AgriSen-COG, a Multicountry, Multitemporal Large-Scale Sentinel-2 Benchmark Dataset for Crop Mapping Using Deep Learning
Abstract
1. Introduction
- We created AgriSen-COG, a large-scale benchmark dataset for crop type mapping, including the largest number of different European countries (five), designed for AI applications.
- We introduce a methodology for LPIS processing to obtain GT for a crop-type dataset, useful for further extensions of the current dataset.
- We incorporate an anomaly detection method based on autoencoders and dynamic time warping (DTW) distance as a preprocessing step to identify mislabeled data from LPIS.
- We experiment with popular DL models and provide a baseline, showing the generalization capabilities of the models on the proposed dataset across space (multicountry) and time (multitemporal).
- We provide the LPIS preprocessing, anomaly detection, training/testing code, and our trained models to enable further development.
2. Crop Datasets for ML/DL Applications
3. Popular DL Methods for Crop Type Mapping
4. Anomaly Detection
5. The AgriSen-COG Dataset
5.1. Input Satellite Data
5.2. LPIS—Crop Type Labels
5.3. Dataset Creation Methodology
- (1)
- LPIS processing
- (2)
- Preparing rasterized data
- (3)
- Improving dataset quality with Anomaly Detection
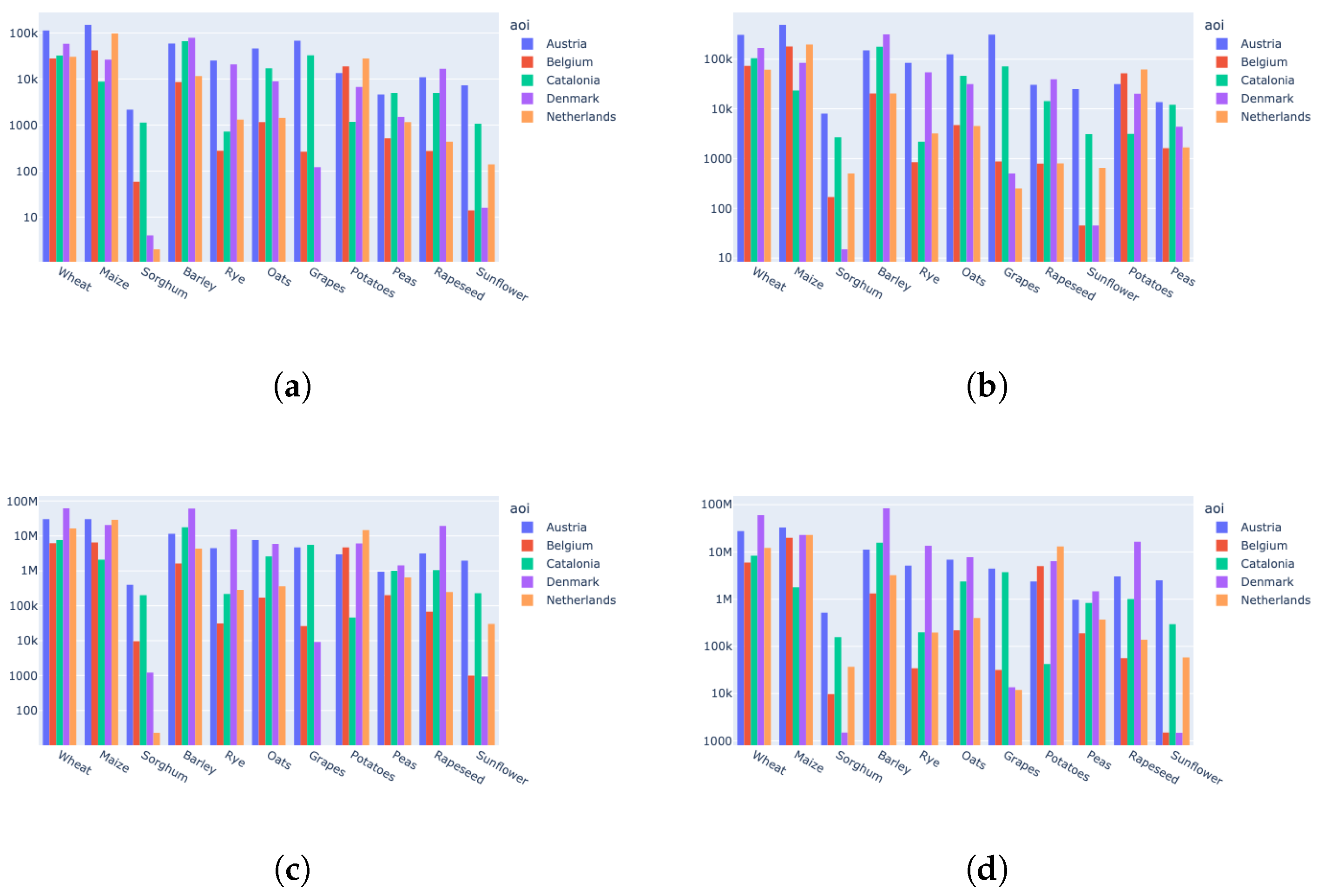
5.4. Dataset Description
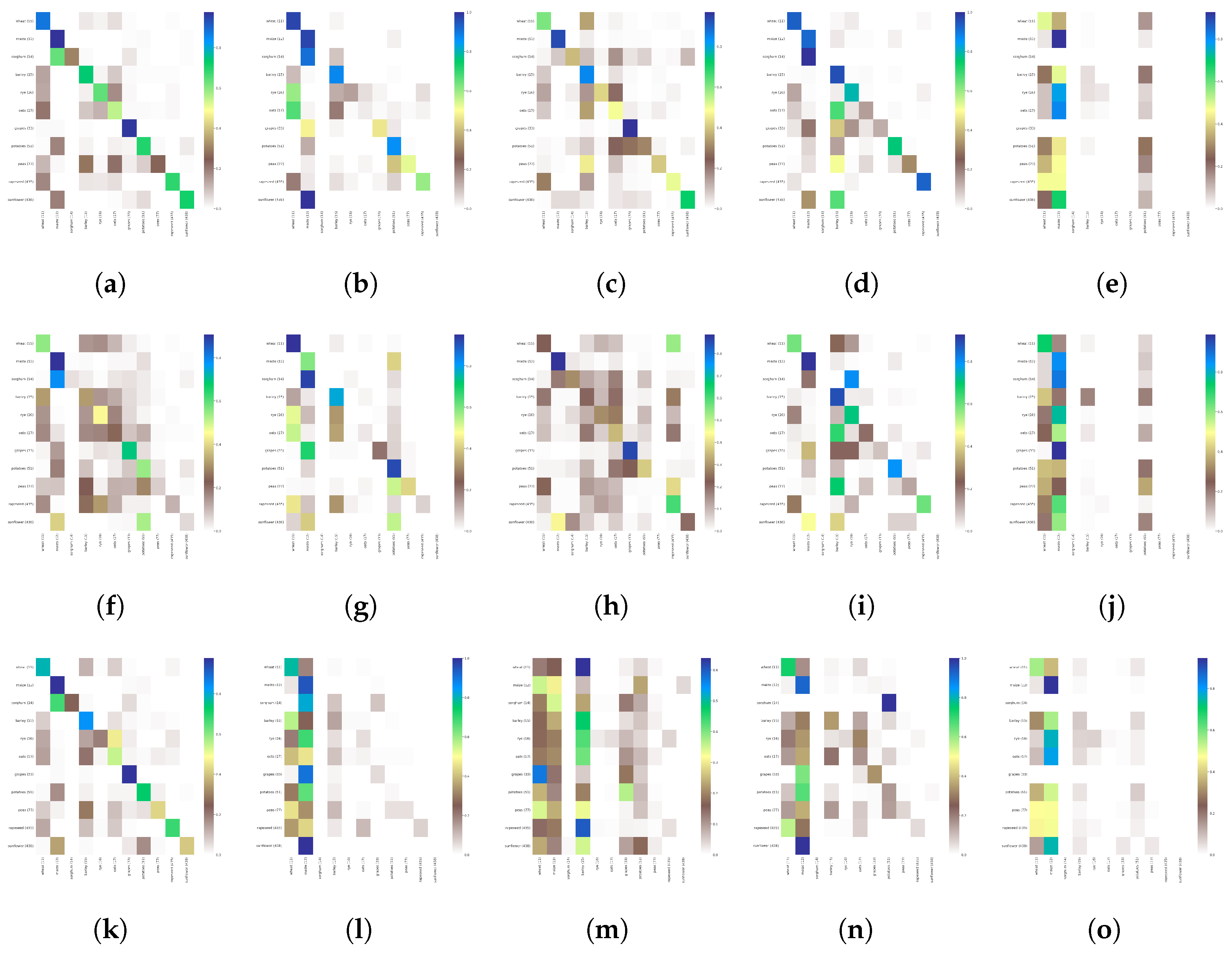
6. Experimental Results
- Experiment Type 1 (anomalies variation): We conduct individual experiments on each AOI for a single year, with one model, to highlight the importance of curated data labels.
- Experiment Type 2 (temporal generalization): We conduct individual experiments on each AOI using the model trained in Experiment Type 1 and predict the instances for 2020.
- Experiment Type 3 (spatial generalization): We conduct several experiments, using data from one year and splitting our data based on regions’ similarity in crop patterns.
- Experiment Type 4 (overall generalization): We train on two AOIs for 2019, with different models (LSTM, Transformer, TempCNN, U-Net, ConvStar) to see the behavior of the proposed dataset.
6.1. Crop Type Classification Experiments—Time-Series Classification
6.2. Crop Mapping Experiments—Semantic Segmentation
7. Discussion
8. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Sustainable Agriculture|Sustainable Development Goals|Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/sustainable-development-goals/overview/fao-and-the-2030-agenda-for-sustainable-development/sustainable-agriculture/en/ (accessed on 20 October 2022).
