Annual Field-Scale Maps of Tall and Short Crops at the Global Scale Using GEDI and Sentinel-2
Abstract
:1. Introduction
2. Datasets
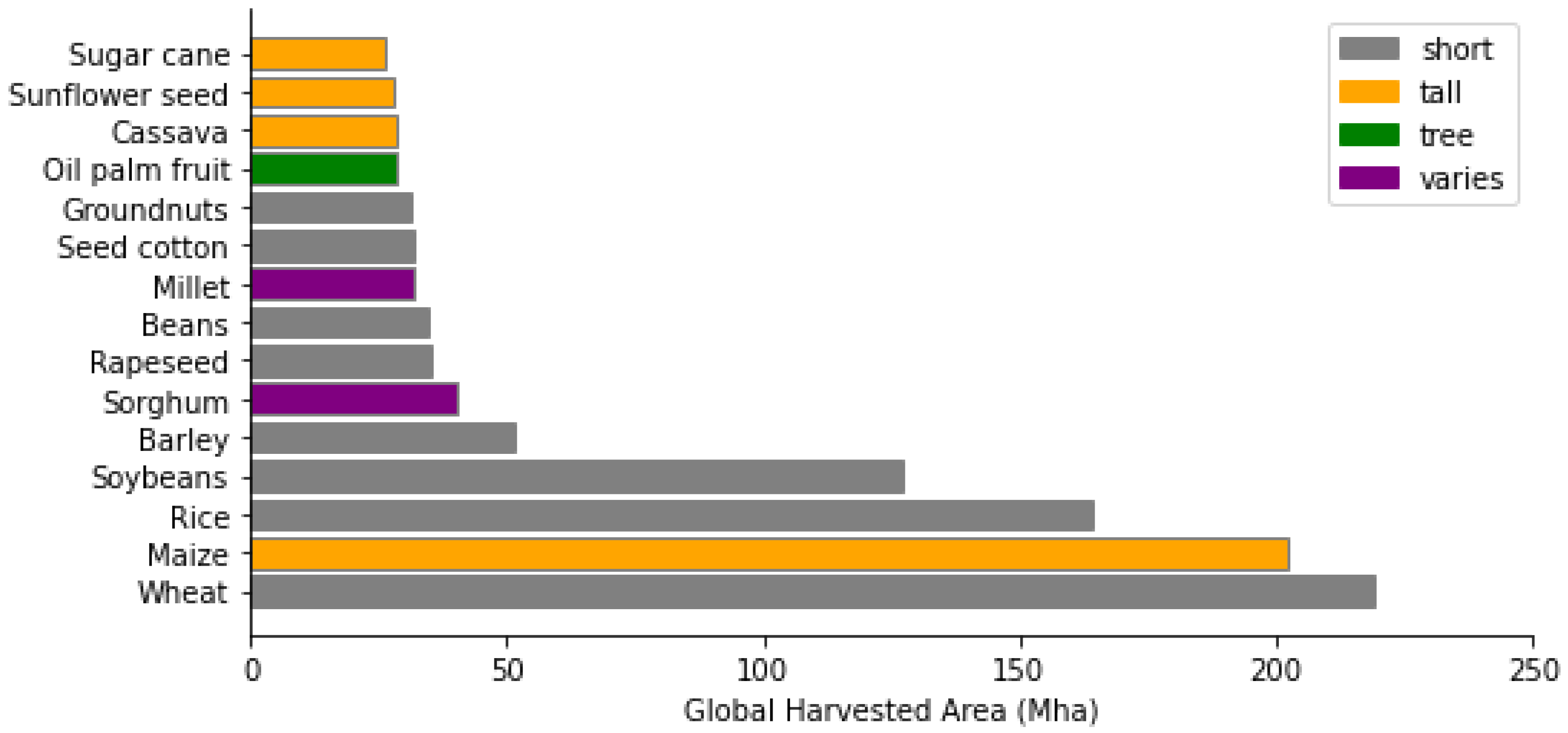
2.1. Crop Mask
2.2. GEDI Data
2.3. Sentinel-2
2.4. GEDI Model Training Dataset
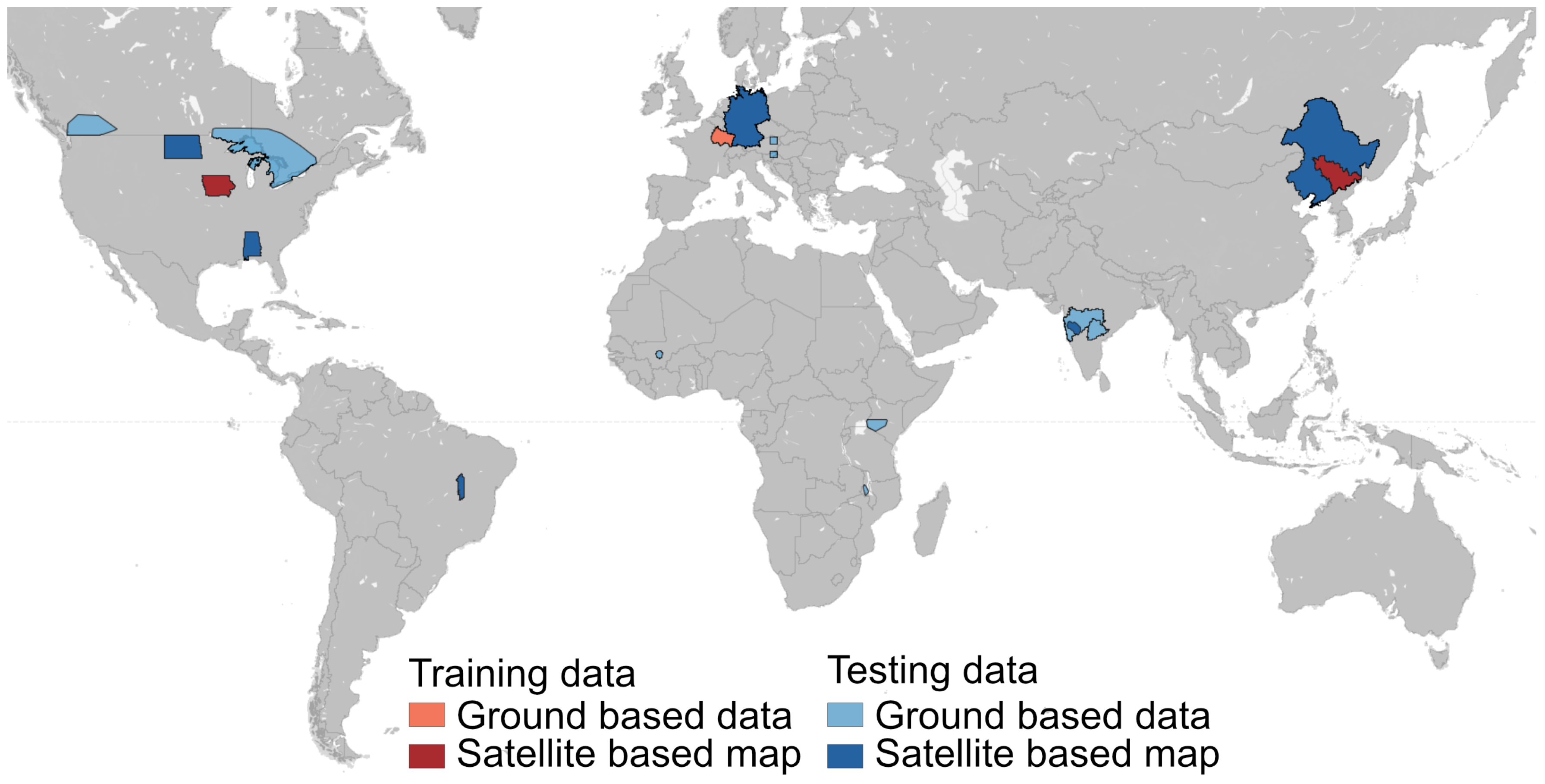
2.5. Evaluation Datasets
2.5.1. Ground-Based Reference Data
Europe
Canada
Malawi
Mali
Kenya
India
2.5.2. Satellite-Based Reference Data
United States
Germany
Brazil
China
India
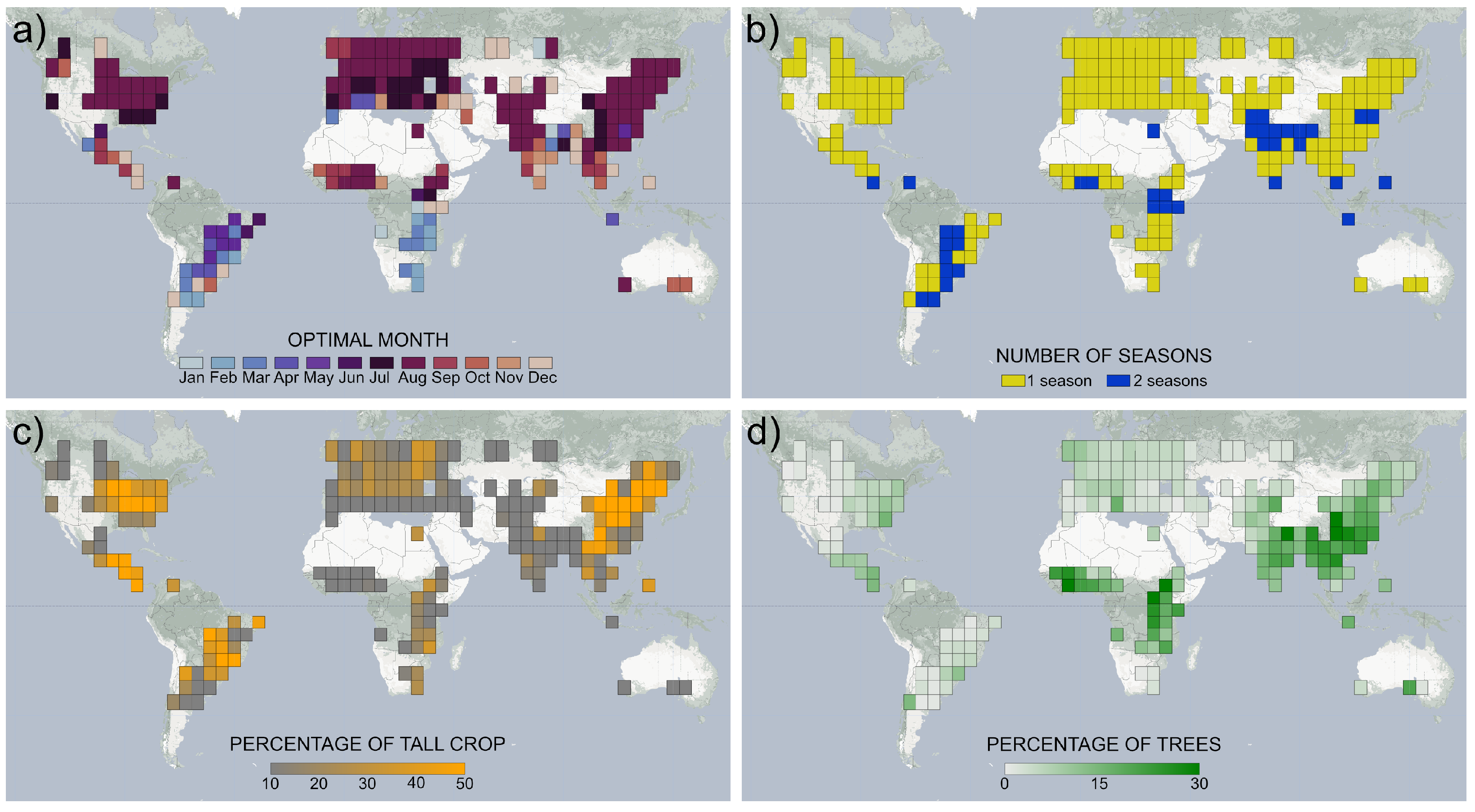
2.6. Number of Growing Seasons per Year
2.7. Digital Elevation Model (DEM)
2.8. Reference Maps for Error Analysis
2.8.1. Canada
2.8.2. Kenya
3. Methods
- 1.
- Train a single model, which we refer to as the GEDI model, that uses GEDI features to classify locations as having short crops, tall crops, or trees;
- 2.
- Apply the GEDI model to GEDI shots acquired from cropland areas globally for three years of 2019–2021;
- 3.
- Tile the globe into grid cells;
- 4.
- Determine the optimal month to predict tall crops for each grid-cell;
- 5.
- Train a local GEDI-S2 model for each grid-cell based on GEDI predictions in the 3-month time window around the optimal month;
- 6.
- Evaluate results against local reference data.
