Crop-Planting Area Prediction from Multi-Source Gaofen Satellite Images Using a Novel Deep Learning Model: A Case Study of Yangling District
Abstract
:1. Introduction
2. Materials
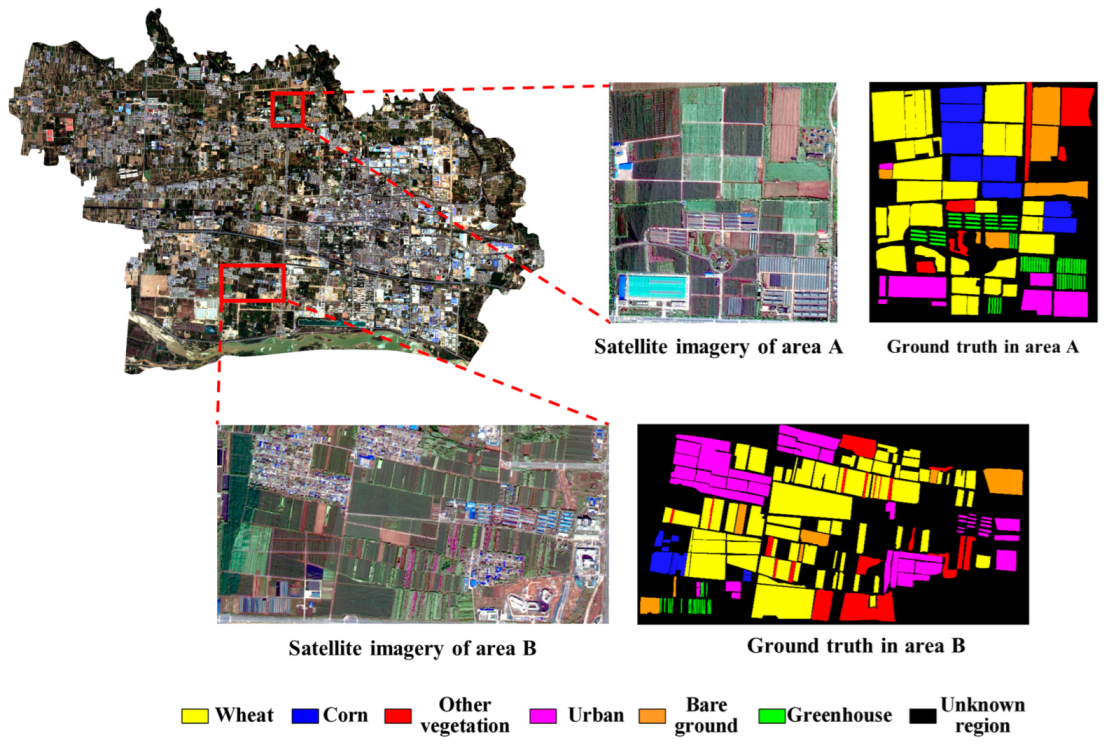
2.1. Study Area
2.2. Dataset Selection and Data Processing
3. Methodology
3.1. Extraction Process of Crop-Planting Area
3.2. Data Dimensionality Reduction Method for Stacked Autoencoder Network
3.3. The Fusion Network Model of Stacked Autoencoder and CNN
4. Experimental Results and Analysis
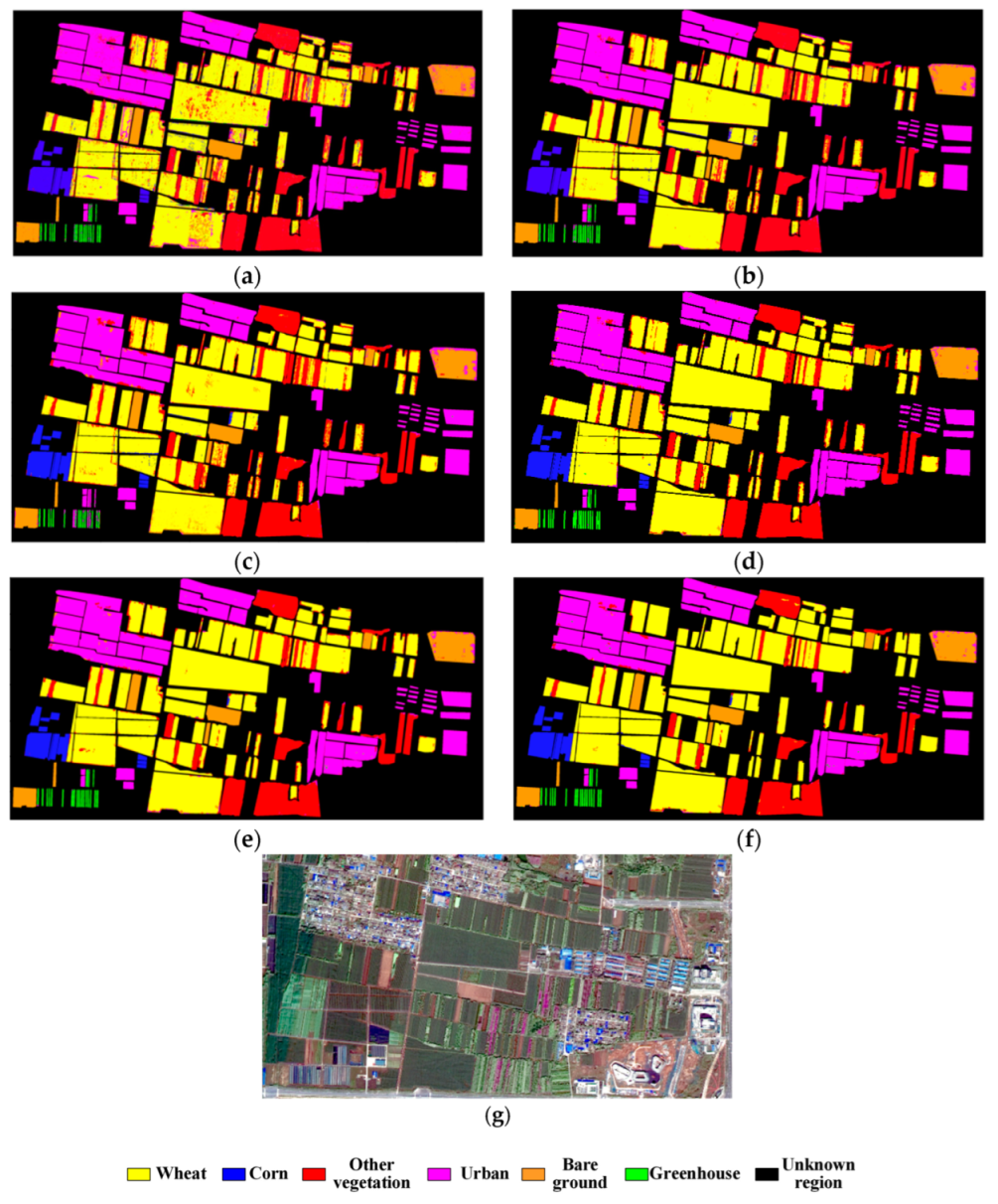
4.1. Condition 1: The Training Samples Belong to the Classification Area
4.2. Condition 2: The Training Samples Do Not Belong to the Classification Area
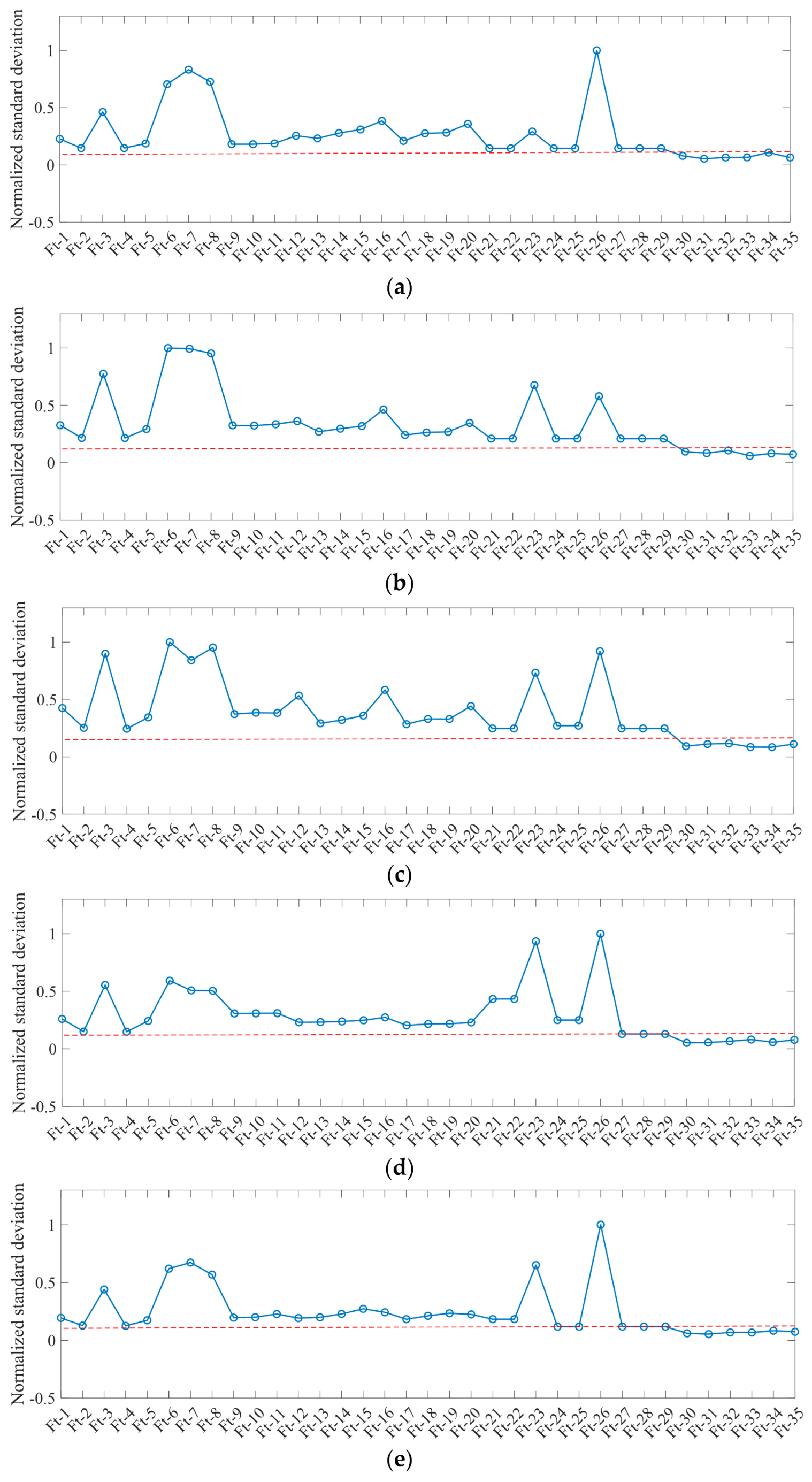
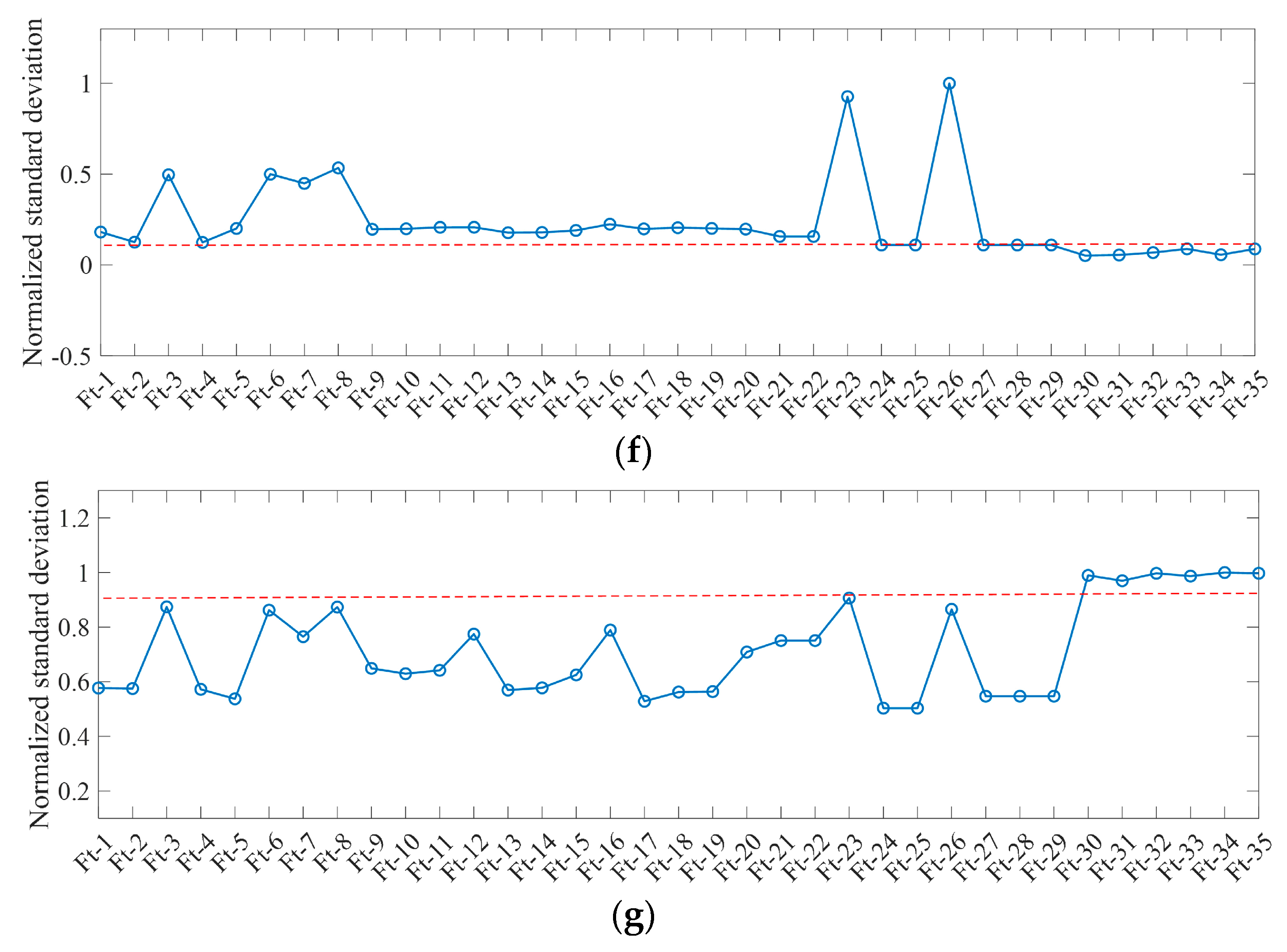
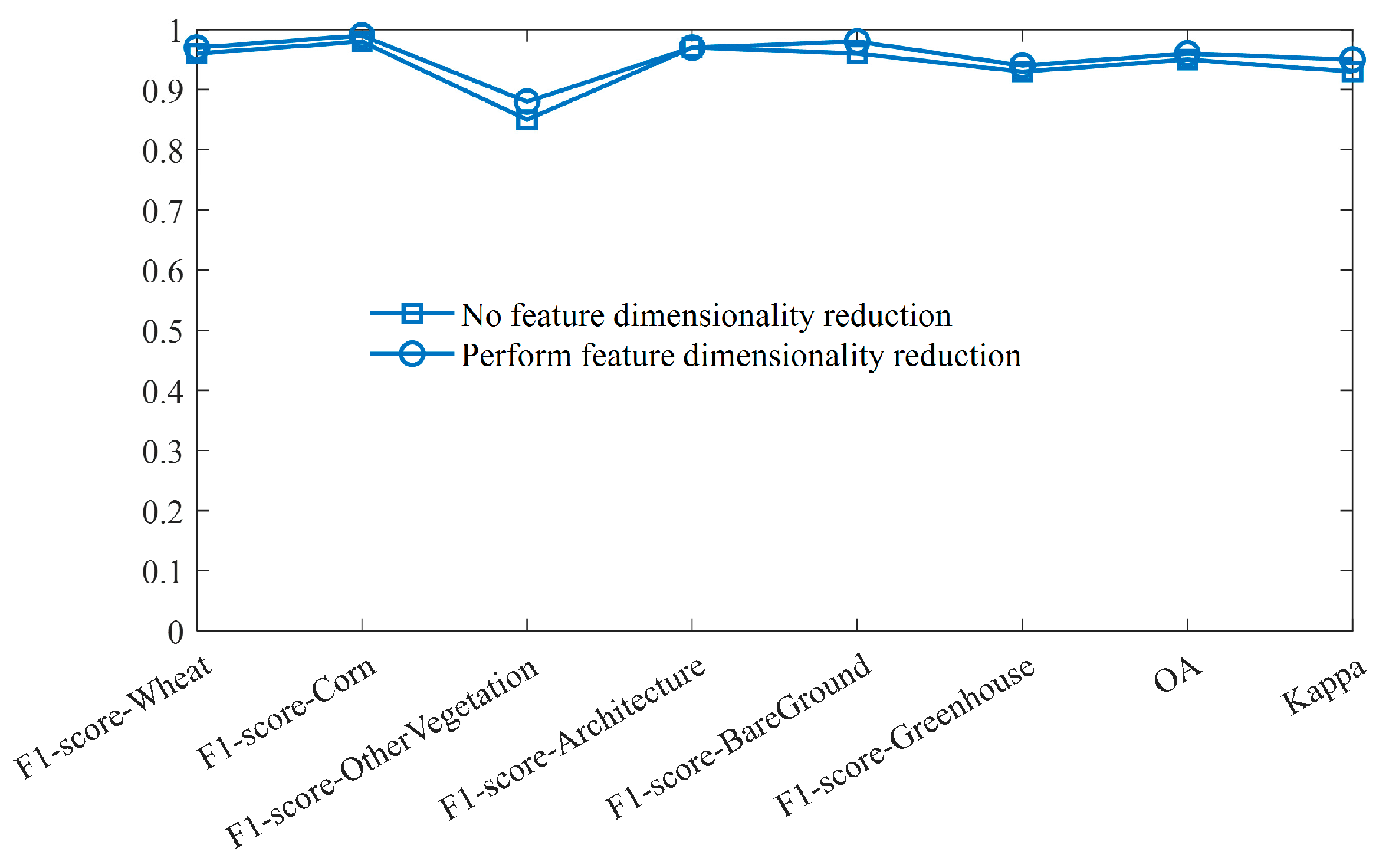
4.3. Performance Analysis of Characteristic Dimensionality Reduction
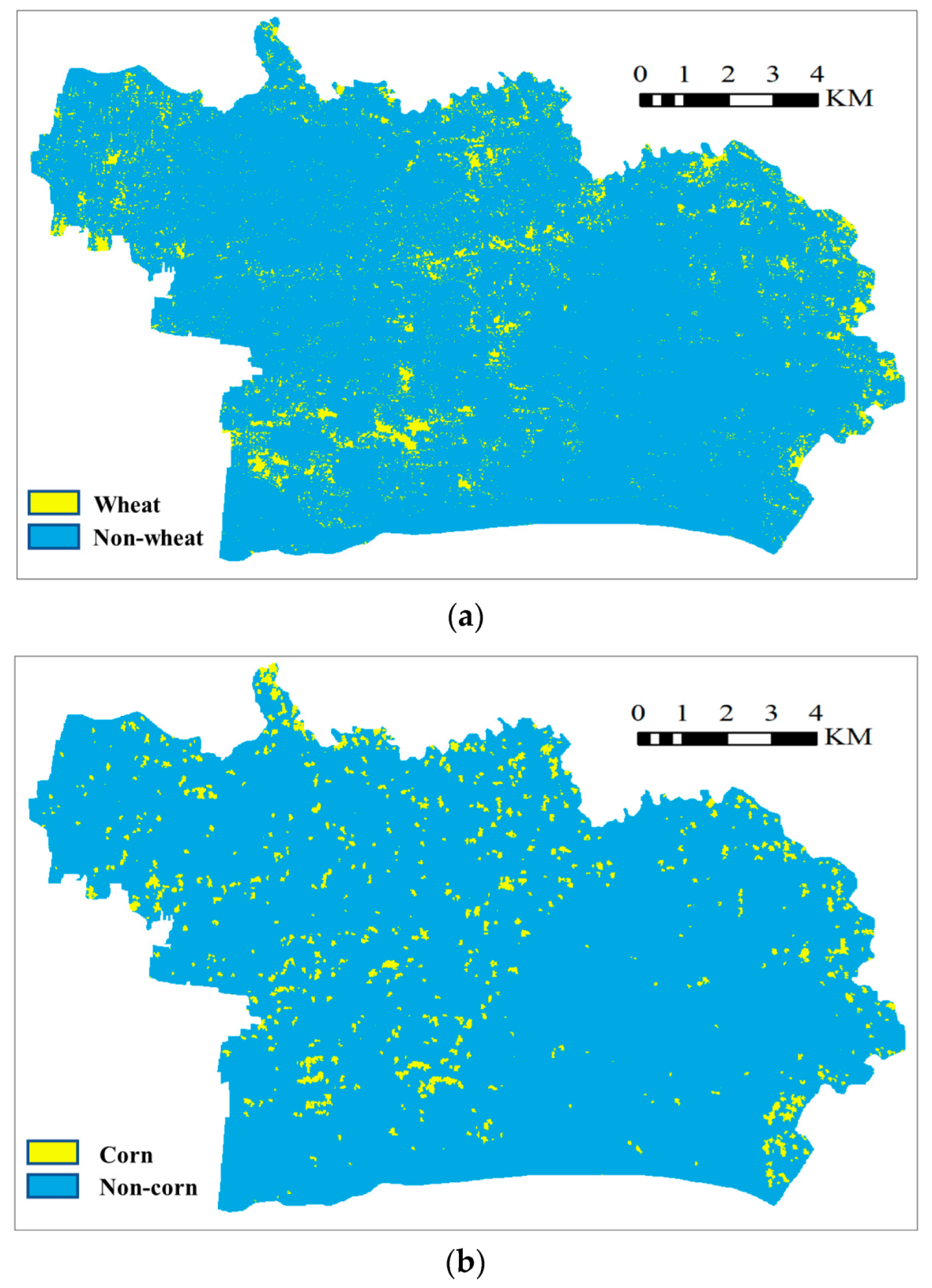
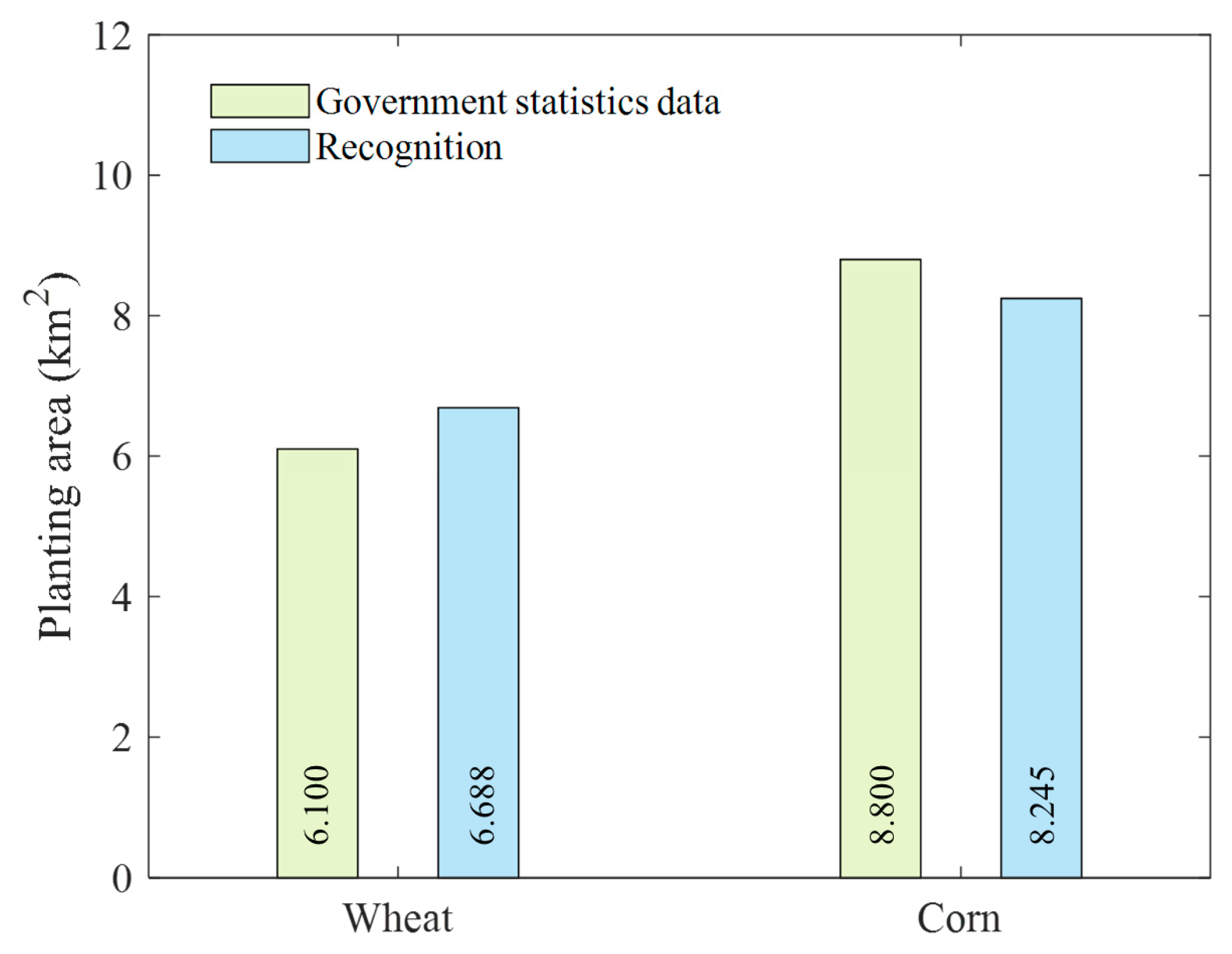
4.4. Extraction of Crop Area in the Entire Region
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, M.-K.; Golzarian, M.R.; Kim, I. A new color index for vegetation segmentation and classification. Precis. Agric. 2021, 22, 179–204. [Google Scholar] [CrossRef]
- Qiu, B.; Jiang, F.; Chen, C.; Tang, Z.; Wu, W.; Berry, J. Phenology-pigment based automated peanut mapping using sentinel-2 images. GIScience Remote Sens. 2021, 58, 1335–1351. [Google Scholar] [CrossRef]
- Bongiovanni, R.; Lowenberg-DeBoer, J. Precision agriculture and sustainability. Precis. Agric. 2004, 5, 359–387. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, C.; Huang, W.; Tang, J.; Li, X.; Zhang, Q. Machine learning-based crop recognition from aerial remote sensing imagery. Front. Earth Sci. 2021, 15, 54–69. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, H.; Wang, L.; Hellwich, O.; Chen, C.; Yang, L.; Liu, T.; Luo, G.; Bao, A.; Chen, X. A joint learning Im-BiLSTM model for incomplete time-series Sentinel-2A data imputation and crop classification. Int. J. Appl. Earth Obs. Geoinf. 2022, 108, 102762. [Google Scholar] [CrossRef]
