UAV-Based Computer Vision System for Orchard Apple Tree Detection and Health Assessment
Abstract
1. Introduction
1.1. Tree Detection
1.2. Tree Health Classification
- We propose a novel framework for automatic apple orchard tree detection and health assessment from UAV images. The proposed framework could be generalized for a wide range of other UAV applications that involve a detection/classification process.
- We adopt a hard negative mining approach for tree detection to improve the performance of the detection model.
- We formulate the tree health assessment problem as a supervised classification task based on vegetation indices calculated from multi-band images.
- We present an extensive experimental analysis covering various aspects of the proposed framework. Our analysis includes an ablation study demonstrating the importance of the HNM technique for tree detection, an exploration of several classification methods for health assessment, and a feature importance analysis within the context of health classification.
2. Materials and Methods
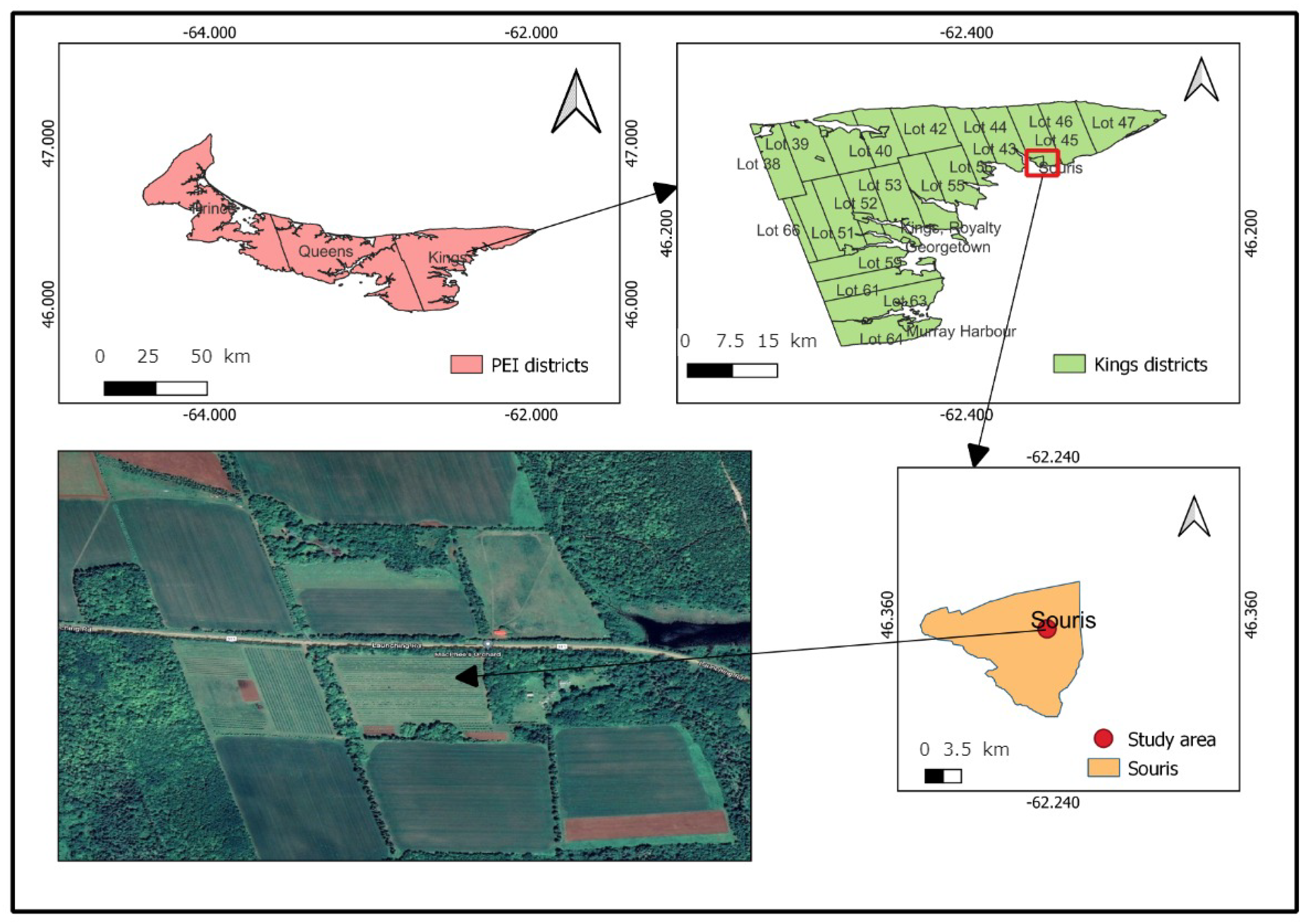
2.1. Study Area
2.2. Dataset Construction
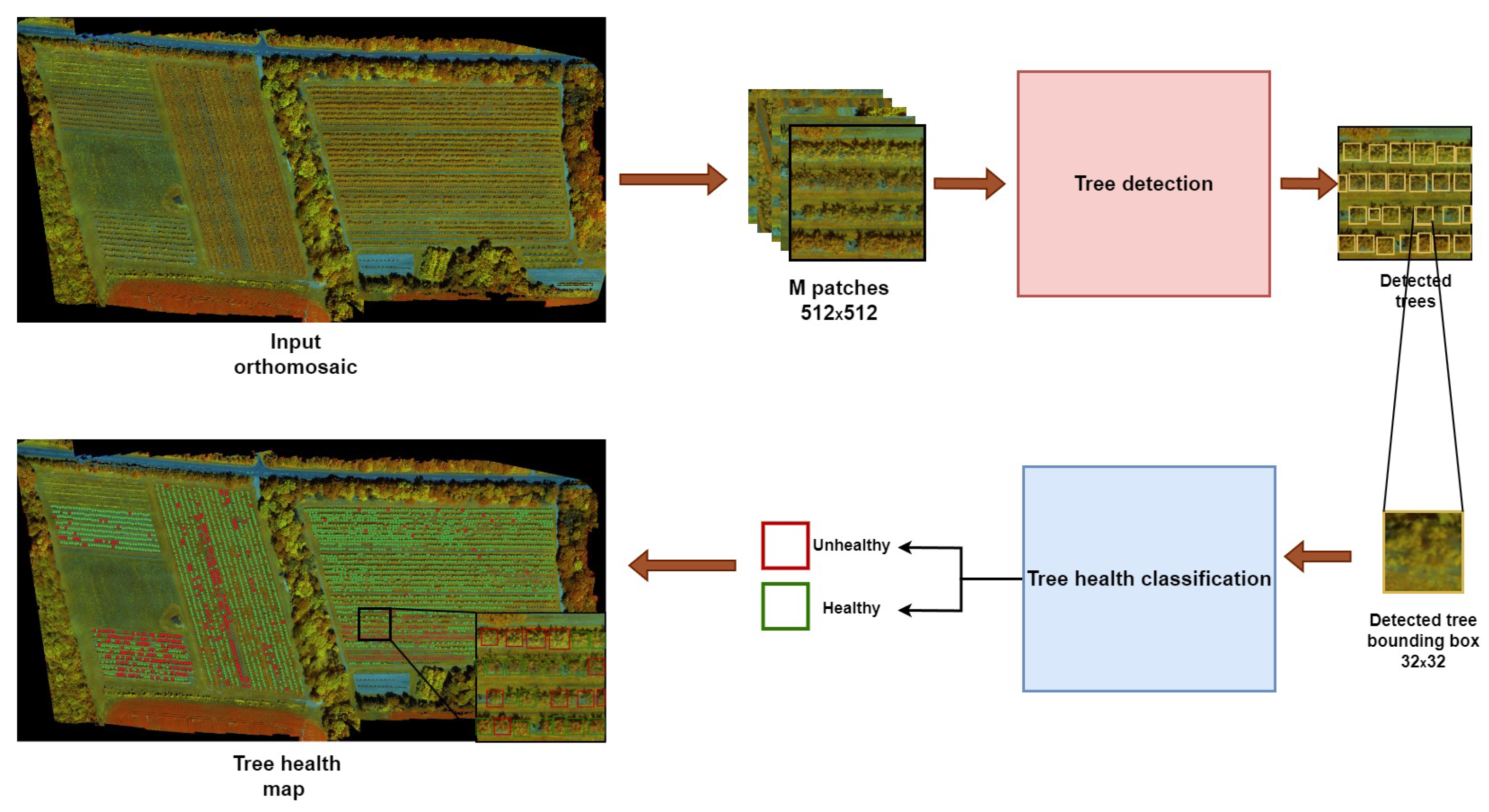
2.3. Proposed Framework
2.3.1. Tree Detection Stage
2.3.2. Tree Health Assessment
2.4. Implementation
2.5. Evaluation Metrics
- Precision (Equation (1)) is the percentage of correct detections among all the detected trees.
- Recall (Equation (2)) is the percentage of correctly detected trees over the total number of trees in the ground truth.
- (Equation (3)) is the harmonic average of precision and recall.
- Precision (Equation (5)) is defined as the ratio of correct classifications for a given class to the total number of classifications made for that class.