3.1. GEDI Model Training
3.2. GEDI Model Predictions
3.3. Model Grids
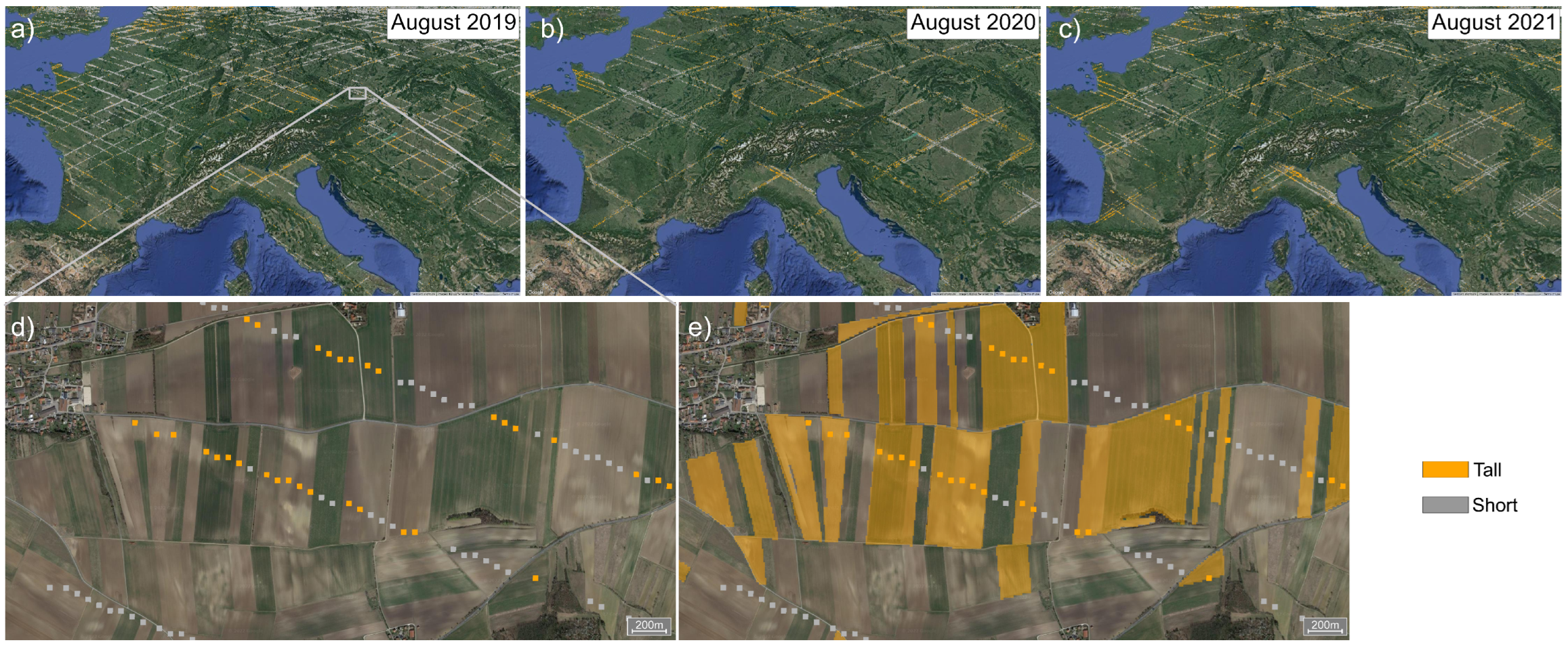
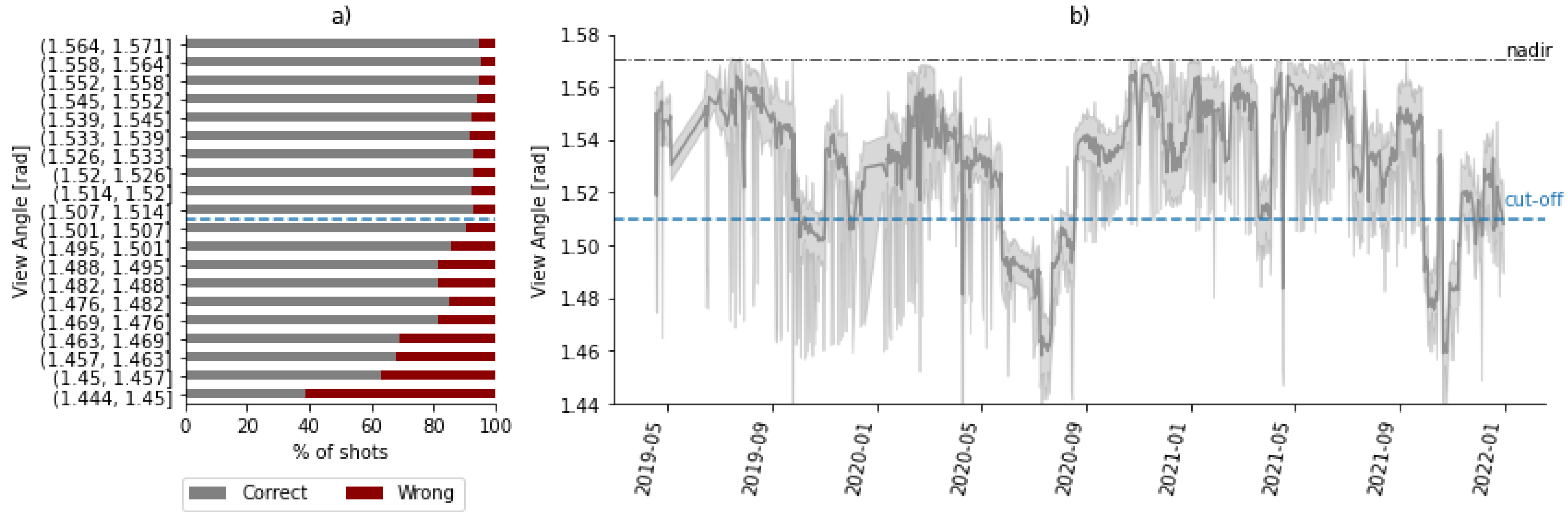
3.4. Optimal Timing
3.5. GEDI-S2 Models
3.6. Evaluation of GEDI-S2 Predictions
- True Positive, , is the number of samples labelled as positive by the model that are actually positive
- False Positive, , is the number of samples labelled as positive by the model that are actually negative
- True Negative, , is the number of samples labelled as negative by the model that are actually negative
- False Negative, , is the number of samples labelled as negative by the model that are actually positive
- is the proportion of observed agreement, i.e., the accuracy achieved by the model
- is the proportion of agreements expected by chance
4. Results
4.1. GEDI Predictions during Optimal Months
4.2. GEDI-S2 Model Training
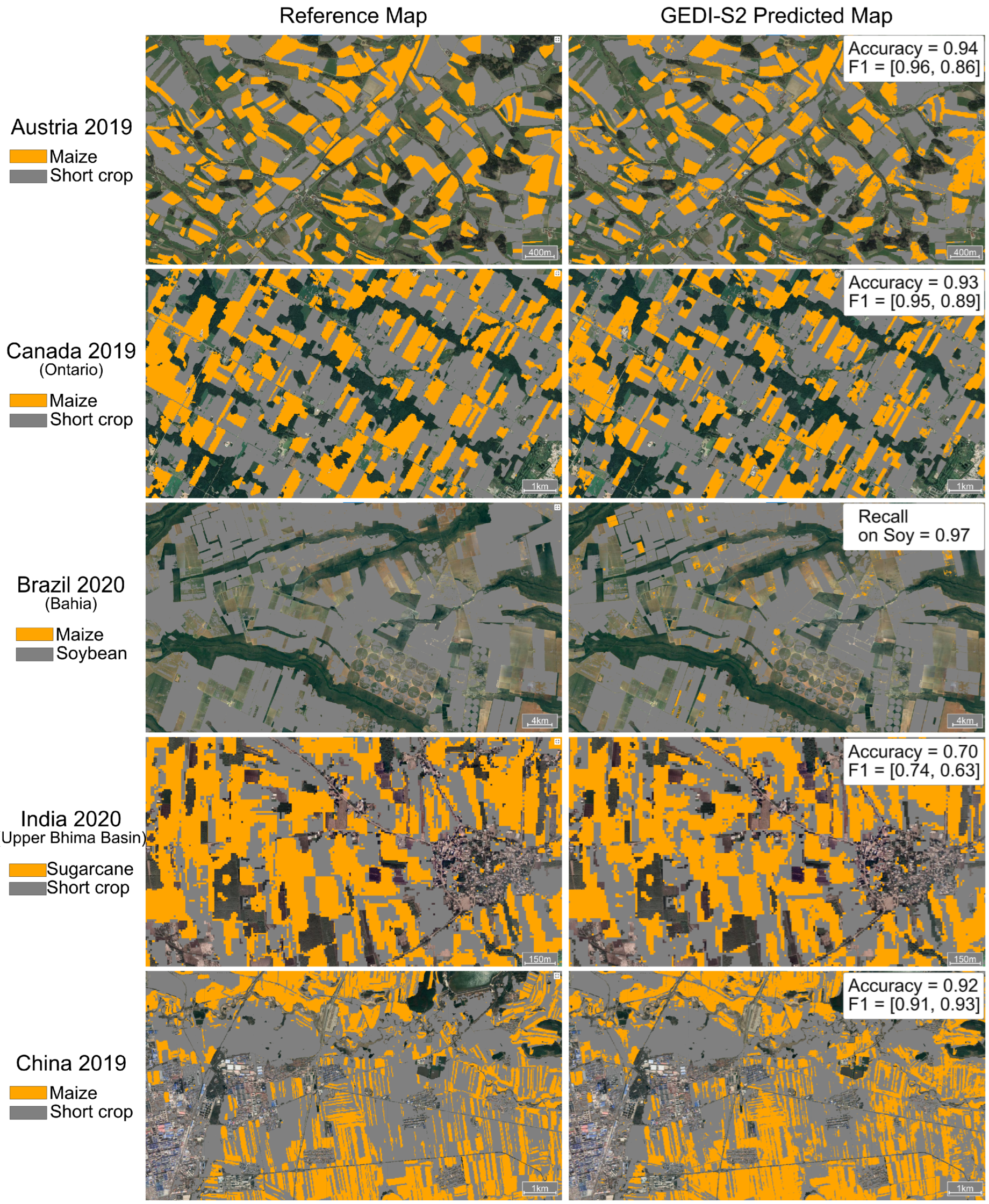