- THE 17 GOALS|Sustainable Development. Available online: https://sdgs.un.org/goals#icons (accessed on 20 October 2022).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Cordts, M.; Omran, M.; Ramos, S.; Scharwächter, T.; Enzweiler, M.; Benenson, R.; Franke, U.; Roth, S.; Schiele, B. The cityscapes dataset. In Proceedings of the CVPR Workshop on the Future of Datasets in Vision, Boston, MA, USA, 7–12 June 2015; Volume 2. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 740–755. [Google Scholar]
- Sumbul, G.; Charfuelan, M.; Demir, B.; Markl, V. Bigearthnet: A large-scale benchmark archive for remote sensing image understanding. In Proceedings of the IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 5901–5904. [Google Scholar]
- Schmitt, M.; Hughes, L.H.; Qiu, C.; Zhu, X.X. SEN12MS–A Curated Dataset of Georeferenced Multi-Spectral Sentinel-1/2 Imagery for Deep Learning and Data Fusion. arXiv 2019, arXiv:1906.07789. [Google Scholar] [CrossRef]
- European Court of Auditors. The Land Parcel Identification System: A Useful Tool to Determine the Eligibility of Agricultural Land—But Its Management Could Be Further Improved; Special Report No 25; Publications Office: Luxembourg, 2016. [CrossRef]
- Rußwurm, M.; Pelletier, C.; Zollner, M.; Lefèvre, S.; Körner, M. BreizhCrops: A Time Series Dataset for Crop Type Mapping. ISPRS Int. Arch. Photogramm. Remote. Sens. Spat. Inf. Sci. 2020, XLIII-B2-2020, 1545–1551. [Google Scholar] [CrossRef]
- Turkoglu, M.O.; D’Aronco, S.; Perich, G.; Liebisch, F.; Streit, C.; Schindler, K.; Wegner, J.D. Crop mapping from image time series: Deep learning with multi-scale label hierarchies. arXiv 2021, arXiv:2102.08820. [Google Scholar] [CrossRef]
- Weikmann, G.; Paris, C.; Bruzzone, L. TimeSen2Crop: A Million Labeled Samples Dataset of Sentinel 2 Image Time Series for Crop-Type Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 4699–4708. [Google Scholar] [CrossRef]
- Kondmann, L.; Toker, A.; Rußwurm, M.; Camero, A.; Peressuti, D.; Milcinski, G.; Mathieu, P.P.; Longépé, N.; Davis, T.; Marchisio, G.; et al. DENETHOR: The DynamicEarthNET dataset for Harmonized, inter-Operable, analysis-Ready, daily crop monitoring from space. In Proceedings of the Thirty-Fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 2), Virtual, 6–14 December 2021. [Google Scholar]
- Sykas, D.; Sdraka, M.; Zografakis, D.; Papoutsis, I. A Sentinel-2 Multiyear, Multicountry Benchmark Dataset for Crop Classification and Segmentation With Deep Learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 3323–3339. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. A System of Integrated Agricultural Censuses and Surveys: World Programme for the Census of Agriculture 2010; Food and Agriculture Organization of the United Nations: Rome, Italy, 2005; Volume 1. [Google Scholar]
- Hoyer, S.; Hamman, J. xarray: N-D labeled arrays and datasets in Python. J. Open Res. Softw. 2017, 5, 10. [Google Scholar] [CrossRef]
- Hoyer, S.; Fitzgerald, C.; Hamman, J.; Akleeman; Kluyver, T.; Roos, M.; Helmus, J.J.; Markel; Cable, P.; Maussion, F.; et al. xarray: V0.8.0. 2016. Available online: https://doi.org/10.5281/zenodo.59499 (accessed on 4 June 2023).
- d’Andrimont, R.; Claverie, M.; Kempeneers, P.; Muraro, D.; Yordanov, M.; Peressutti, D.; Batič, M.; Waldner, F. AI4Boundaries: An open AI-ready dataset to map field boundaries with Sentinel-2 and aerial photography. Earth Syst. Sci. Data Discuss. 2022, 15, 317–329. [Google Scholar] [CrossRef]
- Jordan, M.I. Serial Order: A Parallel Distributed Processing Approach. Technical Report, June 1985–March 1986. 1986. Available online: https://doi.org/10.1016/S0166-4115(97)80111-2 (accessed on 4 June 2023).
- Rumelhart, D.E.; McClelland, J.L. Learning Internal Representations by Error Propagation. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations; U.S. Department of Energy Office of Scientific and Technical Information; MIT Press: Cambridge, MA, USA, 1987; pp. 318–362. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017. Available online: https://proceedings.neurips.cc/paper_files/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf (accessed on 4 June 2023).