- Yan, S.; Yao, X.; Zhu, D.; Liu, D.; Zhang, L.; Yu, G.; Gao, B.; Yang, J.; Yun, W. Large-scale crop mapping from multi-source optical satellite imageries using machine learning with discrete grids. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102485. [Google Scholar] [CrossRef]
- Yuan, Y.; Lin, L.; Liu, Q.; Hang, R.; Zhou, Z.-G. SITS-Former: A pre-trained spatio-spectral-temporal representation model for Sentinel-2 time series classification. Int. J. Appl. Earth Obs. Geoinf. 2022, 106, 102651. [Google Scholar] [CrossRef]
- Amorim, M.T.A.; Silvero, N.E.; Bellinaso, H.; Gómez, A.M.R.; Greschuk, L.T.; Campos, L.R.; Demattê, J.A. Impact of soil types on sugarcane development monitored over time by remote sensing. Precis. Agric. 2022, 23, 1532–1552. [Google Scholar] [CrossRef]
- Martinez, J.A.C.; La Rosa, L.E.C.; Feitosa, R.Q.; Sanches, I.D.A.; Happ, P.N. Fully convolutional recurrent networks for multidate crop recognition from multitemporal image sequences. ISPRS J. Photogramm. Remote Sens. 2021, 171, 188–201. [Google Scholar] [CrossRef]
- Zhou, Y.n.; Luo, J.; Feng, L.; Yang, Y.; Chen, Y.; Wu, W. Long-short-term-memory-based crop classification using high-resolution optical images and multi-temporal SAR data. GIScience Remote Sens. 2019, 56, 1170–1191. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, L.; Sun, W.; Zhang, Z.; Zhang, H.; Yang, G.; Meng, X. Exploring the potential of multi-source unsupervised domain adaptation in crop mapping using Sentinel-2 images. GIScience Remote Sens. 2022, 59, 2247–2265. [Google Scholar] [CrossRef]
- Adrian, J.; Sagan, V.; Maimaitijiang, M. Sentinel SAR-optical fusion for crop type mapping using deep learning and Google Earth Engine. ISPRS J. Photogramm. Remote Sens. 2021, 175, 215–235. [Google Scholar] [CrossRef]