4.3. GEDI-S2 Model Evaluation
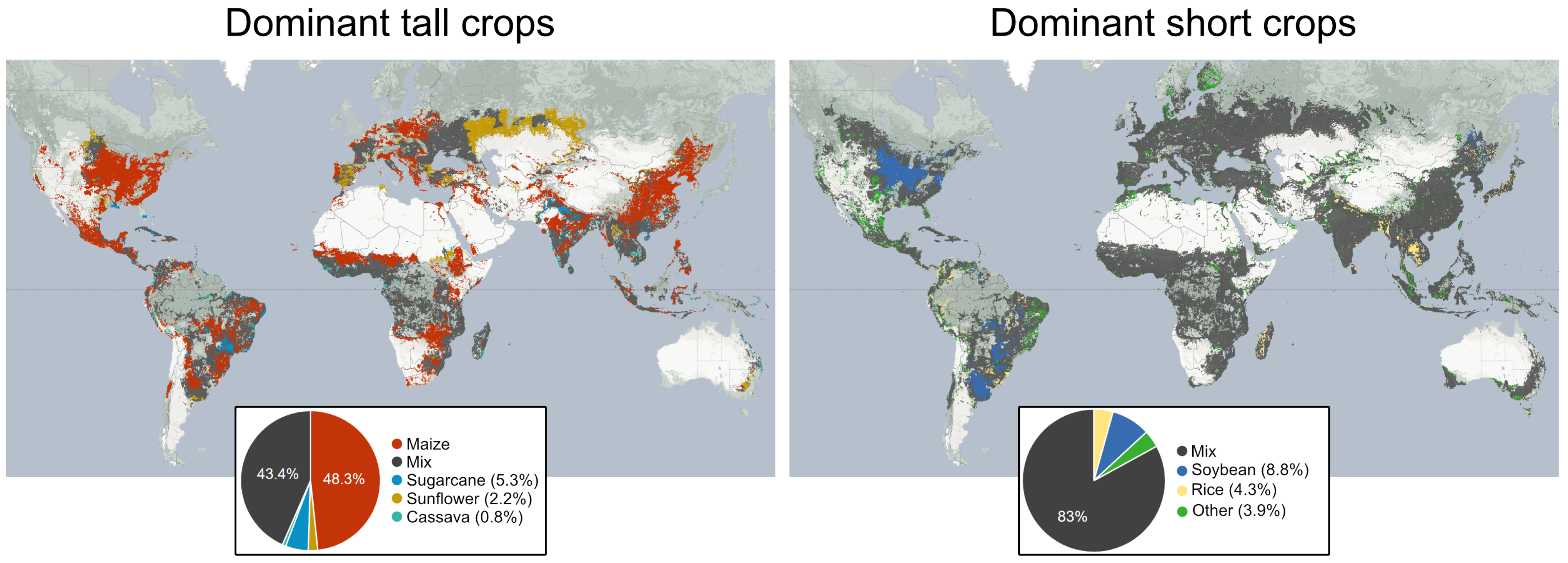
4.4. The Global Distribution of Tall Crops
5. Discussion
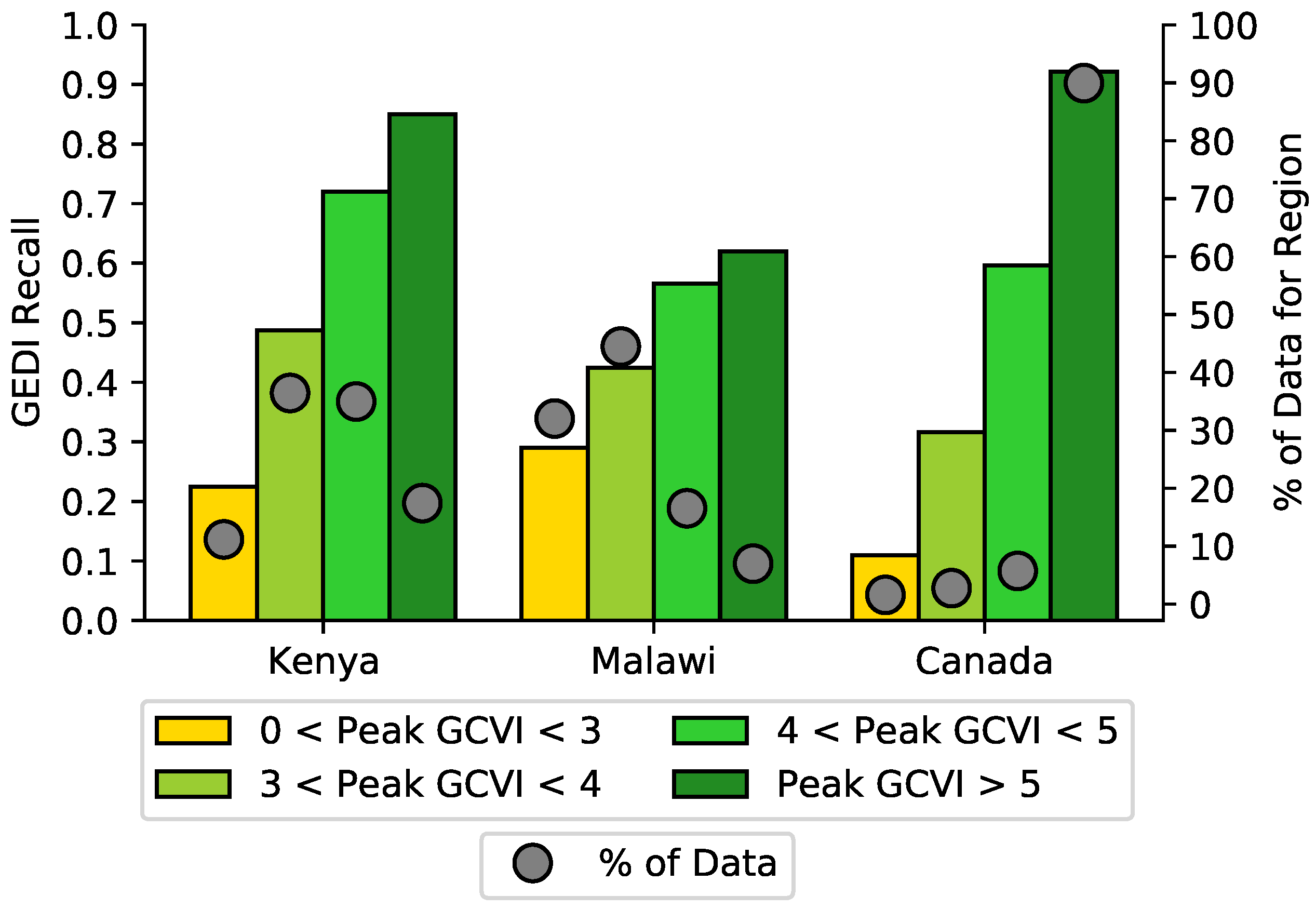
5.1. Sources of Error
5.2. Future Improvements
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bégué, A.; Arvor, D.; Bellon, B.; Betbeder, J.; De Abelleyra, D.; Ferraz, R.P.D.; Lebourgeois, V.; Lelong, C.; Simões, M.; Verón, S.R. Remote sensing and cropping practices: A review. Remote Sens. 2018, 10, 99. [Google Scholar] [CrossRef]
- Kim, K.H.; Doi, Y.; Ramankutty, N.; Iizumi, T. A review of global gridded cropping system data products. Environ. Res. Lett. 2021, 16, 093005. [Google Scholar] [CrossRef]
- Nakalembe, C.; Becker-Reshef, I.; Bonifacio, R.; Hu, G.; Humber, M.L.; Justice, C.J.; Keniston, J.; Mwangi, K.; Rembold, F.; Shukla, S.; et al. A review of satellite-based global agricultural monitoring systems available for Africa. Glob. Food Secur. 2021, 29, 100543. [Google Scholar] [CrossRef]
- Boryan, C.; Yang, Z.; Mueller, R.; Craig, M. Monitoring US agriculture: The US Department of Agriculture, National Agricultural Statistics Service, Cropland Data Layer Program. Geocarto Int. 2011, 26, 341–358. [Google Scholar] [CrossRef]
- Agriculture and Agri-Food Canada. Annual Crop Inventory. Available online: https://open.canada.ca/data/en/dataset/ba2645d5-4458-414d-b196-6303ac06c1c9 (accessed on 1 January 2021).
- Agence de Services et de Paiement. Registre Parcellaire Graphique (RPG): Contours des Parcelles et Îlots Culturaux et Leur Groupe de Cultures Majoritaire. 2019. Available online: https://www.data.gouv.fr/en/datasets/registre-parcellaire-graphique-rpg-contours-des-parcelles-et-ilots-culturaux-et-leur-groupe-de-cultures-majoritaire/ (accessed on 1 January 2021).
