture complex features in the data, leading to a decrease in accuracy. Taken together, networks such as RSSAN and SSTN with fewer parameters than MFERN have lower model accuracy than MFERN, which indicates that MFERN is able to keep the model complexity within a reasonable range while ensuring higher accuracy.
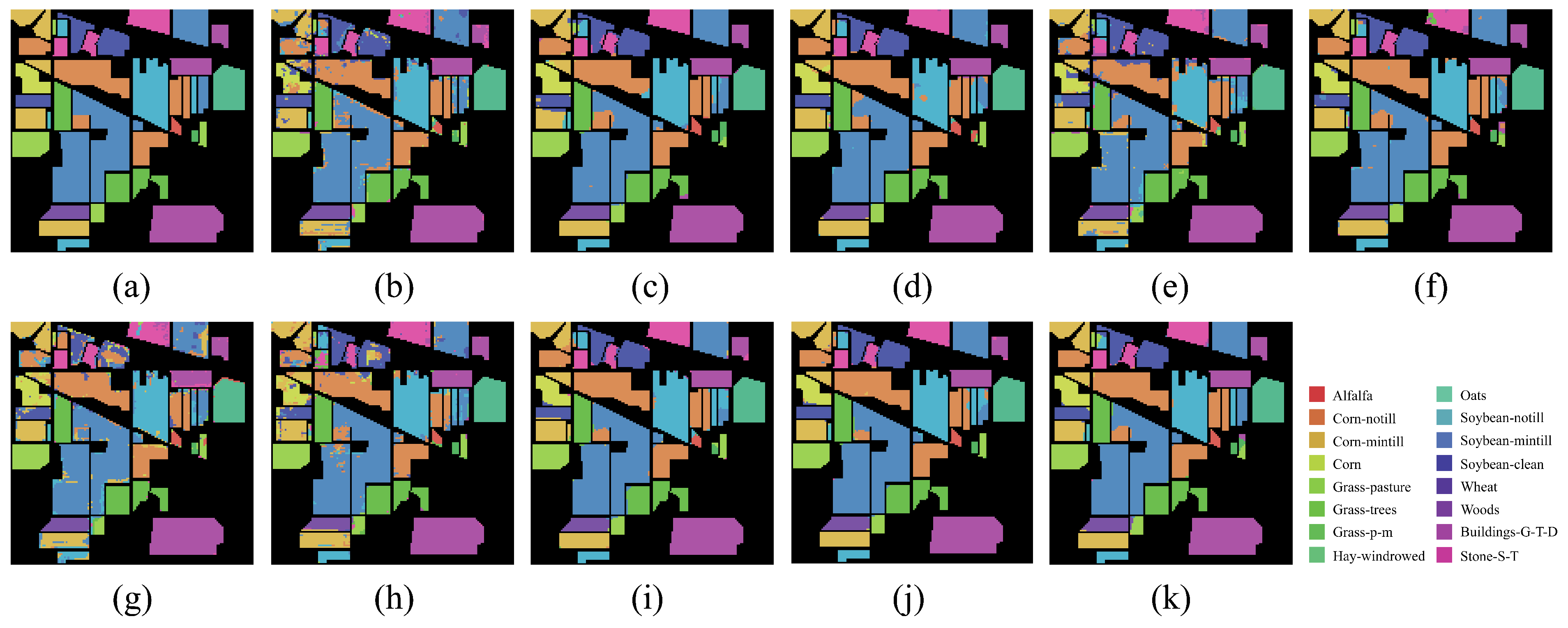
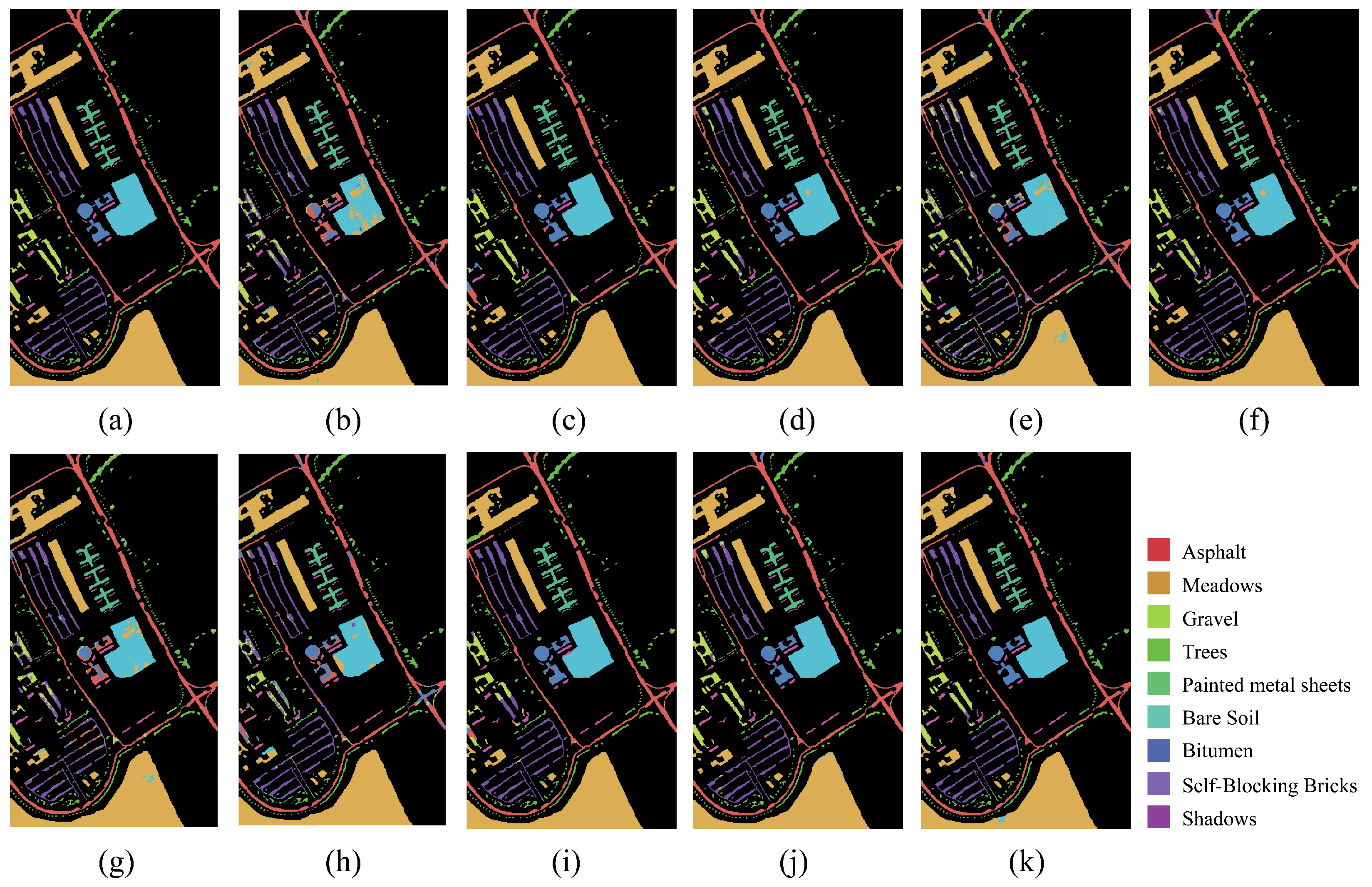
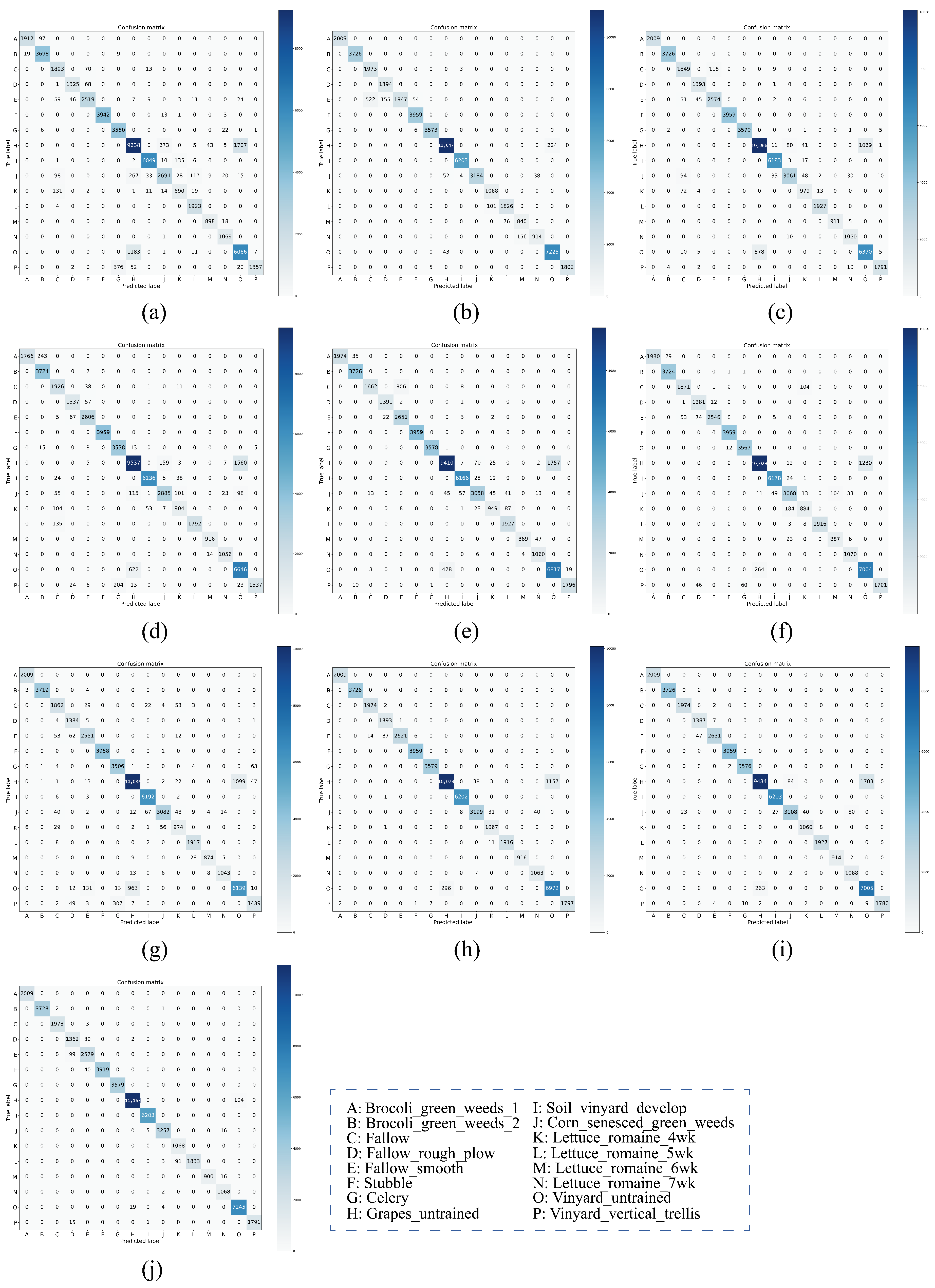
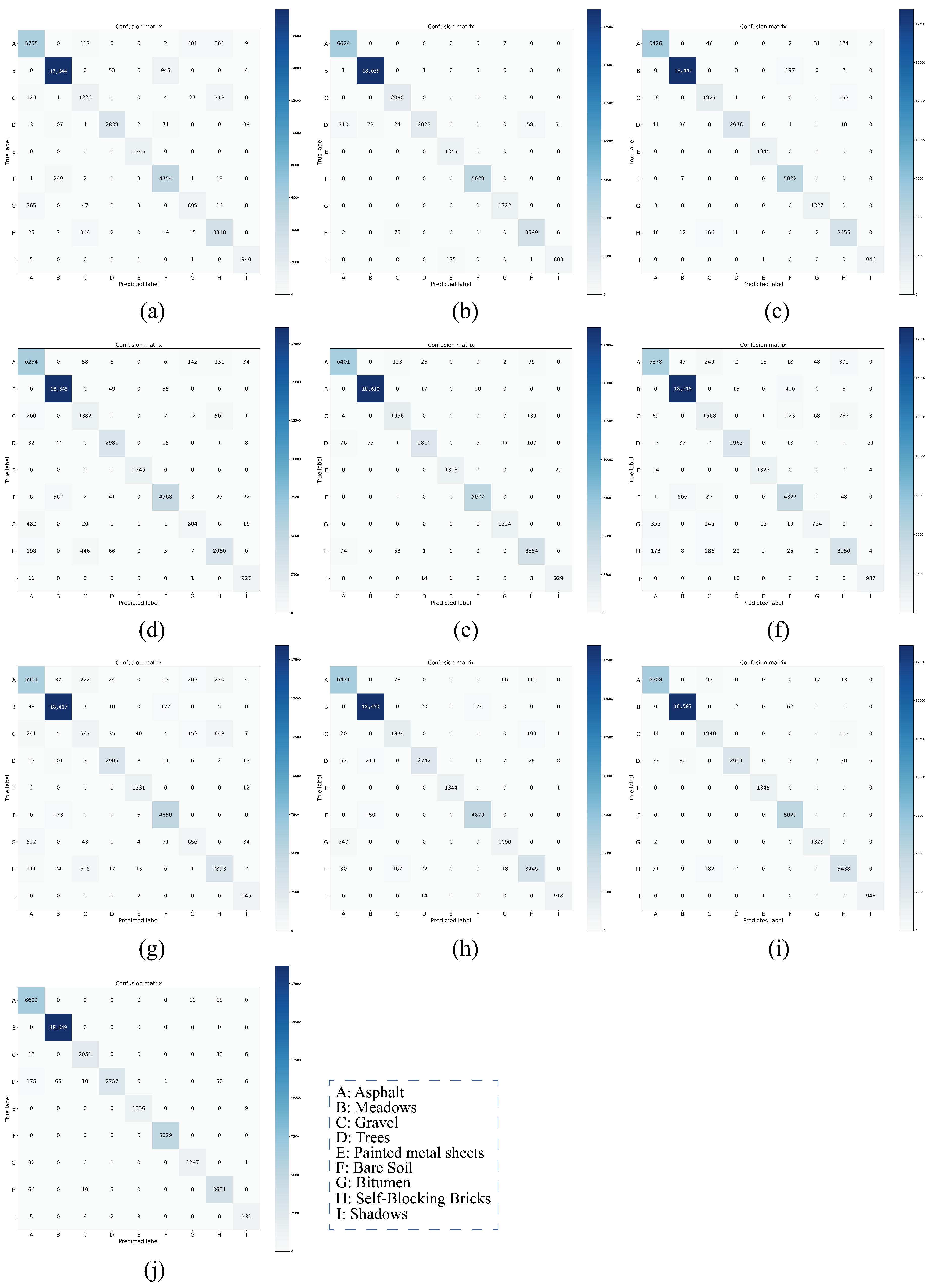
On both IP and PU datasets, compared with the DCRN model, which obtains the next best performance, the DCRN model adopts a dual-channel structure to extract spectral and spatial features of hyperspectral images separately, which may lead