- Jayakumari, R.; Nidamanuri, R.R.; Ramiya, A.M. Object-level classification of vegetable crops in 3D LiDAR point cloud using deep learning convolutional neural networks. Precis. Agric. 2021, 22, 1617–1633. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, X.; Ding, C.; Liu, S.; Wu, C.; Wu, L. Mapping rice paddies in complex landscapes with convolutional neural networks and phenological metrics. GIScience Remote Sens. 2020, 57, 37–48. [Google Scholar] [CrossRef]
- Elango, S.; Haldar, D.; Danodia, A. Discrimination of maize crop in a mixed Kharif crop scenario with synergism of multiparametric SAR and optical data. Geocarto Int. 2022, 37, 5307–5326. [Google Scholar] [CrossRef]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Gumma, M.K.; Thenkabail, P.S.; Teluguntla, P.G.; Oliphant, A.; Xiong, J.; Giri, C.; Pyla, V.; Dixit, S.; Whitbread, A.M. Agricultural cropland extent and areas of South Asia derived using Landsat satellite 30-m time-series big-data using random forest machine learning algorithms on the Google Earth Engine cloud. GIScience Remote Sens. 2020, 57, 302–322. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhao, Z.; Yin, C. Fine crop classification based on UAV hyperspectral images and random forest. ISPRS Int. J. Geo-Inf. 2022, 11, 252. [Google Scholar] [CrossRef]
- Maponya, M.G.; Van Niekerk, A.; Mashimbye, Z.E. Pre-harvest classification of crop types using a Sentinel-2 time-series and machine learning. Comput. Electron. Agric. 2020, 169, 105164. [Google Scholar] [CrossRef]
- Löw, F.; Conrad, C.; Michel, U. Decision fusion and non-parametric classifiers for land use mapping using multi-temporal RapidEye data. ISPRS J. Photogramm. Remote Sens. 2015, 108, 191–204. [Google Scholar] [CrossRef]
