Uncertainty of Partial Dependence Relationship between Climate and Vegetation Growth Calculated by Machine Learning Models
Abstract
1. Introduction
2. Method
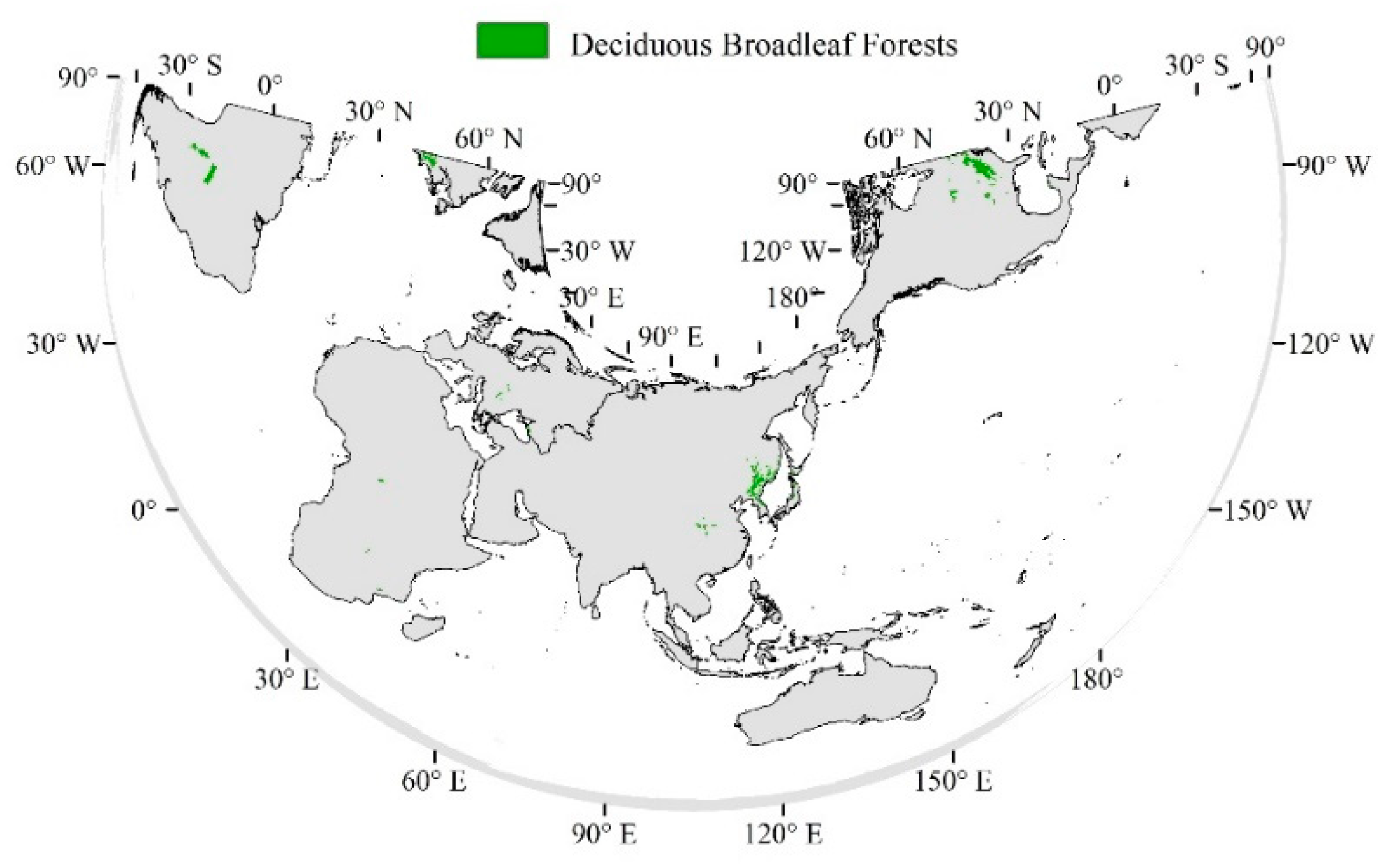
2.1. Data Resources
2.1.1. MODIS NDVI
2.1.2. WFDEI Meteorological Dataset
2.2. Methods for Analysis
3. Results
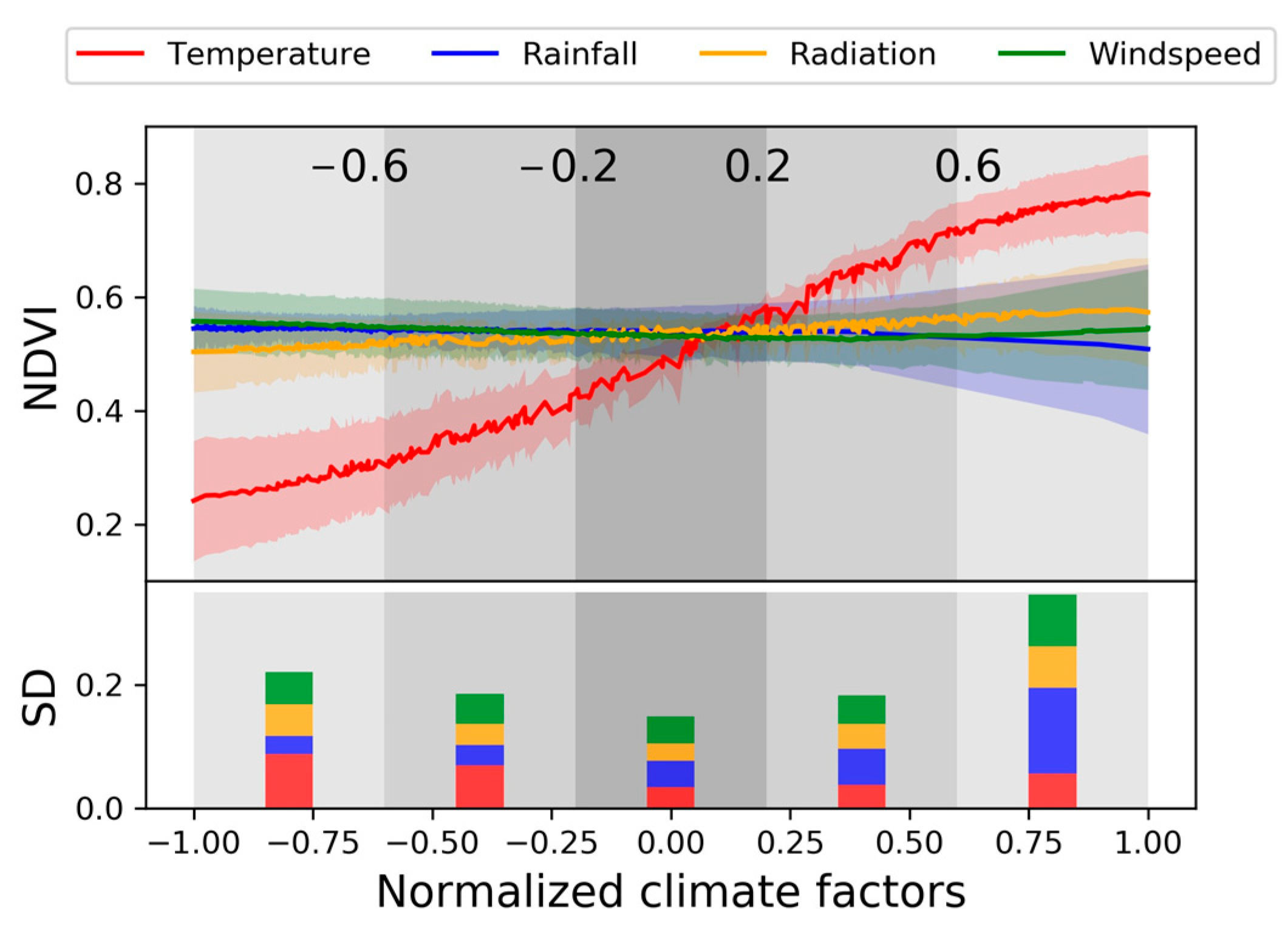
3.1. Overall Variation of the PDPs
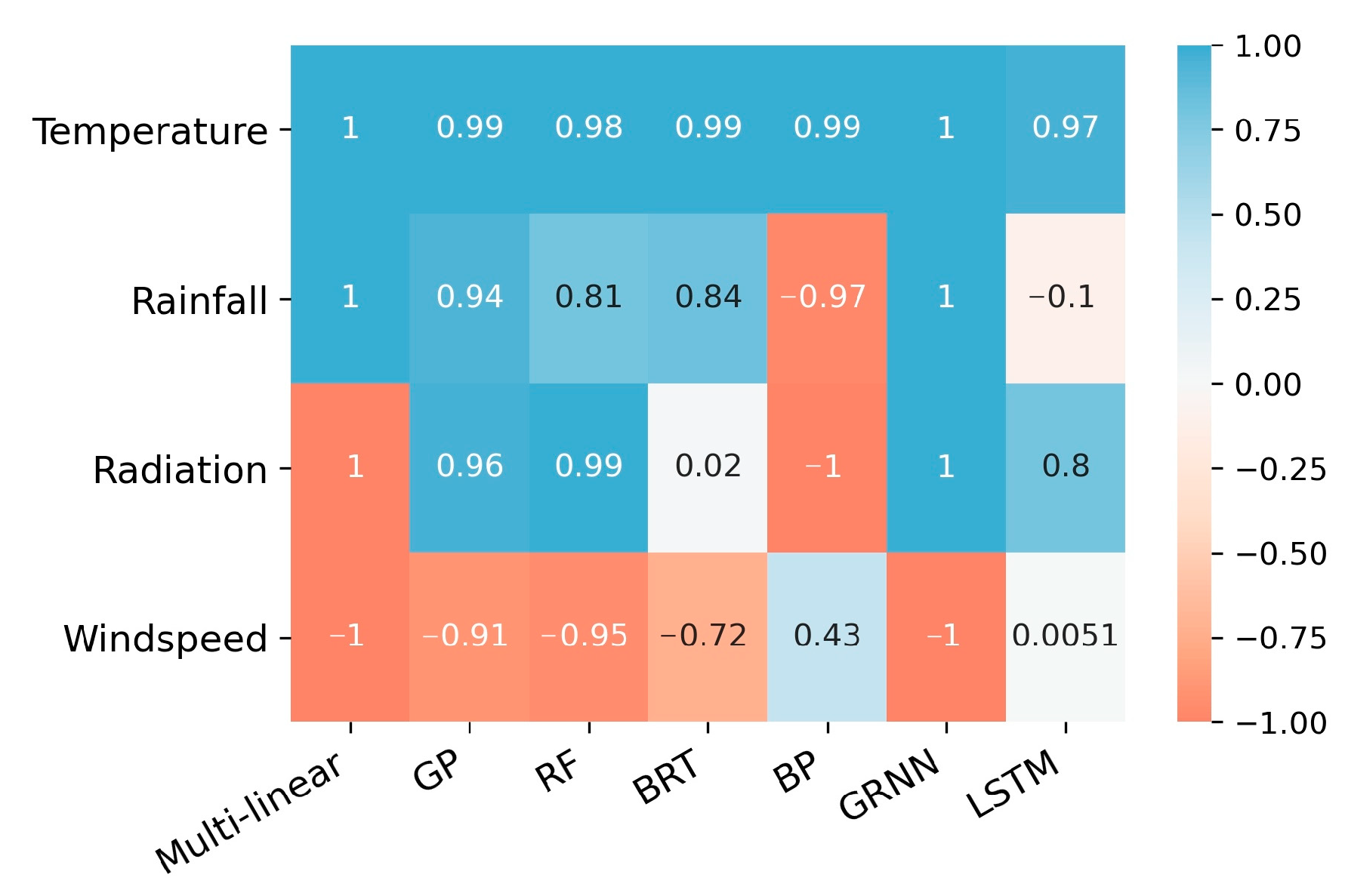
3.2. Linear Trend of PDP
3.3. Characteristics and Influencing Factors of Change Points
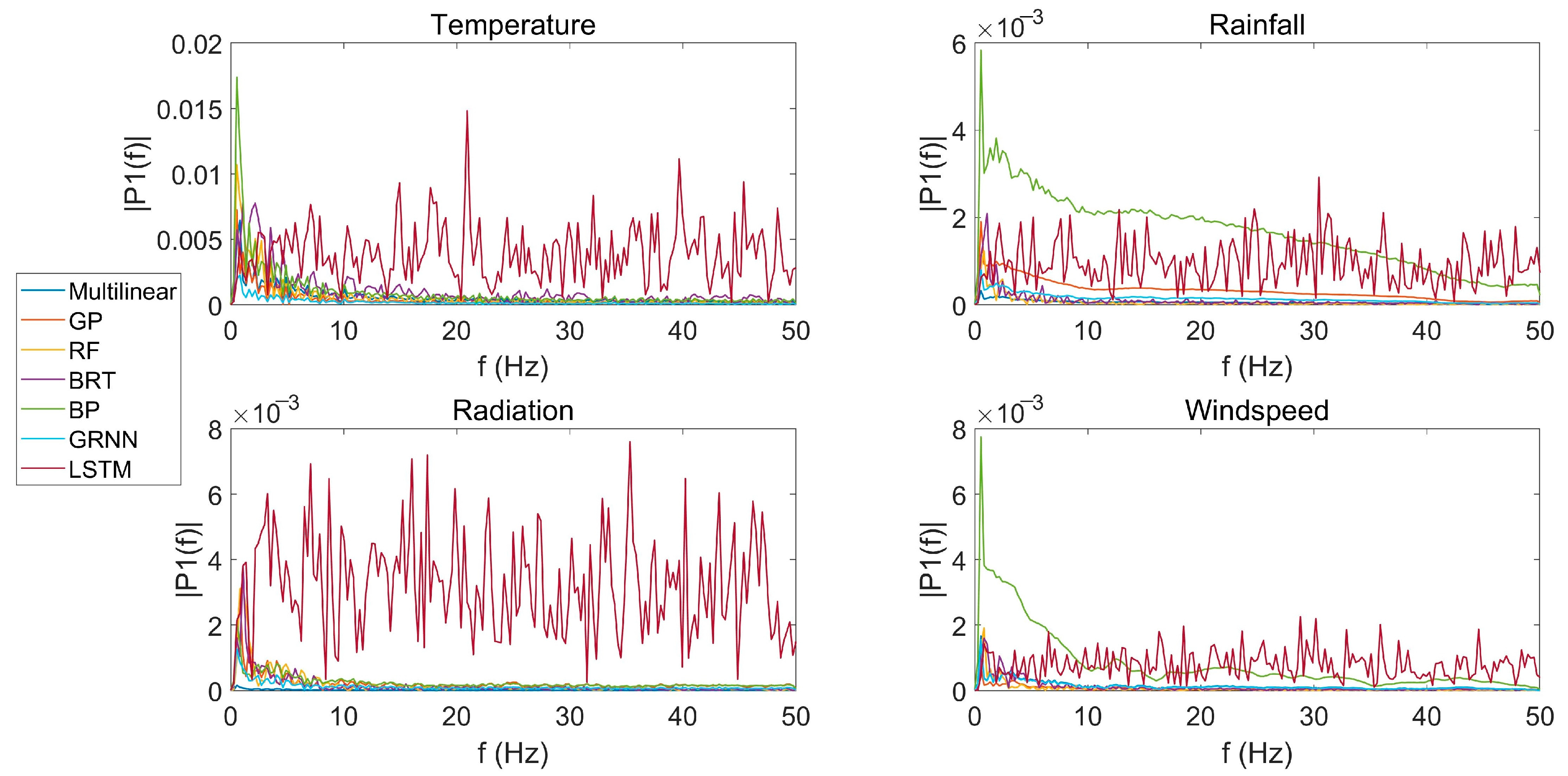
3.4. Fluctuation of PDP in the Frequency Domain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ma, X.; Huete, A.; Moran, S.; Ponce-Campos, G.; Eamus, D. Abrupt shifts in phenology and vegetation productivity under climate extremes. J. Geophys. Res. Biogeosci. 2015, 120, 2036–2052. [Google Scholar] [CrossRef]
- Piao, S.; Wang, X.; Park, T.; Chen, C.; Lian, X.; He, Y.; Bjerke, J.W.; Chen, A.; Ciais, P.; Tømmervik, H. Characteristics, drivers and feedbacks of global greening. Nat. Rev. Earth Environ. 2020, 1, 14–27. [Google Scholar] [CrossRef]
- Liu, Y.; Kumar, M.; Katul, G.G.; Porporato, A. Reduced resilience as an early warning signal of forest mortality. Nat. Clim. Chang. 2019, 9, 880–885. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Schwalm, C.R.; Gentine, P.; Smith, W.K.; Ciais, P.; Kimball, J.S.; Gazol, A.; Kannenberg, S.A.; Chen, A. Widespread spring phenology effects on drought recovery of Northern Hemisphere ecosystems. Nat. Clim. Chang. 2023, 13, 182–188. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhu, Z.; Zeng, H.; Myneni, R.B.; Zhang, Y.; Peñuelas, J.; Piao, S. Seasonal peak photosynthesis is hindered by late canopy development in northern ecosystems. Nat. Plants 2022, 8, 1484–1492. [Google Scholar] [CrossRef]