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Rakhlin, A.; Davydow, A.; Nikolenko, S. Land cover classification from satellite imagery with u-net and lovász-softmax loss. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 262–266. [Google Scholar]
- Solórzano, J.V.; Mas, J.F.; Gao, Y.; Gallardo-Cruz, J.A. Land use land cover classification with U-net: Advantages of combining sentinel-1 and sentinel-2 imagery. Remote Sens. 2021, 13, 3600. [Google Scholar] [CrossRef]
- Wang, J.; Yang, M.; Chen, Z.; Lu, J.; Zhang, L. An MLC and U-Net Integrated Method for Land Use/Land Cover Change Detection Based on Time Series NDVI-Composed Image from PlanetScope Satellite. Water 2022, 14, 3363. [Google Scholar] [CrossRef]
- Zhang, Z.; Iwasaki, A.; Xu, G.; Song, J. Cloud detection on small satellites based on lightweight U-net and image compression. J. Appl. Remote Sens. 2019, 13, 026502. [Google Scholar] [CrossRef]
- Guo, Y.; Cao, X.; Liu, B.; Gao, M. Cloud detection for satellite imagery using attention-based U-Net convolutional neural network. Symmetry 2020, 12, 1056. [Google Scholar] [CrossRef]
- Xing, D.; Hou, J.; Huang, C.; Zhang, W. Spatiotemporal Reconstruction of MODIS Normalized Difference Snow Index Products Using U-Net with Partial Convolutions. Remote Sens. 2022, 14, 1795. [Google Scholar] [CrossRef]
- Ivanovsky, L.; Khryashchev, V.; Pavlov, V.; Ostrovskaya, A. Building detection on aerial images using U-NET neural networks. In Proceedings of the 2019 24th Conference of Open Innovations Association (FRUCT), Moscow, Russia, 8–12 April 2019; pp. 116–122. [Google Scholar]
- Irwansyah, E.; Heryadi, Y.; Gunawan, A.A.S. Semantic image segmentation for building detection in urban area with aerial photograph image using U-Net models. In Proceedings of the 2020 IEEE Asia-Pacific Conference on Geoscience, Electronics and Remote Sensing Technology (AGERS), Jakarta, Indonesia, 7–8 December 2020; pp. 48–51. [Google Scholar]
- Wu, C.; Zhang, F.; Xia, J.; Xu, Y.; Li, G.; Xie, J.; Du, Z.; Liu, R. Building damage detection using U-Net with attention mechanism from pre-and post-disaster remote sensing datasets. Remote Sens. 2021, 13, 905. [Google Scholar] [CrossRef]
- Wei, S.; Zhang, H.; Wang, C.; Wang, Y.; Xu, L. Multi-temporal SAR data large-scale crop mapping based on U-Net model. Remote Sens. 2019, 11, 68. [Google Scholar] [CrossRef]
- Fan, X.; Yan, C.; Fan, J.; Wang, N. Improved U-Net Remote Sensing Classification Algorithm Fusing Attention and Multiscale Features. Remote Sens. 2022, 14, 3591. [Google Scholar] [CrossRef]
- Li, G.; Cui, J.; Han, W.; Zhang, H.; Huang, S.; Chen, H.; Ao, J. Crop type mapping using time-series Sentinel-2 imagery and U-Net in early growth periods in the Hetao irrigation district in China. Comput. Electron. Agric. 2022, 203, 107478. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, H.; Yeung, D.Y.; Wong, W.K.; Woo, W.C. Convolutional LSTM network: A machine learning approach for precipitation nowcasting. Adv. Neural Inf. Process. Syst. 2015, 2015, 802–810. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Farooque, G.; Xiao, L.; Yang, J.; Sargano, A.B. Hyperspectral image classification via a novel spectral–spatial 3D ConvLSTM-CNN. Remote Sens. 2021, 13, 4348. [Google Scholar] [CrossRef]
- Cherif, E.; Hell, M.; Brandmeier, M. DeepForest: Novel Deep Learning Models for Land Use and Land Cover Classification Using Multi-Temporal and-Modal Sentinel Data of the Amazon Basin. Remote Sens. 2022, 14, 5000. [Google Scholar] [CrossRef]
- Meng, X.; Liu, Q.; Shao, F.; Li, S. Spatio–Temporal–Spectral Collaborative Learning for Spatio–Temporal Fusion with Land Cover Changes. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5704116. [Google Scholar] [CrossRef]
- Habiboullah, A.; Louly, M.A. Soil Moisture Prediction Using NDVI and NSMI Satellite Data: ViT-Based Models and ConvLSTM-Based Model. SN Comput. Sci. 2023, 4, 140. [Google Scholar] [CrossRef]
- Park, S.; Im, J.; Han, D.; Rhee, J. Short-term forecasting of satellite-based drought indices using their temporal patterns and numerical model output. Remote Sens. 2020, 12, 3499. [Google Scholar] [CrossRef]
- Yeom, J.M.; Deo, R.C.; Adamowski, J.F.; Park, S.; Lee, C.S. Spatial mapping of short-term solar radiation prediction incorporating geostationary satellite images coupled with deep convolutional LSTM networks for South Korea. Environ. Res. Lett. 2020, 15, 094025. [Google Scholar] [CrossRef]
- Muthukumar, P.; Cocom, E.; Nagrecha, K.; Comer, D.; Burga, I.; Taub, J.; Calvert, C.F.; Holm, J.; Pourhomayoun, M. Predicting PM2. 5 atmospheric air pollution using deep learning with meteorological data and ground-based observations and remote-sensing satellite big data. Air Qual. Atmos. Health 2021, 15, 1221–1234. [Google Scholar] [CrossRef]
- Yaramasu, R.; Bandaru, V.; Pnvr, K. Pre-season crop type mapping using deep neural networks. Comput. Electron. Agric. 2020, 176, 105664. [Google Scholar] [CrossRef]