- Kwak, G.-H.; Park, C.-W.; Lee, K.-D.; Na, S.-I.; Ahn, H.-Y.; Park, N.-W. Potential of hybrid CNN-RF model for early crop mapping with limited input data. Remote Sens. 2021, 13, 1629. [Google Scholar] [CrossRef]
- Yuan, Y.; Lin, L.; Zhou, Z.-G.; Jiang, H.; Liu, Q. Bridging optical and SAR satellite image time series via contrastive feature extraction for crop classification. ISPRS J. Photogramm. Remote Sens. 2023, 195, 222–232. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Revelo, M.Y.; Guachi-Guachi, L.; Gómez-Mendoza, J.B.; Revelo-Fuelagán, J.; Peluffo-Ordóñez, D.H. Enhanced convolutional-neural-network architecture for crop classification. Appl. Sci. 2021, 11, 4292. [Google Scholar] [CrossRef]
- Seydi, S.T.; Amani, M.; Ghorbanian, A. A dual attention convolutional neural network for crop classification using time-series Sentinel-2 imagery. Remote Sens. 2022, 14, 498. [Google Scholar] [CrossRef]
- Sun, H.; Zheng, X.; Lu, X. A supervised segmentation network for hyperspectral image classification. IEEE Trans. Image Process. 2021, 30, 2810–2825. [Google Scholar] [CrossRef]
- Yang, W.; Nigon, T.; Hao, Z.; Paiao, G.D.; Fernández, F.G.; Mulla, D.; Yang, C. Estimation of corn yield based on hyperspectral imagery and convolutional neural network. Comput. Electron. Agric. 2021, 184, 106092. [Google Scholar] [CrossRef]
- Li, B.; Wang, K.C.; Zhang, A.; Yang, E.; Wang, G. Automatic classification of pavement crack using deep convolutional neural network. Int. J. Pavement Eng. 2020, 21, 457–463. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Automation in agriculture by machine and deep learning techniques: A review of recent developments. Precis. Agric. 2021, 22, 2053–2091. [Google Scholar] [CrossRef]