- Recall (Equation (6)) is defined as the ratio of correct classifications for a given class to the total number of instances that actually belong to that class.
- is the harmonic average of and of a given class.
- Accuracy (Equation (7)) is defined as the ratio of the correct classifications to the total number of tree instances classified.
3. Results
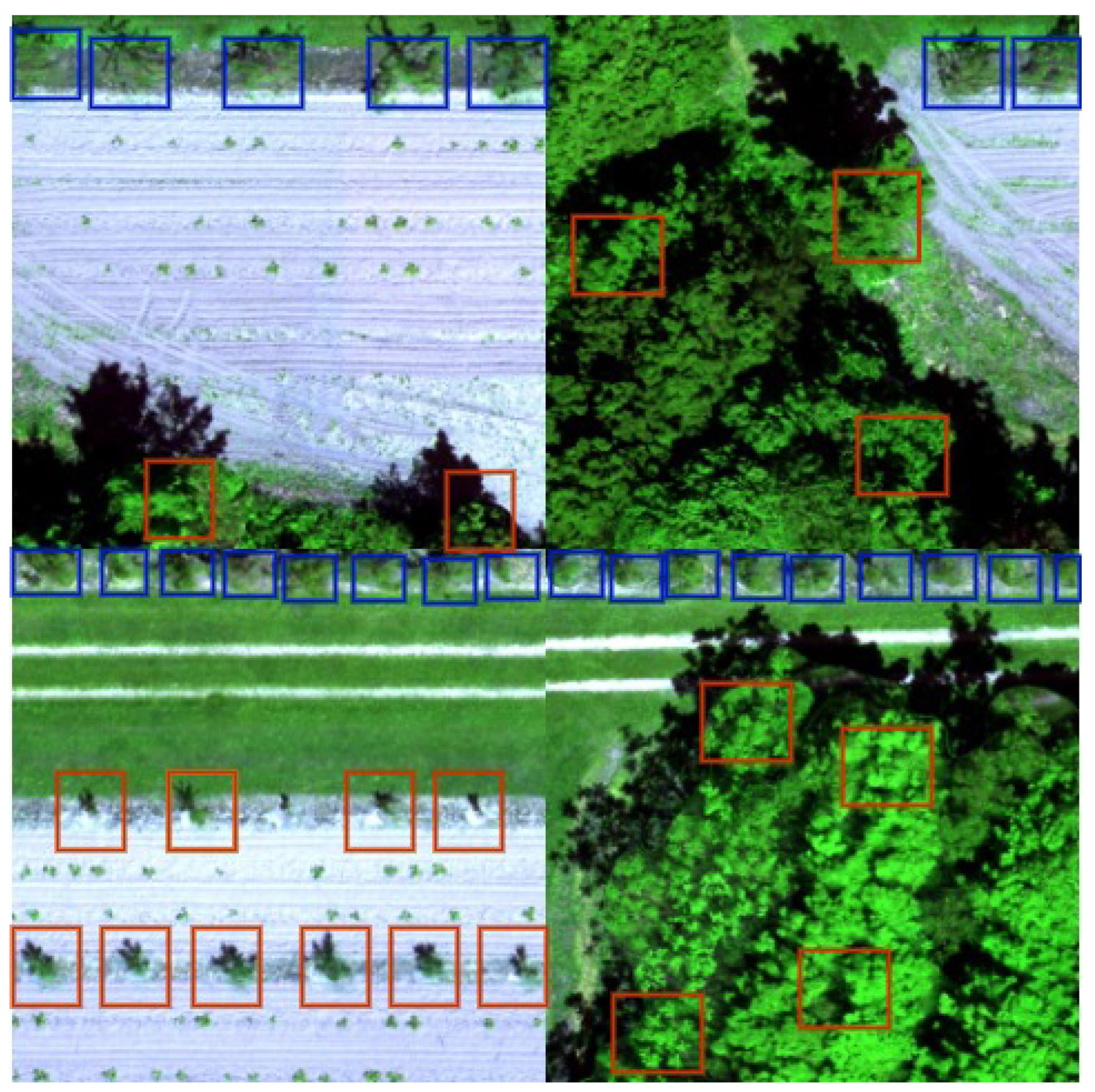
3.1. Tree Detection
3.2. Health Classification
4. Discussion
4.1. Tree Detection
4.2. Trees Health Assessment
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. Brief Description of the Explored Object Detectors Used as Baseline Models in the Hard Negative Mining Approach to Address Tree Detection, That Is, YOLO and DeepForest
Appendix B. Brief Description of the Explored Machine Learning Classifiers for Tree Health Classification
References
- Kim, J.; Kim, S.; Ju, C.; Son, H.I. Unmanned aerial vehicles in agriculture: A review of perspective of platform, control, and applications. IEEE Access 2019, 7, 105100–105115. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the use of unmanned aerial vehicles and imaging sensors for monitoring and assessing plant stresses. Drones 2019, 3, 40. [Google Scholar] [CrossRef]
- Costa, F.G.; Ueyama, J.; Braun, T.; Pessin, G.; Osório, F.S.; Vargas, P.A. The use of unmanned aerial vehicles and wireless sensor network in agricultural applications. In Proceedings of the 2012 IEEE International Geoscience and Remote Sensing Symposium, Munich, Germany, 22–27 July 2012; pp. 5045–5048. [Google Scholar]
- Urbahs, A.; Jonaite, I. Features of the use of unmanned aerial vehicles for agriculture applications. Aviation 2013, 17, 170–175. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Bouachir, W.; Ihou, K.E.; Gueziri, H.E.; Bouguila, N.; Bélanger, N. Computer Vision System for Automatic Counting of Planting Microsites Using UAV Imagery. IEEE Access 2019, 7, 82491–82500. [Google Scholar] [CrossRef]
- Haddadi, A.; Leblon, B.; Patterson, G. Detecting and Counting Orchard Trees on Unmanned Aerial Vehicle (UAV)-Based Images Using Entropy and Ndvi Features. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, 43, 1211–1215. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, G.; Li, M.; Han, S. Automated classification analysis of geological structures based on images data and deep learning model. Appl. Sci. 2018, 8, 2493. [Google Scholar] [CrossRef]
- Geng, L.; Zhang, Y.; Wang, P.; Wang, J.J.; Fuh, J.Y.; Teo, S. UAV surveillance mission planning with gimbaled sensors. In Proceedings of the 11th IEEE International Conference on Control & Automation (ICCA), Taichung, Taiwan, 21 November 2014; pp. 320–325. [Google Scholar]
- Kanistras, K.; Martins, G.; Rutherford, M.J.; Valavanis, K.P. A survey of unmanned aerial vehicles (UAVs) for traffic monitoring. In Proceedings of the 2013 International Conference on Unmanned Aircraft Systems (ICUAS), Atlanta, GA, USA, 28–31 May 2013; pp. 221–234. [Google Scholar]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Sedaghat, A.; Mokhtarzade, M.; Ebadi, H. Uniform robust scale-invariant feature matching for optical remote sensing images. IEEE Trans. Geosci. Remote Sens. 2011, 49, 4516–4527. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the CVPR, San Diego, CA, USA, 20–26 June 2005; pp. 886–893. [Google Scholar]
- Shao, W.; Yang, W.; Liu, G.; Liu, J. Car detection from high-resolution aerial imagery using multiple features. In Proceedings of the 2012 IEEE International Geoscience and Remote Sensing Symposium, Munich, Germany, 22–27 July 2012; pp. 4379–4382. [Google Scholar]