- Chen, L.; Hänninen, H.; Rossi, S.; Smith, N.G.; Pau, S.; Liu, Z.; Feng, G.; Gao, J.; Liu, J. Leaf senescence exhibits stronger climatic responses during warm than during cold autumns. Nat. Clim. Chang. 2020, 10, 777–780. [Google Scholar] [CrossRef]
- Wu, C.; Wang, J.; Ciais, P.; Peñuelas, J.; Zhang, X.; Sonnentag, O.; Tian, F.; Wang, X.; Wang, H.; Liu, R. Widespread decline in winds delayed autumn foliar senescence over high latitudes. Proc. Natl. Acad. Sci. USA 2021, 118, e2015821118. [Google Scholar] [CrossRef]
- Moles, A.T.; Perkins, S.E.; Laffan, S.W.; Flores-Moreno, H.; Awasthy, M.; Tindall, M.L.; Sack, L.; Pitman, A.; Kattge, J.; Aarssen, L.W. Which is a better predictor of plant traits: Temperature or precipitation? J. Veg. Sci. 2014, 25, 1167–1180. [Google Scholar] [CrossRef]
- Collalti, A.; Ibrom, A.; Stockmarr, A.; Cescatti, A.; Alkama, R.; Fernández-Martínez, M.; Matteucci, G.; Sitch, S.; Friedlingstein, P.; Ciais, P.; et al. Forest production efficiency increases with growth temperature. Nat. Commun. 2020, 11, 5322. [Google Scholar] [CrossRef]
- Zellweger, F.; De Frenne, P.; Lenoir, J.; Vangansbeke, P.; Verheyen, K.; Bernhardt-Römermann, M.; Baeten, L.; Hédl, R.; Berki, I.; Brunet, J.; et al. Forest microclimate dynamics drive plant responses to warming. Science 2020, 368, 772–775. [Google Scholar] [CrossRef]
- Murray, K.; Conner, M.M. Methods to quantify variable importance: Implications for the analysis of noisy ecological data. Ecology 2009, 90, 348–355. [Google Scholar] [CrossRef]
- Meyer, H.; Pebesma, E. Machine learning-based global maps of ecological variables and the challenge of assessing them. Nat. Commun. 2022, 13, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Piao, S.; Wang, K.; Wang, X.; Wang, T.; Ciais, P.; Chen, A.; Lian, X.; Peng, S.; Peñuelas, J. Temporal trade-off between gymnosperm resistance and resilience increases forest sensitivity to extreme drought. Nat. Ecol. Evol. 2020, 4, 1075–1083. [Google Scholar] [CrossRef]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N.; Prabhat. Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Shen, H.; Li, T.; Li, Z.; Li, S.; Jiang, Y.; Xu, H.; Tan, W.; Yang, Q.; Wang, J.; et al. Deep learning in environmental remote sensing: Achievements and challenges. Remote Sens. Environ. 2020, 241, 111716. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Förster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of feature reduction algorithms for classifying tree species with hyperspectral data on three central European test sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Kosicki, J.Z. Generalised Additive Models and Random Forest Approach as effective methods for predictive species density and functional species richness. Environ. Ecol. Stat. 2020, 27, 273–292. [Google Scholar] [CrossRef]
- Lucas, T.C.D. A translucent box: Interpretable machine learning in ecology. Ecol. Monogr. 2020, 90, e01422. [Google Scholar] [CrossRef]
- Lipton, Z.C. The Mythos of Model Interpretability: In machine learning, the concept of interpretability is both important and slippery. Queue 2018, 16, 31–57. [Google Scholar] [CrossRef]
- Wu, M.; Hughes, M.; Parbhoo, S.; Zazzi, M.; Roth, V.; Doshi-Velez, F. Beyond sparsity: Tree regularization of deep models for interpretability. In Proceedings of the Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Gevrey, M.; Dimopoulos, I.; Lek, S. Review and comparison of methods to study the contribution of variables in artificial neural network models. Ecol. Model. 2003, 160, 249–264. [Google Scholar] [CrossRef]
- Olden, J.D.; Joy, M.K.; Death, R.G. An accurate comparison of methods for quantifying variable importance in artificial neural networks using simulated data. Ecol. Model. 2004, 178, 389–397. [Google Scholar] [CrossRef]