- Potapov, P.; Turubanova, S.; Hansen, M.C.; Tyukavina, A.; Zalles, V.; Khan, A.; Song, X.P.; Pickens, A.; Shen, Q.; Cortez, J. Global maps of cropland extent and change show accelerated cropland expansion in the twenty-first century. Nat. Food 2022, 3, 19–28. [Google Scholar] [CrossRef]
- Lobell, D.B.; Asner, G.P.; Ortiz-Monasterio, J.I.; Benning, T.L. Remote sensing of regional crop production in the Yaqui Valley, Mexico: Estimates and uncertainties. Agric. Ecosyst. Environ. 2003, 94, 205–220. [Google Scholar] [CrossRef]
- You, N.; Dong, J.; Huang, J.; Du, G.; Zhang, G.; He, Y.; Yang, T.; Di, Y.; Xiao, X. The 10-m crop type maps in Northeast China during 2017–2019. Sci. Data 2021, 8, 41. [Google Scholar] [CrossRef]
- Han, J.; Zhang, Z.; Luo, Y.; Cao, J.; Zhang, L.; Cheng, F.; Zhuang, H.; Zhang, J. AsiaRiceMap10m: High-resolution annual paddy rice maps for Southeast and Northeast Asia from 2017 to 2019. Earth Syst. Sci. Data Discuss 2021, 211, 1–27. [Google Scholar]
- Song, X.P.; Hansen, M.C.; Potapov, P.; Adusei, B.; Pickering, J.; Adami, M.; Lima, A.; Zalles, V.; Stehman, S.V.; Di Bella, C.M.; et al. Massive soybean expansion in South America since 2000 and implications for conservation. Nat. Sustain. 2021, 4, 784–792. [Google Scholar] [CrossRef]
- Blickensdörfer, L.; Schwieder, M.; Pflugmacher, D.; Nendel, C.; Erasmi, S.; Hostert, P. Mapping of crop types and crop sequences with combined time series of Sentinel-1, Sentinel-2 and Landsat 8 data for Germany. Remote Sens. Environ. 2022, 269, 112831. [Google Scholar] [CrossRef]
- Kluger, D.M.; Wang, S.; Lobell, D.B. Two shifts for crop mapping: Leveraging aggregate crop statistics to improve satellite-based maps in new regions. Remote Sens. Environ. 2021, 262, 112488. [Google Scholar] [CrossRef]
- Lin, C.; Zhong, L.; Song, X.P.; Dong, J.; Lobell, D.B.; Jin, Z. Early-and in-season crop type mapping without current-year ground truth: Generating labels from historical information via a topology-based approach. Remote Sens. Environ. 2022, 274, 112994. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Z.; Zhang, L.; Han, J.; Cao, J.; Zhang, J. Developing High-Resolution Crop Maps for Major Crops in the European Union Based on Transductive Transfer Learning and Limited Ground Data. Remote Sens. 2022, 14, 1809. [Google Scholar] [CrossRef]
- Di Tommaso, S.; Wang, S.; Lobell, D.B. Combining GEDI and Sentinel-2 for wall-to-wall mapping of tall and short crops. Environ. Res. Lett. 2021, 16, 125002. [Google Scholar] [CrossRef]
- Dubayah, R.; Blair, J.B.; Goetz, S.; Fatoyinbo, L.; Hansen, M.; Healey, S.; Hofton, M.; Hurtt, G.; Kellner, J.; Luthcke, S.; et al. The Global Ecosystem Dynamics Investigation: High-resolution laser ranging of the Earth’s forests and topography. Sci. Remote Sens. 2020, 1, 100002. [Google Scholar] [CrossRef]
- International Food Policy Research Institute. Global Spatially-Disaggregated Crop Production Statistics Data for 2010; Version 2.0; Harvard Library: Cambridge, MA, USA, 2019. [Google Scholar] [CrossRef]
- Zanaga, D.; Van De Kerchove, R.; De Keersmaecker, W.; Souverijns, N.; Brockmann, C.; Quast, R.; Wevers, J.; Grosu, A.; Paccini, A.; Vergnaud, S.; et al. ESA WorldCover 10 m 2020 v100; Zenodo: Geneve, Switzerland, 2021. [Google Scholar]
- Karra, K.; Kontgis, C.; Statman-Weil, Z.; Mazzariello, J.C.; Mathis, M.; Brumby, S.P. Global land use/land cover with Sentinel 2 and deep learning. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 4704–4707. [Google Scholar]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Roy, S.; Swetnam, T.; Robitaille, A.; Trochim, E.; Pasquarella, V. Samapriya/Awesome—Gee—Community—Datasets: Community Catalog (1.0.1); Zenodo: Geneve, Switzerland, 2022. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Bailly, J.S.; Frappart, F.; Zribi, M. Analysis of GEDI elevation data accuracy for inland waterbodies altimetry. Remote Sens. 2020, 12, 2714. [Google Scholar] [CrossRef]
- Healey, S.P.; Yang, Z.; Gorelick, N.; Ilyushchenko, S. Highly local model calibration with a new GEDI LiDAR asset on Google Earth Engine reduces landsat forest height signal saturation. Remote Sens. 2020, 12, 2840. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Vina, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef]
- Wang, S.; Azzari, G.; Lobell, D.B. Crop type mapping without field-level labels: Random forest transfer and unsupervised clustering techniques. Remote Sens. Environ. 2019, 222, 303–317. [Google Scholar] [CrossRef]
- Song, X.P.; Huang, W.; Hansen, M.C.; Potapov, P. An evaluation of Landsat, Sentinel-2, Sentinel-1 and MODIS data for crop type mapping. Sci. Remote Sens. 2021, 3, 100018. [Google Scholar] [CrossRef]
- Schneider, M.; Broszeit, A.; Körner, M. Eurocrops: A pan-european dataset for time series crop type classification. arXiv 2021, arXiv:2106.08151. [Google Scholar]
- Agriculture and Agri-Food Canada. Annual Crop Inventory Ground Truth Data. 2021. Available online: https://open.canada.ca/data/en/dataset/503a3113-e435-49f4-850c-d70056788632 (accessed on 15 November 2022).
- Tseng, G.; Zvonkov, I.; Nakalembe, C.; Kerner, H. CropHarvest: A global dataset for crop-type classification. In Proceedings of the Neural Information Processing Systems Track on Datasets and Benchmarks, Paris, France, 8 December 2021; Volume 1. [Google Scholar]
- European Commission, Joint Research Centre (JRC). Kenya AOI. European Commission, Joint Research Centre (JRC) [Dataset]. 2021. Available online: https://data.jrc.ec.europa.eu/dataset/5b6245d3-e561-4f6c-8c09-627888063d11 (accessed on 15 November 2022).
- Wang, S.; Di Tommaso, S.; Faulkner, J.; Friedel, T.; Kennepohl, A.; Strey, R.; Lobell, D.B. Mapping Crop Types in Southeast India with Smartphone Crowdsourcing and Deep Learning. Remote Sens. 2020, 12, 2957. [Google Scholar] [CrossRef]
- Blickensdörfer, L.; Schwieder, M.; Pflugmacher, D.; Nendel, C.; Erasmi, S.; Hostert, P. National-Scale Crop Type Maps for Germany from Combined Time Series of Sentinel-1, Sentinel-2 and Landsat 8 Data (2017, 2018 and 2019); Zenodo: Geneve, Switzerland, 2021. [Google Scholar] [CrossRef]
- Schwieder, M.; Erasmi, S.; Nendel, C.; Hostert, P. National-Scale Crop Type Maps for Germany from Combined Time Series of Sentinel-1, Sentinel-2 and Landsat 8 Data (2020); Zenodo: Geneve, Switzerland, 2022. [Google Scholar] [CrossRef]
- Lee, J.Y.; Wang, S.; Figueroa, A.J.; Strey, R.; Lobell, D.B.; Naylor, R.L.; Gorelick, S.M. Mapping Sugarcane in Central India with Smartphone Crowdsourcing. Remote Sens. 2022, 14, 703. [Google Scholar] [CrossRef]
- Rembold, F.; Meroni, M.; Urbano, F.; Csak, G.; Kerdiles, H.; Perez-Hoyos, A.; Lemoine, G.; Leo, O.; Negre, T. ASAP: A new global early warning system to detect anomaly hot spots of agricultural production for food security analysis. Agric. Syst. 2019, 168, 247–257. [Google Scholar] [CrossRef] [PubMed]
- European Commission, Joint Research Centre (JRC). Global Land Surface Phenology—Number of Growing Seasons [Dataset]. 2018. Available online: http://data.europa.eu/89h/jrc-10112-10008 (accessed on 15 November 2022).