- Chang, Y.L.; Tan, T.H.; Chen, T.H.; Chuah, J.H.; Chang, L.; Wu, M.C.; Tatini, N.B.; Ma, S.C.; Alkhaleefah, M. Spatial-temporal neural network for rice field classification from SAR images. Remote Sens. 2022, 14, 1929. [Google Scholar] [CrossRef]
- Ienco, D.; Interdonato, R.; Gaetano, R.; Minh, D.H.T. Combining Sentinel-1 and Sentinel-2 Satellite Image Time Series for land cover mapping via a multi-source deep learning architecture. ISPRS J. Photogramm. Remote Sens. 2019, 158, 11–22. [Google Scholar] [CrossRef]
- Turkoglu, M.O.; D’Aronco, S.; Wegner, J.D.; Schindler, K. Gating revisited: Deep multi-layer rnns that can be trained. arXiv 2019, arXiv:1911.11033. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal convolutional neural network for the classification of satellite image time series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef]
- Mitra, P.; Akhiyarov, D.; Araya-Polo, M.; Byrd, D. Machine Learning-based Anomaly Detection with Magnetic Data. Preprints.org 2020. [Google Scholar] [CrossRef]
- Sontowski, S.; Lawrence, N.; Deka, D.; Gupta, M. Detecting Anomalies using Overlapping Electrical Measurements in Smart Power Grids. In Proceedings of the 2021 IEEE International Conference on Big Data (Big Data), Orlando, FL, USA, 15–18 December 2021; pp. 2434–2441. [Google Scholar]
- Wagner, N.; Antoine, V.; Koko, J.; Mialon, M.M.; Lardy, R.; Veissier, I. Comparison of machine learning methods to detect anomalies in the activity of dairy cows. In Proceedings of the International Symposium on Methodologies for Intelligent Systems, Graz, Austria, 23–25 September 2020; pp. 342–351. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Ballard, D.H. Modular learning in neural networks. In Proceedings of the AAAI, Seattle, WA, USA, 13 July 1987; Volume 647, pp. 279–284. [Google Scholar]
- Petitjean, F.; Ketterlin, A.; Gançarski, P. A global averaging method for dynamic time warping, with applications to clustering. Pattern Recognit. 2011, 44, 678–693. [Google Scholar] [CrossRef]
- Avolio, C.; Tricomi, A.; Zavagli, M.; De Vendictis, L.; Volpe, F.; Costantini, M. Automatic Detection of Anomalous Time Trends from Satellite Image Series to Support Agricultural Monitoring. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 6524–6527. [Google Scholar]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A commentary review on the use of normalized difference vegetation index (NDVI) in the era of popular remote sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Castillo-Villamor, L.; Hardy, A.; Bunting, P.; Llanos-Peralta, W.; Zamora, M.; Rodriguez, Y.; Gomez-Latorre, D.A. The Earth Observation-based Anomaly Detection (EOAD) system: A simple, scalable approach to mapping in-field and farm-scale anomalies using widely available satellite imagery. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102535. [Google Scholar] [CrossRef]
- Komisarenko, V.; Voormansik, K.; Elshawi, R.; Sakr, S. Exploiting time series of Sentinel-1 and Sentinel-2 to detect grassland mowing events using deep learning with reject region. Sci. Rep. 2022, 12, 983. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Ma, T.; Wang, X.; Wang, G. Anomaly Detection for Internet of Things Time Series Data Using Generative Adversarial Networks With Attention Mechanism in Smart Agriculture. Front. Plant Sci. 2022, 13, 890563. [Google Scholar] [CrossRef]
- Cui, L.; Zhang, Q.; Shi, Y.; Yang, L.; Wang, Y.; Wang, J.; Bai, C. A method for satellite time series anomaly detection based on fast-DTW and improved-KNN. Chin. J. Aeronaut. 2022, 36, 149–159. [Google Scholar] [CrossRef]
- Diab, D.M.; AsSadhan, B.; Binsalleeh, H.; Lambotharan, S.; Kyriakopoulos, K.G.; Ghafir, I. Anomaly detection using dynamic time warping. In Proceedings of the 2019 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), New York, NY, USA, 1–3 August 2019; pp. 193–198. [Google Scholar]
- Di Martino, T.; Guinvarc’h, R.; Thirion-Lefevre, L.; Colin, E. FARMSAR: Fixing AgRicultural Mislabels Using Sentinel-1 Time Series and AutoencodeRs. Remote Sens. 2022, 15, 35. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man, Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s optical high-resolution mission for GMES operational services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- PaperdJuly, W. ESRI shapefile technical description. Comput. Stat. 1998, 16, 370–371. [Google Scholar]
- Yutzler, J. OGC® GeoPackage Encoding Standard-with Corrigendum, Version 1.2. 175. 2018. Available online: https://www.geopackage.org/spec121/ (accessed on 4 June 2023).
- Zeiler, M. Modeling Our World: The ESRI Guide to Geodatabase Design; ESRI, Inc.: Redlands, CA, USA, 1999; Volume 40. [Google Scholar]
- Butler, H.; Daly, M.; Doyle, A.; Gillies, S.; Hagen, S.; Schaub, T. The Geojson Format. Technical Report. 2016. Available online: https://www.rfc-editor.org/rfc/rfc7946 (accessed on 4 June 2023).