- Maillard, P.; Gomes, M.F. Detection and counting of orchard trees from vhr images using a geometrical-optical model and marked template matching. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 3, 75. [Google Scholar] [CrossRef]
- Malek, S.; Bazi, Y.; Alajlan, N.; AlHichri, H.; Melgani, F. Efficient framework for palm tree detection in UAV images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 4692–4703. [Google Scholar] [CrossRef]
- Bazi, Y.; Malek, S.; Alajlan, N.A.; Alhichri, H.S. An automatic approach for palm tree counting in UAV images. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 537–540. [Google Scholar]
- Wang, Y.; Zhu, X.; Wu, B. Automatic detection of individual oil palm trees from UAV images using HOG features and an SVM classifier. Int. J. Remote Sens. 2019, 40, 7356–7370. [Google Scholar] [CrossRef]
- Manandhar, A.; Hoegner, L.; Stilla, U. Palm tree detection using circular autocorrelation of polar shape matrix. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 3, 465. [Google Scholar] [CrossRef]
- Mansoori, S.A.; Kunhu, A.; Ahmad, H.A. Automatic palm trees detection from multispectral UAV data using normalized difference vegetation index and circular Hough transform. Remote Sens. 2018, 10792, 11–19. [Google Scholar]
- Hassaan, O.; Nasir, A.K.; Roth, H.; Khan, M.F. Precision forestry: Trees counting in urban areas using visible imagery based on an unmanned aerial vehicle. IFAC-PapersOnLine 2016, 49, 16–21. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of citrus trees from unmanned aerial vehicle imagery using convolutional neural networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef]
- Li, W.; Fu, H.; Yu, L. Deep convolutional neural network based large-scale oil palm tree detection for high-resolution remote sensing images. In Proceedings of the 2017 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), New York, NY, USA, 11–13 December 2017; pp. 846–849. [Google Scholar]
- Jintasuttisak, T.; Edirisinghe, E.; Elbattay, A. Deep neural network based date palm tree detection in drone imagery. Comput. Electron. Agric. 2022, 192, 106560. [Google Scholar] [CrossRef]
- Jemaa, H.; Bouachir, W.; Leblon, B.; Bouguila, N. Computer vision system for detecting orchard trees from UAV images. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, 43, 661–668. [Google Scholar] [CrossRef]
- Santos, A.A.D.; Marcato Junior, J.; Araújo, M.S.; Di Martini, D.R.; Tetila, E.C.; Siqueira, H.L.; Aoki, C.; Eltner, A.; Matsubara, E.T.; Pistori, H.; et al. Assessment of CNN-based methods for individual tree detection on images captured by RGB cameras attached to UAVs. Sensors 2019, 19, 3595. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.B.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 42, 318–327. [Google Scholar] [CrossRef]
- Fromm, M.; Schubert, M.; Castilla, G.; Linke, J.; McDermid, G. Automated detection of conifer seedlings in drone imagery using convolutional neural networks. Remote Sens. 2019, 11, 2585. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.E.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Proceedings of the European Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 21–37. [Google Scholar]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-fcn: Object detection via region-based fully convolutional networks. Adv. Neural Inf. Process. Syst. 2016, 29, 379–387. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.J.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Hoiem, D.; Divvala, S.K.; Hays, J.H. Pascal VOC 2008 challenge. World Lit. Today 2009, 24. [Google Scholar]