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H. Deep convolutional neural networks for hyperspectral image classification. J. Sens. 2015, 2015, 258619. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Wang, J.; Xie, Q.; Baz, A.A.; Huang, X.; Shang, J.; He, Y. Synergistic use of multi-temporal RADARSAT-2 and VENµS data for crop classification based on 1D convolutional neural network. Remote Sens. 2020, 12, 832. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, W.; Zhang, L. Seamless and automated rapeseed mapping for large cloudy regions using time-series optical satellite imagery. ISPRS J. Photogramm. Remote Sens. 2022, 184, 45–62. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, Z.; Zhang, C.; Wei, S.; Lu, M.; Duan, Y. Learning discriminative spatiotemporal features for precise crop classification from multi-temporal satellite images. Int. J. Remote Sens. 2020, 41, 3162–3174. [Google Scholar] [CrossRef]
- Zhu, J.; Pan, Z.; Wang, H.; Huang, P.; Sun, J.; Qin, F.; Liu, Z. An improved multi-temporal and multi-feature tea plantation identification method using Sentinel-2 imagery. Sensors 2019, 19, 2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Gu, L.; Li, X.; Jiang, T.; Ren, R. Crop classification method based on optimal feature selection and hybrid CNN-RF networks for multi-temporal remote sensing imagery. Remote Sens. 2020, 12, 3119. [Google Scholar] [CrossRef]
- Kyere, I.; Astor, T.; Graß, R.; Wachendorf, M. Agricultural crop discrimination in a heterogeneous low-mountain range region based on multi-temporal and multi-sensor satellite data. Comput. Electron. Agric. 2020, 179, 105864. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, H.; Du, W.; Lyu, X. Crop Identification Based on Multi-Temporal Active and Passive Remote Sensing Images. ISPRS Int. J. Geo-Inf. 2022, 11, 388. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Quan, T.; Peng, Q.; Yu, X.; Liu, L. A model-based collaborate filtering algorithm based on stacked AutoEncoder. Neural Comput. Appl. 2022, 34, 2503–2511. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Wang, Z.; Wang, Z.J.; Ward, R.K.; Wang, X. Deep learning for pixel-level image fusion: Recent advances and future prospects. Inf. Fusion 2018, 42, 158–173. [Google Scholar] [CrossRef]