- Farr, T.G.; Rosen, P.A.; Caro, E.; Crippen, R.; Duren, R.; Hensley, S.; Kobrick, M.; Paller, M.; Rodriguez, E.; Roth, L.; et al. The shuttle radar topography mission. Rev. Geophys. 2007, 45, RG2004. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Frappart, F. Comparative Analysis of GEDI’s Elevation Accuracy from the First and Second Data Product Releases over Inland Waterbodies. Remote Sens. 2022, 14, 340. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Alcarde Alvares, C.; Stape, J.L.; Bailly, J.S.; Scolforo, H.F.; Cegatta, I.R.; Zribi, M.; Le Maire, G. Terrain slope effect on forest height and wood volume estimation from GEDI data. Remote Sens. 2021, 13, 2136. [Google Scholar] [CrossRef]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E.; et al. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sens. Environ. 2021, 253, 112165. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for multi-class classification: An overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in RGB imagery using semi-supervised deep learning neural networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Wu, H.; Prasad, S. Semi-supervised deep learning using pseudo labels for hyperspectral image classification. IEEE Trans. Image Process. 2017, 27, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Huang, Y.; Chen, C.; Tang, Z.; Zou, F. Mapping spatiotemporal dynamics of maize in China from 2005 to 2017 through designing leaf moisture based indicator from Normalized Multi-band Drought Index. Comput. Electron. Agric. 2018, 153, 82–93. [Google Scholar] [CrossRef]
- Veloso, A.; Mermoz, S.; Bouvet, A.; Le Toan, T.; Planells, M.; Dejoux, J.F.; Ceschia, E. Understanding the temporal behavior of crops using Sentinel-1 and Sentinel-2-like data for agricultural applications. Remote Sens. Environ. 2017, 199, 415–426. [Google Scholar] [CrossRef]
- Nelson, A.; Setiyono, T.; Rala, A.B.; Quicho, E.D.; Raviz, J.V.; Abonete, P.J.; Maunahan, A.A.; Garcia, C.A.; Bhatti, H.Z.M.; Villano, L.S.; et al. Towards an operational SAR-based rice monitoring system in Asia: Examples from 13 demonstration sites across Asia in the RIICE project. Remote Sens. 2014, 6, 10773–10812. [Google Scholar] [CrossRef]
- Singha, M.; Dong, J.; Zhang, G.; Xiao, X. High resolution paddy rice maps in cloud-prone Bangladesh and Northeast India using Sentinel-1 data. Sci. Data 2019, 6, 26. [Google Scholar] [CrossRef]




| Region | Year | Type | Samples | % Tall Crop | Main Labels |
|---|---|---|---|---|---|
| Labels | |||||
| Austria | 2019 | polygons | 159,528 | 16.1 | maize, pasture, wheat |
| Slovenia | 2019 | polygons | 122,792 | 40 | maize, wheat, barley |
| Germany | 2019 | map | 2278 | 24.1 | maize, wheat, barley |
| Germany | 2020 | map | 2864 | 15.1 | maize, wheat, barley |
| Canada (BC) | 2019 | points | 704 | 31.3 | mixed forage, maize |
| Canada (ON) | 2019 | points | 29,960 | 33.9 | soybean, maize, mixed forage |
| Canada (BC) | 2020 | points | 871 | 39.6 | mixed forage, maize, alfalfa |
| Canada (ON) | 2020 | points | 14,960 | 31.2 | soybean, maize, mixed forage |
| Canada (BC) | 2021 | points | 15,384 | 31.2 | mixed forage, maize, alfalfa |
| US (ND) | 2019 | map | 1847 | 18.8 | soybean, wheat, maize |
| US (ND) | 2020 | map | 1860 | 11.4 | soybean, wheat, maize |
| US (ND) | 2021 | map | 1882 | 22.4 | soybean, wheat, maize |
| US (AL) | 2019 | map | 1085 | 24.1 | cotton, maize, soybean |
| US (AL) | 2020 | map | 1088 | 24.7 | cotton, maize, soybean |
| US (AL) | 2021 | map | 1078 | 25 | cotton, maize, soybean |
| Brazil (BA) | 2020 | map | 1992 | 0 | soybean |
| China | 2019 | map | 2736 | 56.5 | maize, soybean, rice |
| India (U.B.B.) | 2020 | map | 1211 | 50.6 | sugarcane, cotton, rice |
| India (TG) | 2020 | points | 4844 | 4.6 | rice, cotton, peanut, maize |
| India (TG) | 2021 | points | 28,562 | 4.9 | rice, cotton, peanut, maize |
| India (MH) | 2020 | points | 8639 | 27.7 | cotton, maize, rice, sugarcane |
| Malawi | 2021 | polygons | 719 | 31.4 | groundnut, maize, soybean |
| Mali | 2019 | polygons | 73 | 26 | sorghum, millet, maize, rice |
| Kenya | 2021 | points | 1423 | 58.1 | maize, tea, sugarcane |