- Zhang, L.; Wang, Y.; Huo, Y. Object detection in high-resolution remote sensing images based on a hard-example-mining network. IEEE Trans. Geosci. Remote Sens. 2020, 59, 8768–8780. [Google Scholar] [CrossRef]
- Xia, G.S.; Bai, X.; Ding, J.; Zhu, Z.; Belongie, S.J.; Luo, J.; Datcu, M.; Pelillo, M.; Zhang, L. DOTA: A Large-Scale Dataset for Object Detection in Aerial Images. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3974–3983. [Google Scholar]
- Jin, S.; RoyChowdhury, A.; Jiang, H.; Singh, A.; Prasad, A.; Chakraborty, D.; Learned-Miller, E.G. Unsupervised Hard Example Mining from Videos for Improved Object Detection. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 307–324. [Google Scholar]
- Shrivastava, A.; Gupta, A.K.; Girshick, R.B. Training Region-Based Object Detectors with Online Hard Example Mining. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 761–769. [Google Scholar]
- Wan, S.; Chen, Z.; Tao, Z.; Zhang, B.; kat Wong, K. Bootstrapping Face Detection with Hard Negative Examples. arXiv 2016, arXiv:1608.02236. [Google Scholar]
- Liu, Y. An Improved Faster R-CNN for Object Detection. In Proceedings of the 2018 11th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 8–9 December 2018; Volume 2, pp. 119–123. [Google Scholar]
- Sun, X.; Wu, P.; Hoi, S.C. Face detection using deep learning: An improved faster RCNN approach. Neurocomputing 2018, 299, 42–50. [Google Scholar] [CrossRef]
- Yang, S.; Luo, P.; Loy, C.C.; Tang, X. WIDER FACE: A Face Detection Benchmark. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 5525–5533. [Google Scholar]
- Zhang, L.; Lin, L.; Liang, X.; He, K. Is Faster R-CNN Doing Well for Pedestrian Detection? arXiv 2016, arXiv:1607.07032. [Google Scholar]
- Wang, X.; Shrivastava, A.; Gupta, A.K. A-Fast-RCNN: Hard Positive Generation via Adversary for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3039–3048. [Google Scholar]
- Ravi, N.; El-Sharkawy, M. Improved Single Shot Detector with Enhanced Hard Negative Mining Approach. In Proceedings of the 2022 International Conference on Advanced Computer Science and Information Systems (ICACSIS), Depok, Indonesia, 1–3 October 2022; pp. 25–30. [Google Scholar]
- Albetis, J.; Jacquin, A.; Goulard, M.; Poilvé, H.; Rousseau, J.; Clenet, H.; Dedieu, G.; Duthoit, S. On the potentiality of UAV multispectral imagery to detect Flavescence dorée and Grapevine Trunk Diseases. Remote Sens. 2018, 11, 23. [Google Scholar] [CrossRef]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the spatial variability of Botrytis bunch rot risk in vineyards using UAV multispectral imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- Chang, A.; Yeom, J.; Jung, J.; Landivar, J. Comparison of canopy shape and vegetation indices of citrus trees derived from UAV multispectral images for characterization of citrus greening disease. Remote Sens. 2020, 12, 4122. [Google Scholar] [CrossRef]
- Șandric, I.; Irimia, R.; Petropoulos, G.P.; Anand, A.; Srivastava, P.K.; Pleșoianu, A.; Faraslis, I.; Stateras, D.; Kalivas, D. Tree’s detection & health’s assessment from ultra-high resolution UAV imagery and deep learning. Geocarto Int. 2022, 37, 10459–10479. [Google Scholar]
- Solano, F.; Di Fazio, S.; Modica, G. A methodology based on GEOBIA and WorldView-3 imagery to derive vegetation indices at tree crown detail in olive orchards. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101912. [Google Scholar] [CrossRef]