to a certain degree of information loss, and the correlation and interplay between spatial and spectral features may be lost when extracting both separately, which may limit the expressive power of the features. In contrast, our SSRM module captures richer feature information by designing local and global feature extraction branches, where the local branch focuses on fine-grained local structural and textural features, which can perceive the subtle changes and local information of the target object, thus enhancing the robustness of the model against noise, occlusion, and other disturbing factors. The global branch, on the other hand, can obtain the overall contextual information, providing a grasp of the overall features of the image and improving the classification accuracy and robustness. On the SA dataset, the DFFN network is deeper and extracts deeper features compared to the DFFN model that obtains the next best performance but ignores the fact that different sizes of features are subject to different RF and only uses a fixed-size RF for feature extraction, which restricts the learning weight of the model. Our MSFE module solves this problem well, and the multi-scale feature extraction branch provides different sizes of receptive fields to extract feature information at different scales and also considers the spatial relationship between pixels through the receptive fields at different scales, which helps the model to better understand the spatial distribution characteristics of the target object and improve the classification accuracy. Therefore, MFERN is more robust, does not need to stack too many blocks to fully extract the feature information, ensures high accuracy in the case of small samples, and keeps the model complexity at a low level. To check the classification performance more visually, we plotted the classification maps and confusion matrix plots produced using different methodologies on the three datasets, as shown in
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18 and
Figure 19. On all three datasets, MFERN has a high classification accuracy with a low noise level.