- Greenwell, B.M. pdp: An R Package for constructing partial dependence plots. R J. 2017, 9, 421. [Google Scholar] [CrossRef]
- Shi, H.; Yang, N.; Yang, X.; Tang, H. Clarifying Relationship between PM2. 5 Concentrations and Spatiotemporal Predictors Using Multi-Way Partial Dependence Plots. Remote Sens. 2023, 15, 358. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, Y.; Zhou, S.; Song, J.; Fu, B. Soil moisture determines the recovery time of ecosystems from drought. Glob. Chang. Biol. 2023, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.K.F.; Lantz, T.C.; Fraser, R.H.; Hogan, D. High Arctic vegetation change mediated by hydrological conditions. Ecosyst. 2021, 24, 106–121. [Google Scholar] [CrossRef]
- Zhang, Y.; Keenan, T.F.; Zhou, S. Exacerbated drought impacts on global ecosystems due to structural overshoot. Nat. Ecol. Evol. 2021, 5, 1490–1498. [Google Scholar] [CrossRef]
- Wu, X.; Li, X.; Chen, Y.; Bai, Y.; Tong, Y.; Wang, P.; Liu, H.; Wang, M.; Shi, F.; Zhang, C. Atmospheric water demand dominates daily variations in water use efficiency in alpine meadows, northeastern Tibetan Plateau. J. Geophys. Res. Biogeosci. 2019, 124, 2174–2185. [Google Scholar] [CrossRef]
- Schaffers, A.P. Soil, biomass, and management of semi-natural vegetation–Part II. Factors controlling species diversity. Plant Ecol. 2002, 158, 247–268. [Google Scholar] [CrossRef]
- Ingram, M.; Vukcevic, D.; Golding, N. Multi-output Gaussian processes for species distribution modelling. Methods Ecol. Evol. 2020, 11, 1587–1598. [Google Scholar] [CrossRef]
- Peters, J.; Verhoest, N.E.C.; Samson, R.; Boeckx, P.; De Baets, B. Wetland vegetation distribution modelling for the identification of constraining environmental variables. Landsc. Ecol. 2008, 23, 1049–1065. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Oppel, S.; Strobl, C.; Huettmann, F. Alternative Methods to Quantify Variable Importance in Ecology; University of Munich: Munich, Germany, 2009. [Google Scholar]