- Lv, Z.; Qiao, L.; Li, J.; Song, H. Deep-learning-enabled security issues in the internet of things. IEEE Internet Things J. 2020, 8, 9531–9538. [Google Scholar] [CrossRef]
- Chen, L.; Letu, H.; Fan, M.; Shang, H.; Tao, J.; Wu, L.; Zhang, Y.; Yu, C.; Gu, J.; Zhang, N. An Introduction to the Chinese High-Resolution Earth Observation System: Gaofen-1~7 Civilian Satellites. J. Remote Sens. 2022, 2022, 9769536. [Google Scholar] [CrossRef]
- Shi, L.; Huang, X.; Zhong, T.; Taubenböck, H. Mapping plastic greenhouses using spectral metrics derived from GaoFen-2 satellite data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 13, 49–59. [Google Scholar] [CrossRef]
- Tong, X.; Zhao, W.; Xing, J.; Fu, W. Status and development of China high-resolution earth observation system and application. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3738–3741. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. In IEEE Transactions on Systems, Man, and Cybernetics; IEEE: New York, NY, USA, 1973; pp. 610–621. [Google Scholar]
- Tian, F.; Fensholt, R.; Verbesselt, J.; Grogan, K.; Horion, S.; Wang, Y. Evaluating temporal consistency of long-term global NDVI datasets for trend analysis. Remote Sens. Environ. 2015, 163, 326–340. [Google Scholar] [CrossRef]
- Yan, K.; Gao, S.; Chi, H.; Qi, J.; Song, W.; Tong, Y.; Mu, X.; Yan, G. Evaluation of the vegetation-index-based dimidiate pixel model for fractional vegetation cover estimation. IEEE Trans. Geosci. Remote Sens. 2021, 60, 21382349. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Fang, P.; Zhang, X.; Wei, P.; Wang, Y.; Zhang, H.; Liu, F.; Zhao, J. The classification performance and mechanism of machine learning algorithms in winter wheat mapping using Sentinel-2 10 m resolution imagery. Appl. Sci. 2020, 10, 5075. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; Van Den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef] [PubMed]