- Moyroud, N.; Portet, F. Introduction to QGIS. QGIS Generic Tools 2018, 1, 1–17. [Google Scholar]
- Vohra, D. Apache parquet. In Practical Hadoop Ecosystem: A Definitive Guide to Hadoop-Related Frameworks and Tools; Apress: Berkley, CA, USA, 2016; pp. 325–335. [Google Scholar]
- Trakas, A.; McKee, L. OGC standards and the space community—Processes, application and value. In Proceedings of the 2011 2nd International Conference on Space Technology, Athens, Greece, 15–17 September 2011; pp. 1–5. [Google Scholar] [CrossRef]
- Durbin, C.; Quinn, P.; Shum, D. Task 51-Cloud-Optimized Format Study; Technical Report; NTRS: Chicago, IL, USA, 2020. [Google Scholar]
- Sanchez, A.H.; Picoli, M.C.A.; Camara, G.; Andrade, P.R.; Chaves, M.E.D.; Lechler, S.; Soares, A.R.; Marujo, R.F.; Simões, R.E.O.; Ferreira, K.R.; et al. Comparison of Cloud cover detection algorithms on sentinel–2 images of the amazon tropical forest. Remote Sens. 2020, 12, 1284. [Google Scholar] [CrossRef]
- AgrarMarkt Austria InVeKoS Strikes Austria. Available online: https://www.data.gv.at/ (accessed on 9 February 2023).
- Department of Agriculture and Fisheries Flemish Government. Available online: https://data.gov.be/en (accessed on 9 February 2023).
- Government of Catalonia Department of Agriculture Livestock Fisheries and Food. Available online: https://analisi.transparenciacatalunya.cat (accessed on 9 February 2023).
- The Danish Agency for Agriculture. Available online: https://lbst.dk/landbrug/ (accessed on 9 February 2023).
- Netherlands Enterprise Agency. Available online: https://nationaalgeoregister.nl/geonetwork/srv/dut/catalog.search#/home (accessed on 9 February 2023).
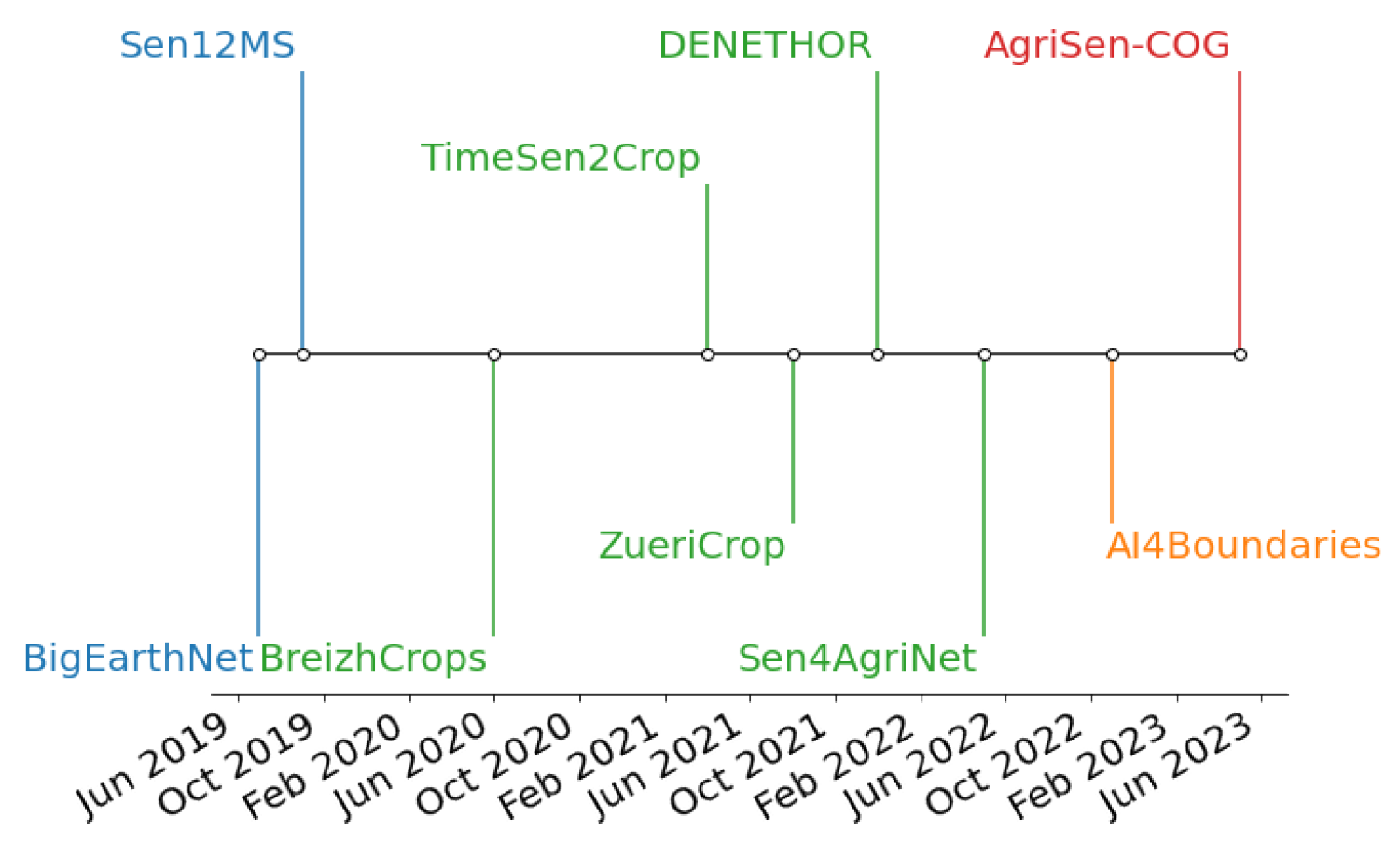








| Dataset | Nr. of Samples | Sample Size | Data Source | Short Summary |
|---|---|---|---|---|
| BigEarthNet | 590,326 patches | up to | Sentinel-2 L2A, CLC | land cover multiclass classification; nonoverlapping patches; 10 EU regions; June 2017–May 2018 |
| SEN12MS | 541,986 patches | Sentinel-1, Sentinel-2, MODIS Land Cover | land cover semantic segmentation; patches overlap with a stride of 128; global coverage; December 2016–November 2017 | |
| BreizhCrops | 768,000 fields | mean for each parcel band/timeframe | Sentinel-2, LPIS France | crop classification; covers Brittany region (France); January 2017–December 2018 |
| TimeSen2Crop | 1,200,000 fields | monthly medians | Sentinel-2 L2A, LPIS Austria | crop type mapping; covers Austria; September 2017–August 2018 |
| ZueriCrop | 28,000 patches | Sentinel-2 L2A, LPIS Switzerland | crop type mapping; covers Swiss Cantons of Zurich and Thurgau; January 2019–December 2019 | |
| DENETHOR | 4500 fields | Planet, Sentinel-1, Sentinel-2, LPIS Germany | pixel level; crop type classification; covers Northern Germany; January 2018–December 2019 | |
| Sen4AgriNet | 5000 patches | up to | Sentinel-2 L1C, LPIS France and Catalonia | crop type mapping; covers Catalonia and France for two years (2019 and 2020); ICC labels; pixel and parcel aggregated time series (mean and standard deviation) |