- Vidal-Macua, J.J.; Nicolau, J.M.; Vicente, E.; Moreno-de Las Heras, M. Assessing vegetation recovery in reclaimed opencast mines of the Teruel coalfield (Spain) using Landsat time series and boosted regression trees. Sci. Total Environ. 2020, 717, 137250. [Google Scholar] [CrossRef] [PubMed]
- Zhi, J.; Zhou, Z.; Cao, X. Exploring the determinants and distribution patterns of soil mattic horizon thickness in a typical alpine environment using boosted regression trees. Ecol. Indic. 2021, 133, 108373. [Google Scholar] [CrossRef]
- Li, M.-Y.; Lai, X.-J. Evaluation on ecological security of urban land based on BP neural network-a case study of Guangzhou. Econ. Geogr. 2011, 31, 289–293. [Google Scholar]
- Xu, B.; Zhang, H.; Wang, Z.; Wang, H.; Zhang, Y. Model and algorithm of BP neural network based on expanded multichain quantum optimization. Math. Probl. Eng. 2015, 2015, 362150. [Google Scholar] [CrossRef]
- Li, J.; Cheng, J.-h.; Shi, J.-y.; Huang, F. Brief introduction of back propagation (BP) neural network algorithm and its improvement. In Advances in Computer Science and Information Engineering; Springer: Berlin/Heidelberg, Germany, 2012; pp. 553–558. [Google Scholar]
- Jia, K.; Liang, S.; Liu, S.; Li, Y.; Xiao, Z.; Yao, Y.; Jiang, B.; Zhao, X.; Wang, X.; Xu, S. Global land surface fractional vegetation cover estimation using general regression neural networks from MODIS surface reflectance. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4787–4796. [Google Scholar] [CrossRef]
- Specht, D.F. A general regression neural network. IEEE Trans. Neural Netw. 1991, 2, 568–576. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, H.; Xu, C.; Wu, X.; Liang, B.; Cao, J.; Chen, D. Modeling vegetation greenness and its climate sensitivity with deep-learning technology. Ecol. Evol. 2021, 11, 7335–7345. [Google Scholar] [CrossRef]