| Region | Year | S2 Local | GEDI− S2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | F1 | Precision | Recall | K-Score | Accuracy | F1 | Precision | Recall | K-Score | ||
| Austria | 2019 | 0.96 | 0.97, 0.90 | 0.97, 0.89 | 0.97, 0.91 | 0.87 | 0.94 | 0.96, 0.86 | 0.95, 0.89 | 0.97, 0.83 | 0.82 |
| Slovenia | 2019 | 0.9 | 0.92, 0.87 | 0.93, 0.87 | 0.91, 0.88 | 0.79 | 0.88 | 0.90, 0.83 | 0.88, 0.86 | 0.91, 0.80 | 0.73 |
| Germany | 2019 | 0.97 | 0.98, 0.94 | 0.98, 0.92 | 0.97, 0.95 | 0.92 | 0.96 | 0.97, 0.92 | 0.97, 0.95 | 0.98, 0.89 | 0.89 |
| Germany | 2020 | 0.96 | 0.97, 0.85 | 0.97, 0.88 | 0.98, 0.81 | 0.82 | 0.94 | 0.96, 0.83 | 0.99, 0.74 | 0.94, 0.94 | 0.79 |
| Canada (BC) | 2019 | 0.97 | 0.98, 0.94 | 0.97, 0.96 | 0.99, 0.92 | 0.92 | 0.97 | 0.98, 0.92 | 0.96, 0.98 | 0.99, 0.88 | 0.9 |
| Canada (ON) | 2019 | 0.94 | 0.96, 0.90 | 0.94, 0.95 | 0.98, 0.86 | 0.86 | 0.93 | 0.95, 0.89 | 0.93, 0.95 | 0.98, 0.84 | 0.84 |
| Canada (BC) | 2020 | 0.93 | 0.92, 0.88 | 0.90, 0.88 | 0.94, 0.89 | 0.8 | 0.9 | 0.88, 0.85 | 0.83, 0.95 | 0.98, 0.78 | 0.75 |
| Canada (ON) | 2020 | 0.92 | 0.94, 0.85 | 0.93, 0.88 | 0.95, 0.83 | 0.8 | 0.88 | 0.92, 0.80 | 0.91, 0.81 | 0.92, 0.79 | 0.71 |
| Canada (BC) | 2021 | 0.94 | 0.96, 0.90 | 0.94, 0.93 | 0.97, 0.87 | 0.86 | 0.94 | 0.95, 0.90 | 0.96, 0.88 | 0.94, 0.92 | 0.85 |
| US (ND) | 2019 | 0.94 | 0.96, 0.81 | 0.94, 0.93 | 0.99, 0.73 | 0.78 | 0.96 | 0.97, 0.87 | 0.96, 0.94 | 0.99, 0.81 | 0.84 |
| US (ND) | 2020 | 0.95 | 0.97, 0.65 | 0.95, 0.91 | 0.99, 0.52 | 0.63 | 0.95 | 0.97, 0.70 | 0.97, 0.73 | 0.97, 0.68 | 0.68 |
| US (ND) | 2021 | 0.9 | 0.94, 0.70 | 0.90, 0.88 | 0.98, 0.58 | 0.64 | 0.91 | 0.94, 0.75 | 0.92, 0.86 | 0.97, 0.66 | 0.69 |
| US (AL) | 2019 | 0.93 | 0.95, 0.85 | 0.94, 0.91 | 0.97, 0.80 | 0.81 | 0.94 | 0.96, 0.86 | 0.94, 0.95 | 0.99, 0.80 | 0.83 |
| US (AL) | 2020 | 0.95 | 0.97, 0.89 | 0.96, 0.92 | 0.98, 0.86 | 0.85 | 0.87 | 0.91, 0.76 | 0.96, 0.68 | 0.86, 0.88 | 0.68 |
| US (AL) | 2021 | 0.94 | 0.96, 0.88 | 0.95, 0.92 | 0.97, 0.85 | 0.84 | 0.94 | 0.96, 0.88 | 0.94, 0.96 | 0.99, 0.82 | 0.85 |
| Brazil (BA) | 2020 | 0.97 | |||||||||
| China | 2019 | 0.91 | 0.89, 0.92 | 0.88, 0.92 | 0.90, 0.91 | 0.81 | 0.92 | 0.91, 0.93 | 0.90, 0.94 | 0.92, 0.93 | 0.84 |
| India (U.B.B.) | 2020 | 0.87 | 0.85, 0.87 | 0.84, 0.88 | 0.87, 0.87 | 0.73 | 0.7 | 0.74, 0.63 | 0.62, 0.87 | 0.92, 0.50 | 0.41 |
| India (TG) | 2020 | 0.96 | 0.98, 0.16 | 0.96, 0.38 | 0.99, 0.10 | 0.15 | 0.93 | 0.96, 0.01 | 0.96, 0.01 | 0.97, 0.01 | −0.02 |
| India (TG) | 2021 | 0.94 | 0.97, 0.19 | 0.94, 0.69 | 0.99, 0.11 | 0.18 | 0.82 | 0.90, 0.03 | 0.93, 0.02 | 0.87, 0.04 | −0.06 |
| India (MH) | 2020 | 0.84 | 0.89, 0.68 | 0.87, 0.71 | 0.90, 0.65 | 0.57 | 0.6 | 0.74, 0.15 | 0.70, 0.19 | 0.77, 0.13 | −0.1 |
| Malawi | 2021 | 0.72 | 0.82, 0.43 | 0.76, 0.59 | 0.89, 0.36 | 0.27 | 0.7 | 0.78, 0.46 | 0.77, 0.53 | 0.81, 0.43 | 0.26 |
| Mali | 2019 | 0.74 | 0.83, 0.31 | 0.78, 0.30 | 0.91, 0.36 | 0.23 | 0.73 | 0.84, 0.07 | 0.73, 0.18 | 0.99, 0.05 | 0.04 |
| Kenya | 2021 | 0.62 | 0.40, 0.71 | 0.51, 0.65 | 0.35, 0.80 | 0.15 | 0.42 | 0.50, 0.30 | 0.39, 0.55 | 0.74, 0.21 | −0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Tommaso, S.; Wang, S.; Vajipey, V.; Gorelick, N.; Strey, R.; Lobell, D.B. Annual Field-Scale Maps of Tall and Short Crops at the Global Scale Using GEDI and Sentinel-2. Remote Sens. 2023, 15, 4123. https://doi.org/10.3390/rs15174123
Di Tommaso S, Wang S, Vajipey V, Gorelick N, Strey R, Lobell DB. Annual Field-Scale Maps of Tall and Short Crops at the Global Scale Using GEDI and Sentinel-2. Remote Sensing. 2023; 15(17):4123. https://doi.org/10.3390/rs15174123
Chicago/Turabian StyleDi Tommaso, Stefania, Sherrie Wang, Vivek Vajipey, Noel Gorelick, Rob Strey, and David B. Lobell. 2023. "Annual Field-Scale Maps of Tall and Short Crops at the Global Scale Using GEDI and Sentinel-2" Remote Sensing 15, no. 17: 4123. https://doi.org/10.3390/rs15174123
APA StyleDi Tommaso, S., Wang, S., Vajipey, V., Gorelick, N., Strey, R., & Lobell, D. B. (2023). Annual Field-Scale Maps of Tall and Short Crops at the Global Scale Using GEDI and Sentinel-2. Remote Sensing, 15(17), 4123. https://doi.org/10.3390/rs15174123