- Garcia-Ruiz, F.; Sankaran, S.; Maja, J.M.; Lee, W.S.; Rasmussen, J.; Ehsani, R. Comparison of two aerial imaging platforms for identification of Huanglongbing-infected citrus trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Navrozidis, I.; Haugommard, A.; Kasampalis, D.; Alexandridis, T.; Castel, F.; Moshou, D.; Ovakoglou, G.; Pantazi, X.E.; Tamouridou, A.A.; Lagopodi, A.L.; et al. Assessing Olive Trees Health Using Vegetation Indices and Mundi Web Services for Sentinel-2 Images. In Proceedings of the Hellenic Association on Information and Communication Technologies in Agriculture, Food & Environment, Thessaloniki, Greece, 24–27 September 2020; pp. 130–136. [Google Scholar]
- Dutta, A.; Zisserman, A. The VIA annotation software for images, audio and video. In Proceedings of the 27th ACM International Conference on Multimedia, Nice, France, 21–25 October 2019; pp. 2276–2279. [Google Scholar]
- Zarco-Tejada, P.J.; Miller, J.R.; Mohammed, G.; Noland, T.L.; Sampson, P. Vegetation stress detection through chlorophyll a+ b estimation and fluorescence effects on hyperspectral imagery. J. Environ. Qual. 2002, 31, 1433–1441. [Google Scholar] [CrossRef]
- Barry, K.M.; Stone, C.; Mohammed, C. Crown-scale evaluation of spectral indices for defoliated and discoloured eucalypts. Int. J. Remote Sens. 2008, 29, 47–69. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.K.; Girshick, R.B.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Weinstein, B.G.; Marconi, S.; Aubry-Kientz, M.; Vincent, G.; Senyondo, H.; White, E.P. DeepForest: A Python package for RGB deep learning tree crown delineation. Methods Ecol. Evol. 2020, 11, 1743–1751. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; Singh, A.; Graves, S.J.; White, E.P. A remote sensing derived data set of 100 million individual tree crowns for the National Ecological Observatory Network. eLife 2021, 10, e62922. [Google Scholar] [CrossRef]
- Kobayashi, N.; Tani, H.; Wang, X.; Sonobe, R. Crop classification using spectral indices derived from Sentinel-2A imagery. J. Inf. Telecommun. 2020, 4, 67–90. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Cogato, A.; Pagay, V.; Marinello, F.; Meggio, F.; Grace, P.; De Antoni Migliorati, M. Assessing the feasibility of using sentinel-2 imagery to quantify the impact of heatwaves on irrigated vineyards. Remote Sens. 2019, 11, 2869. [Google Scholar] [CrossRef]
- Hawryło, P.; Bednarz, B.; Wężyk, P.; Szostak, M. Estimating defoliation of Scots pine stands using machine learning methods and vegetation indices of Sentinel-2. Eur. J. Remote Sens. 2018, 51, 194–204. [Google Scholar] [CrossRef]
- Oumar, Z.; Mutanga, O. Using WorldView-2 bands and indices to predict bronze bug (Thaumastocoris peregrinus) damage in plantation forests. Int. J. Remote Sens. 2013, 34, 2236–2249. [Google Scholar] [CrossRef]
- Verbesselt, J.; Robinson, A.; Stone, C.; Culvenor, D. Forecasting tree mortality using change metrics derived from MODIS satellite data. For. Ecol. Manag. 2009, 258, 1166–1173. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of chlorophyll a, chlorophyll b, chlorophyll a+ b, and total carotenoid content in eucalyptus leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Deng, X.; Guo, S.; Sun, L.; Chen, J. Identification of short-rotation eucalyptus plantation at large scale using multi-satellite imageries and cloud computing platform. Remote Sens. 2020, 12, 2153. [Google Scholar] [CrossRef]
- Bajwa, S.G.; Tian, L. Multispectral CIR image calibration for cloud shadow and soil background influence using intensity normalization. Appl. Eng. Agric. 2002, 18, 627. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Sripada, R.P.; Heiniger, R.W.; White, J.G.; Meijer, A.D. Aerial color infrared photography for determining early in-season nitrogen requirements in corn. Agron. J. 2006, 98, 968–977. [Google Scholar] [CrossRef]