- Chen, Z.-T.; Liu, H.-Y.; Xu, C.-Y.; Wu, X.-C.; Liang, B.-Y.; Cao, J.; Chen, D. Deep learning projects future warming-induced vegetation growth changes under SSP scenarios. Adv. Clim. Chang. Res. 2022, 13, 251–257. [Google Scholar] [CrossRef]
- Sulla-Menashe, D.; Friedl, M.A. User Guide to Collection 6 MODIS Land Cover (MCD12Q1 and MCD12C1) Product; USGS: Rest, VA, USA, 2018. [Google Scholar]
- Didan, K. MOD13C1 MODIS/Terra Vegetation Indices 16-Day L3 Global 0.05 Deg CMG V006 [Dataset]. In NASA EOSDIS Land Process; DAAC: Greenbelt, MD, USA, 2015. [Google Scholar]
- Guo, X.; Zhang, H.; Wu, Z.; Zhao, J.; Zhang, Z. Comparison and evaluation of annual NDVI time series in China derived from the NOAA AVHRR LTDR and Terra MODIS MOD13C1 products. Sensors 2017, 17, 1298. [Google Scholar] [CrossRef]
- Weedon, G.P.; Balsamo, G.; Bellouin, N.; Gomes, S.; Best, M.J.; Viterbo, P. The WFDEI meteorological forcing data set: WATCH Forcing Data methodology applied to ERA-Interim reanalysis data. Water Resour. Res. 2014, 50, 7505–7514. [Google Scholar] [CrossRef]
- Molnar, C.; Freiesleben, T.; König, G.; Casalicchio, G.; Wright, M.N.; Bischl, B. Relating the partial dependence plot and permutation feature importance to the data generating process. arXiv 2021, arXiv:2109.01433. [Google Scholar]
- Moosbauer, J.; Herbinger, J.; Casalicchio, G.; Lindauer, M.; Bischl, B. Explaining hyperparameter optimization via partial dependence plots. Adv. Neural Inf. Process Syst. 2021, 34, 2280–2291. [Google Scholar]
- Hiura, T.; Go, S.; Iijima, H. Long-term forest dynamics in response to climate change in northern mixed forests in Japan: A 38-year individual-based approach. For. Ecol. Manag. 2019, 449, 117469. [Google Scholar] [CrossRef]