We further assessed the performance variation of the different methods when using different training sample percentages; specifically, for the IP dataset, we explored the variation of OA values for each method when the training sample size was taken from 6–10%, and for the SA dataset and PU dataset, we explored the variation of OA values for each method when the training sample size was taken from 1–5%.
Figure 20 displays the experimental results.
It can be seen that HybridSN performs poorly on the IP dataset, ResNet gives a poor performance on the SA dataset, and RSSAN gives poor classification accuracy on the PU dataset, while DFFN and DCRN have a more stable performance on all three datasets. The method in this study significantly outperforms the other comparative methods even with a small training sample set, which shows that MFERN can fully extract the spatial and spectral features of hyperspectral images, making it have high classification accuracy even with a small sample size. The performance gap between the methods gradually closes as the training sample size increases, yet our suggested method consistently outperforms the other approaches on the three datasets. This demonstrates the strong robustness of our proposed MFERN approach.
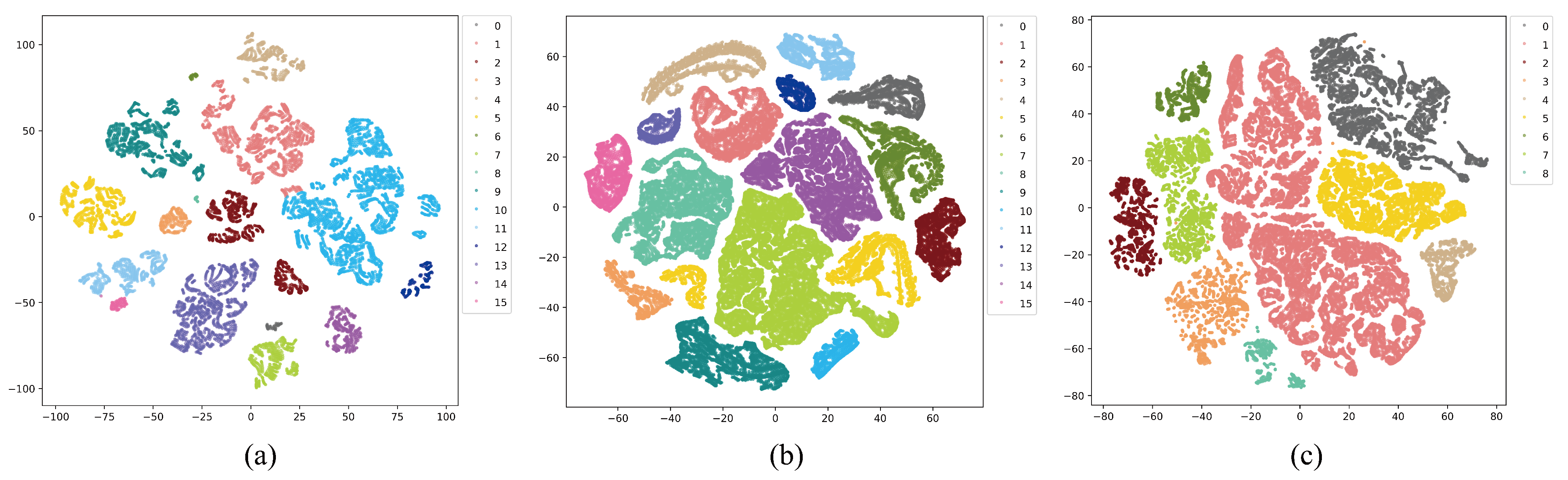
To validate the feature extraction capability of our proposed MSFE and SSRM and the representation capability of our trained models, we visualized the 2D spectral space features proposed by the test samples in the three datasets through the t-SNE algorithm [
48], as depicted in
Figure 21. In the figure, samples from the same class are clustered into one group, while samples from different classes are kept apart, and samples from different classes are shown using different colors are shown. The figure shows that on the three datasets, the samples from various classes are more clearly distinguished, thus indicating that our MSFE extracts feature more adequately at the fine- and coarse-grained levels and that our MFERN model can learn abstract representations of spectral space features well and with high classification accuracy.