- Buschmann, C.; Nagel, E. In vivo spectroscopy and internal optics of leaves as basis for remote sensing of vegetation. Int. J. Remote Sens. 1993, 14, 711–722. [Google Scholar] [CrossRef]
- Villa, P.; Mousivand, A.; Bresciani, M. Aquatic vegetation indices assessment through radiative transfer modeling and linear mixture simulation. Int. J. Appl. Earth Obs. Geoinf. 2014, 30, 113–127. [Google Scholar] [CrossRef]
- Barnes, E.; Clarke, T.; Richards, S.; Colaizzi, P.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident detection of crop water stress, nitrogen status and canopy density using ground based multispectral data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000; Volume 1619, p. 6. [Google Scholar]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Loh, W.Y. Classification and regression trees. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Puliti, S.; Astrup, R. Automatic detection of snow breakage at single tree level using YOLOv5 applied to UAV imagery. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102946. [Google Scholar] [CrossRef]
- Nembrini, S.; König, I.R.; Wright, M.N. The revival of the Gini importance? Bioinformatics 2018, 34, 3711–3718. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Silva, C.A.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.T.; Dia, M. Individual tree detection from unmanned aerial vehicle (UAV) derived canopy height model in an open canopy mixed conifer forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, Y.; Zhang, X. Extraction of tree crowns damaged by Dendrolimus tabulaeformis Tsai et Liu via spectral-spatial classification using UAV-based hyperspectral images. Plant Methods 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Iordache, M.D.; Mantas, V.; Baltazar, E.; Pauly, K.; Lewyckyj, N. A machine learning approach to detecting pine wilt disease using airborne spectral imagery. Remote Sens. 2020, 12, 2280. [Google Scholar] [CrossRef]
- Ortiz, S.M.; Breidenbach, J.; Kändler, G. Early detection of bark beetle green attack using TerraSAR-X and RapidEye data. Remote Sens. 2013, 5, 1912–1931. [Google Scholar] [CrossRef]
- Gago, J.; Douthe, C.; Coopman, R.E.; Gallego, P.P.; Ribas-Carbo, M.; Flexas, J.; Escalona, J.; Medrano, H. UAVs challenge to assess water stress for sustainable agriculture. Agric. Water Manag. 2015, 153, 9–19. [Google Scholar] [CrossRef]
- Berni, J.; Zarco-Tejada, P.; Sepulcre-Cantó, G.; Fereres, E.; Villalobos, F. Mapping canopy conductance and CWSI in olive orchards using high resolution thermal remote sensing imagery. Remote Sens. Environ. 2009, 113, 2380–2388. [Google Scholar] [CrossRef]
- Moriondo, M.; Maselli, F.; Bindi, M. A simple model of regional wheat yield based on NDVI data. Eur. J. Agron. 2007, 26, 266–274. [Google Scholar] [CrossRef]
- Yang, M.; Hassan, M.A.; Xu, K.; Zheng, C.; Rasheed, A.; Zhang, Y.; Jin, X.; Xia, X.; Xiao, Y.; He, Z. Assessment of water and nitrogen use efficiencies through UAV-based multispectral phenotyping in winter wheat. Front. Plant Sci. 2020, 11, 927. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef]
- Michez, A.; Piégay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Environ. Monit. Assess. 2016, 188, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Smigaj, M.; Gaulton, R.; Barr, S.L.; Suárez, J.C. UAV-borne Thermal Imaging for Forest Health Monitoring: Detection of Disease-Induced Canopy Temperature Increase. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, XL-3/W3, 349–354. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.B.; He, K.; Hariharan, B.; Belongie, S.J. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef] [PubMed]