- Jin, G.; Liu, D. Mid-Holocene climate change in North China, and the effect on cultural development. Chin. Sci. Bull. 2002, 47, 408–413. [Google Scholar] [CrossRef]
- Pichler, M.; Boreux, V.; Klein, A.M.; Schleuning, M.; Hartig, F. Machine learning algorithms to infer trait-matching and predict species interactions in ecological networks. Methods Ecol. Evol. 2020, 11, 281–293. [Google Scholar] [CrossRef]
- Ryo, M.; Angelov, B.; Mammola, S.; Kass, J.M.; Benito, B.M.; Hartig, F. Explainable artificial intelligence enhances the ecological interpretability of black-box species distribution models. Ecography 2021, 44, 199–205. [Google Scholar] [CrossRef]
- Visani, G.; Bagli, E.; Chesani, F.; Poluzzi, A.; Capuzzo, D. Statistical stability indices for LIME: Obtaining reliable explanations for machine learning models. J. Oper. Res. Soc. 2022, 73, 91–101. [Google Scholar] [CrossRef]
- Bowen, D.; Ungar, L. Generalized SHAP: Generating multiple types of explanations in machine learning. arXiv 2020, arXiv:2006.07155. [Google Scholar]
- Mangalathu, S.; Hwang, S.-H.; Jeon, J.-S. Failure mode and effects analysis of RC members based on machine-learning-based SHapley Additive exPlanations (SHAP) approach. Eng. Struct. 2020, 219, 110927. [Google Scholar] [CrossRef]
- Maynard, D.S.; Bialic-Murphy, L.; Zohner, C.M.; Averill, C.; van den Hoogen, J.; Ma, H.; Mo, L.; Smith, G.R.; Acosta, A.T.R.; Aubin, I.; et al. Global relationships in tree functional traits. Nat. Commun. 2022, 13, 3185. [Google Scholar] [CrossRef]
- Bellot, S.; Lu, Y.; Antonelli, A.; Baker, W.J.; Dransfield, J.; Forest, F.; Kissling, W.D.; Leitch, I.J.; Nic Lughadha, E.; Ondo, I.; et al. The likely extinction of hundreds of palm species threatens their contributions to people and ecosystems. Nat. Ecol. Evol. 2022, 6, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Webb, E.E.; Liljedahl, A.K.; Cordeiro, J.A.; Loranty, M.M.; Witharana, C.; Lichstein, J.W. Permafrost thaw drives surface water decline across lake-rich regions of the Arctic. Nat. Clim. Chang. 2022, 12, 841–846. [Google Scholar] [CrossRef]