| AI4Boundaries | 7831 fields | (S2), (aerial) | Sentinel-2 L1C, aerial orthophoto at 1 m | crop field boundaries; covers seven regions (Austria, Catalonia, France, Luxembourg, the Netherlands, Slovenia, and Sweden); monthly composite March 2019–August 2019 |
| AgriSen-COG | 6,972,485 fields 41,100 patches | Sentinel-2 L2A, LPIS of 5 EU countries | crop type mapping; crop classification; parcel-based time series aggregated using barycenters; covers five regions (Austria, Belgium, Catalonia, Denmark, the Netherlands); 2019 and 2020 |
| Layer | Input Size | Hidden Size | Number of Recurrent Layers | Output Size |
|---|---|---|---|---|
| Encoder | ||||
| Input time series | [batch_size, seq_len, n_features] | - | - | - |
| LSTM Layer | 1 | 256 | 1 | [batch_size, seq_len, 256] |
| LSTM Layer | 256 | 128 | 1 | [batch_size, seq_len, 128] |
| Decoder | ||||
| LSTM Layer | 128 | 128 | 1 | [batch_size, seq_len, 128] |
| LSTM Layer | 128 | 256 | 1 | [batch_size, seq_len, 256] |
| FC Layer | 256 | - | - | [batch_size, seq_len, 1] |
| AOI | Source | Number of Parcels | Number of Unique Labels |
|---|---|---|---|
| Austria | LPIS Austria [74] | 5,144,532 | 220 |
| Belgium | LPIS Belgium [75] | 1,046,725 | 300 |
| Catalonia | LPIS Catalonia [76] | 1,283,820 | 176 |
| Denmark | LPIS Denmark [77] | 1,171,409 | 320 |
| Netherlands | LPIS Netherlands [78] | 1,592,285 | 377 |
| AOI | S2 Tiles | Number of Parcels with Area >= 0.1 ha (% Kept from the Original Data) | Number of Parcels after Anomaly Detection (% Kept from the Data with Area >= 0.1 ha) | Final Number of Patches |
|---|---|---|---|---|
| Austria | 20 | 4,115,899 (80%) | 3,788,326 (92%) | 16,563 |
| Belgium | 5 | 898,542 (85.8%) | 611,743 (68%) | 2607 |
| Catalonia | 10 | 1,006,573 (78.4%) | 719,631 (71.5%) | 5166 |
| Denmark | 17 | 1,061,070 (90.6%) | 924,914 (87%) | 9685 |
| Netherlands | 10 | 1,457,810 (91.5%) | 927,871 (63.64%) | 7079 |
| Total | 62 | 8,539,894 (83.47%) | 6,972,485 (81.65%) | 41,100 |
| Score | Version | Exp. | Model | Austria | Belgium | Catalonia | Denmark | Netherlands |
|---|---|---|---|---|---|---|---|---|
| Acc. W. (%) | v0 2019 | Type 1 | LSTM | 0.819 | 0.917 | 0.747 | 0.862 | 0.642 |
| v1 2019 | Type 1 | LSTM | 0.841 | 0.924 | 0.762 | 0.885 | 0.659 | |
| v1 2020 | Type 2 | LSTM | 0.614 | 0.719 | 0.369 | 0.752 | 0.657 | |
| v1 2019 | Type 3 | LSTM | 0.805 | 0.637 | 0.263 | 0.454 | 0.652 | |
| v1 2019 | Type 4 | Transformer | - | - | 0.667 | 0.791 | - | |
| v1 2020 | Type 4 | Transformer | - | - | 0.402 | 0.688 | - | |
| v1 2019 | Type 4 | TempCNN | - | - | 0.782 | 0.899 | - | |
| v1 2020 | Type 4 | TempCNN | - | - | 0.331 | 0.713 | - | |
| Prec. W (%) | v0 2019 | Type 1 | LSTM | 0.815 | 0.907 | 0.739 | 0.851 | 0.580 |
| v1 2019 | Type 1 | LSTM | 0.837 | 0.915 | 0.758 | 0.873 | 0.600 | |
| v1 2020 | Type 2 | LSTM | 0.640 | 0.830 | 0.549 | 0.755 | 0.624 | |
| v1 2019 | Type 3 | LSTM | 0.813 | 0.604 | 0.350 | 0.632 | 0.592 | |
| v1 2019 | Type 4 | Transformer | - | - | 0.640 | 0.836 | - | |