| Vegetation Index | Equation | Reference |
|---|---|---|
| Difference Vegetation Index | DVI = Near-infrared (NIR)-Red | [70] |
| Generalized Difference Vegetation Index | GDVI = NIR − Green | [71] |
| Green Normalized Difference Vegetation Index | GNDVI = (NIR − Green)/(NIR + Green) | [72] |
| Green-Red Vegetation Index | GRVI = NIR/Green | [71] |
| Normalized Difference Aquatic Vegetation Index | NDAVI = (NIR − Blue)/(NIR + Blue) | [73] |
| Normalized Difference Vegetation Index | NDVI = (NIR − Red)/(NIR + Red) | [70] |
| Normalized Difference Red-Edge | NDRE = (NIR − RedEdge)/(NIR + RedEdge) | [74] |
| Normalized Green | NG = Green/(NIR + Red + Green) | [69] |
| Normalized Red | NR = Red/(NIR + Red + Green) | [69] |
| Normalized NIR | NNIR = NIR/(NIR + Red + Green) | [69] |
| Red simple ratio Vegetation Index | RVI = NIR/Red | [75] |
| Water Adjusted Vegetation Index | WAVI = (1.5*(NIR − Blue))/((NIR + Blue) + 0.5) | [73] |
| Baseline 1: DeepForest | Baseline 2: YOLO | |||||
|---|---|---|---|---|---|---|
| Fold | (%) | (%) | (%) | (%) | (%) | (%) |
| Fold 1 | 82.25 | 87.24 | 84.67 | 83.89 | 87.27 | 85.15 |
| Fold 2 | 87.57 | 88.06 | 87.82 | 79.12 | 92.35 | 84.91 |
| Fold 3 | 87.87 | 84.73 | 86.27 | 83.09 | 87.45 | 84.38 |
| Average | 85.85 | 86.67 | 86.24 | 82.01 | 88.99 | 84.81 |
| Baseline Model | (%) | (%) | (%) |
|---|---|---|---|
| Without HNM | |||
| Baseline 1: DeepForest | 84.82 | 86.18 | 85.46 |
| Baseline 2: YOLO | 79.40 | 88.05 | 82.64 |
| With HNM | |||
| Baseline 1: DeepForest | 85.85 | 86.67 | 86.24 |
| Baseline 2: YOLO | 82.01 | 88.99 | 84.81 |
| Health Status | (%) | (%) | (%) | Accuracy (%) |
|---|---|---|---|---|
| Random Forest Classifier (RF) | ||||
| Healthy | 98.8 | 98.8 | 98.8 | 97.52 |
| Unhealthy | 95.5 | 95.5 | 95.5 | |
| Light Gradient Boosting Machine (LightGBM) | ||||
| Healthy | 99 | 98.8 | 98.9 | 97.47 |
| Unhealthy | 95.6 | 96.1 | 95.8 | |
| K-nearest neighbors (KNN) | ||||
| Healthy | 98.6 | 98.1 | 98.4 | 97.07 |
| Unhealthy | 92.9 | 95 | 93.9 | |
| Support Vector Machine (SVM) | ||||
| Healthy | 99.4 | 95.8 | 97.6 | 96.31 |
| Unhealthy | 86.2 | 97.8 | 91.6 | |
| Decision Tree Classifier (DT) | ||||
| Healthy | 98.4 | 97.9 | 98.1 | 95.91 |
| Unhealthy | 92.3 | 93.9 | 93.1 | |
| Ranking | Feature | Contribution |
|---|---|---|
| 1 | NR | 34% |
| 2 | NNIR | 25% |
| 3 | RVI | 16% |
| 4 | NDAVI | 15% |
| 5 | DVI | 10% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jemaa, H.; Bouachir, W.; Leblon, B.; LaRocque, A.; Haddadi, A.; Bouguila, N. UAV-Based Computer Vision System for Orchard Apple Tree Detection and Health Assessment. Remote Sens. 2023, 15, 3558. https://doi.org/10.3390/rs15143558
Jemaa H, Bouachir W, Leblon B, LaRocque A, Haddadi A, Bouguila N. UAV-Based Computer Vision System for Orchard Apple Tree Detection and Health Assessment. Remote Sensing. 2023; 15(14):3558. https://doi.org/10.3390/rs15143558
Chicago/Turabian StyleJemaa, Hela, Wassim Bouachir, Brigitte Leblon, Armand LaRocque, Ata Haddadi, and Nizar Bouguila. 2023. "UAV-Based Computer Vision System for Orchard Apple Tree Detection and Health Assessment" Remote Sensing 15, no. 14: 3558. https://doi.org/10.3390/rs15143558
APA StyleJemaa, H., Bouachir, W., Leblon, B., LaRocque, A., Haddadi, A., & Bouguila, N. (2023). UAV-Based Computer Vision System for Orchard Apple Tree Detection and Health Assessment. Remote Sensing, 15(14), 3558. https://doi.org/10.3390/rs15143558