| Satellite | Spatial Resolution/m | Band | Acquisition Date |
|---|---|---|---|
| GF-1 | 2 | Panchromatic | 18 May 2021 |
| 8 | Multispectral | ||
| GF-2 | 0.8 | Panchromatic | 4 May 2021 |
| 4 | Multispectral | ||
| GF-6 | 2 | Panchromatic | 14 April 2021 |
| 8 | Multispectral |
| Feature Name | Identifier of Feature |
|---|---|
| GLCM texture (mean, variance, homogeneity, contrast, dissimilarity, entropy, angle second-order matrix, correlation) from GF-2 | Ft-1~Ft-8 |
| Multispectral features (red, green, blue, near-infrared) from GF-2 | Ft-9~Ft-12 |
| Multispectral features (red, green, blue, near-infrared) from GF-1 | Ft-13~Ft-16 |
| Multispectral features (red, green, blue, near-infrared) from GF-6 | Ft-17~Ft-20 |
| NDVI, RVI, ARI2 from GF-2 | Ft-21~Ft-23 |
| NDVI, RVI, ARI2 from GF-1 | Ft-24~Ft-26 |
| NDVI, RVI, ARI2 from GF-6 | Ft-27~Ft-29 |
| Method | F1-Score | OA (%) | Kappa | |||||
|---|---|---|---|---|---|---|---|---|
| Wheat | Corn | Other Vegetation | Urban | Bare Ground | Greenhouse | |||
| DT | 0.92 | 0.95 | 0.74 | 0.91 | 0.95 | 0.89 | 90.62 | 0.83 |
| RF | 0.94 | 0.98 | 0.79 | 0.94 | 0.99 | 0.95 | 93.88 | 0.88 |
| SVM | 0.94 | 0.97 | 0.79 | 0.91 | 0.97 | 0.88 | 92.89 | 0.87 |
| HICCNN | 0.98 | 0.99 | 0.92 | 0.98 | 0.97 | 0.96 | 97.36 | 0.95 |
| CSCNN | 0.99 | 0.98 | 0.94 | 0.98 | 0.98 | 0.96 | 97.47 | 0.96 |
| FSACNN | 0.99 | 0.99 | 0.95 | 0.98 | 0.99 | 0.98 | 98.57 | 0.97 |
| Method | F1-Score | OA (%) | Kappa | |||||
|---|---|---|---|---|---|---|---|---|
| Wheat | Corn | Other Vegetation | Urban | Bare Ground | Greenhouse | |||
| DT | 0.89 | 0.83 | 0.76 | 0.94 | 0.89 | 0.78 | 88.04 | 0.74 |
| RF | 0.93 | 0.92 | 0.79 | 0.97 | 0.94 | 0.94 | 92.02 | 0.80 |
| SVM | 0.93 | 0.93 | 0.79 | 0.96 | 0.89 | 0.76 | 91.23 | 0.79 |