| v1 2020 | Type 4 | Transformer | - | - | 0.489 | 0.727 | - | |
| v1 2019 | Type 4 | TempCNN | - | - | 0.776 | 0.894 | - | |
| v1 2020 | Type 4 | TempCNN | - | - | 0.489 | 0.757 | - | |
| F1 W(%) | v0 2019 | Type 1 | LSTM | 0.816 | 0.911 | 0.739 | 0.851 | 0.593 |
| v1 2019 | Type 1 | LSTM | 0.838 | 0.918 | 0.755 | 0.874 | 0.613 | |
| v1 2020 | Type 2 | LSTM | 0.618 | 0.733 | 0.401 | 0.745 | 0.625 | |
| v1 2019 | Type 3 | LSTM | 0.796 | 0.548 | 0.267 | 0.439 | 0.586 | |
| v1 2019 | Type 4 | Transformer | - | - | 0.626 | 0.779 | - | |
| v1 2020 | Type 4 | Transformer | - | - | 0.420 | 0.664 | - | |
| v1 2019 | Type 4 | TempCNN | - | - | 0.777 | 0.894 | - | |
| v1 2020 | Type 4 | TempCNN | - | - | 0.349 | 0.716 | - |
| Score | Version | Exp. | Model | Austria | Belgium | Catalonia | Denmark | Netherlands |
|---|---|---|---|---|---|---|---|---|
| Acc. W. (%) | v0 2019 | Type 1 | U-Net | 0.967 | 0.979 | 0.887 | 0.974 | 0.771 |
| v1 2019 | Type 1 | U-Net | 0.969 | 0.981 | 0.894 | 0.982 | 0.783 | |
| v1 2020 | Type 2 | U-Net | 0.904 | 0.883 | 0.799 | 0.901 | 0.786 | |
| v1 2019 | Type 3 | U-Net | 0.954 | 0.801 | 0.756 | 0.761 | 0.775 | |
| v1 2019 | Type 4 | ConvStar | - | - | 0.882 | 0.973 | - | |
| v1 2020 | Type 4 | ConvStar | - | - | 0.804 | 0.893 | - | |
| Prec. W (%) | v0 2019 | Type 1 | U-Net | 0.879 | 0.956 | 0.787 | 0.938 | 0.581 |
| v1 2019 | Type 1 | U-Net | 0.888 | 0.961 | 0.814 | 0.951 | 0.585 | |
| v1 2020 | Type 2 | U-Net | 0.702 | 0.857 | 0.658 | 0.823 | 0.655 | |
| v1 2019 | Type 3 | U-Net | 0.851 | 0.618 | 0.520 | 0.526 | 0.588 | |
| v1 2019 | Type 4 | ConvStar | - | - | 0.790 | 0.939 | - | |
| v1 2020 | Type 4 | ConvStar | - | - | 0.687 | 0.871 | - | |
| F1 W(%) | v0 2019 | Type 1 | U-Net | 0.880 | 0.952 | 0.767 | 0.936 | 0.523 |
| v1 2019 | Type 1 | U-Net | 0.8886 | 0.956 | 0.793 | 0.951 | 0.541 | |
| v1 2020 | Type 2 | U-Net | 0.664 | 0.762 | 0.584 | 0.768 | 0.556 | |
| v1 2019 | Type 3 | U-Net | 0.835 | 0.607 | 0.444 | 0.329 | 0.561 | |
| v1 2019 | Type 4 | ConvStar | - | - | 0.758 | 0.928 | - | |
| v1 2020 | Type 4 | ConvStar | - | - | 0.550 | 0.779 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selea, T. AgriSen-COG, a Multicountry, Multitemporal Large-Scale Sentinel-2 Benchmark Dataset for Crop Mapping Using Deep Learning. Remote Sens. 2023, 15, 2980. https://doi.org/10.3390/rs15122980
Selea T. AgriSen-COG, a Multicountry, Multitemporal Large-Scale Sentinel-2 Benchmark Dataset for Crop Mapping Using Deep Learning. Remote Sensing. 2023; 15(12):2980. https://doi.org/10.3390/rs15122980
Chicago/Turabian StyleSelea, Teodora. 2023. "AgriSen-COG, a Multicountry, Multitemporal Large-Scale Sentinel-2 Benchmark Dataset for Crop Mapping Using Deep Learning" Remote Sensing 15, no. 12: 2980. https://doi.org/10.3390/rs15122980
APA StyleSelea, T. (2023). AgriSen-COG, a Multicountry, Multitemporal Large-Scale Sentinel-2 Benchmark Dataset for Crop Mapping Using Deep Learning. Remote Sensing, 15(12), 2980. https://doi.org/10.3390/rs15122980