| HICCNN | 0.96 | 0.99 | 0.86 | 0.98 | 0.96 | 0.98 | 95.26 | 0.88 |
| CSCNN | 0.98 | 0.98 | 0.92 | 0.96 | 0.96 | 0.97 | 95.88 | 0.92 |
| FSACNN | 0.98 | 0.98 | 0.94 | 0.98 | 0.96 | 0.97 | 97.76 | 0.94 |
| Method | F1-Score | OA (%) | Kappa | |||||
|---|---|---|---|---|---|---|---|---|
| Wheat | Corn | Other Vegetation | Urban | Bare Ground | Greenhouse | |||
| DT | 0.86 | 0.73 | 0.34 | 0.66 | 0.74 | 0.72 | 72.92 | 0.62 |
| RF | 0.90 | 0.75 | 0.51 | 0.80 | 0.88 | 0.66 | 78.52 | 0.71 |
| SVM | 0.92 | 0.77 | 0.59 | 0.40 | 0.77 | 0.53 | 78.06 | 0.70 |
| HICCNN | 0.91 | 0.82 | 0.50 | 0.73 | 0.79 | 0.69 | 79.01 | 0.72 |
| CSCNN | 0.88 | 0.90 | 0.55 | 0.79 | 0.89 | 0.71 | 83.26 | 0.79 |
| FSACNN | 0.97 | 0.87 | 0.57 | 0.80 | 0.88 | 0.75 | 87.15 | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, X.; Guo, J.; Bai, J.; Geng, H.; Wang, H. Crop-Planting Area Prediction from Multi-Source Gaofen Satellite Images Using a Novel Deep Learning Model: A Case Study of Yangling District. Remote Sens. 2023, 15, 3792. https://doi.org/10.3390/rs15153792
Kuang X, Guo J, Bai J, Geng H, Wang H. Crop-Planting Area Prediction from Multi-Source Gaofen Satellite Images Using a Novel Deep Learning Model: A Case Study of Yangling District. Remote Sensing. 2023; 15(15):3792. https://doi.org/10.3390/rs15153792
Chicago/Turabian StyleKuang, Xiaofei, Jiao Guo, Jingyuan Bai, Hongsuo Geng, and Hui Wang. 2023. "Crop-Planting Area Prediction from Multi-Source Gaofen Satellite Images Using a Novel Deep Learning Model: A Case Study of Yangling District" Remote Sensing 15, no. 15: 3792. https://doi.org/10.3390/rs15153792
APA StyleKuang, X., Guo, J., Bai, J., Geng, H., & Wang, H. (2023). Crop-Planting Area Prediction from Multi-Source Gaofen Satellite Images Using a Novel Deep Learning Model: A Case Study of Yangling District. Remote Sensing, 15(15), 3792. https://doi.org/10.3390/rs15153792