- Hamida, S.; El Gannour, O.; Cherradi, B.; Ouajji, H.; Raihani, A. Optimization of machine learning algorithms hyper-parameters for improving the prediction of patients infected with COVID-19. In Proceedings of the 2020 IEEE 2nd International Conference on Electronics, Control, Optimization and Computer Science (Icecocs), Kenitra, Morocco, 2–3 December 2020; pp. 1–6. [Google Scholar]
- Subramanian, M.; Shanmugavadivel, K.; Nandhini, P. On fine-tuning deep learning models using transfer learning and hyper-parameters optimization for disease identification in maize leaves. Neural Comput. Appl. 2022, 34, 13951–13968. [Google Scholar] [CrossRef]
| Category | Model Name | General Equation | Key Parameterization | Reference |
|---|---|---|---|---|
| Linear Model | Multivariate linear model | - | [6,28,29] | |
| Non-parametric model | Gaussian process model (GP) | Default kernel function with parameters | [30] | |
| Regression tree models | Random forest (RF) | - | A total of 200 trees; minimum number of observations per tree leaf is 5 | [31,32,33] |
| Boosted regression tree (BRT) | - | Minimum number of observations per tree leaf is 5; learning rate is 0.01 | [34,35] | |
| Artificial neural network models | Back propagation (BP) neural network | - | Single hidden layer with 10 neurons; learning rate is 0.01 | [36,37,38] |
| General regression neural network (GRNN) | - | Spread of radial basis functions is 1 | [39,40] | |
| Long short-term memory (LSTM) neural network | - | One LSTM layer with 500 hidden units and one fully connected layer; learning rate is 0.008 | [41,42] |
| Independent Factor of PDP | |||||
|---|---|---|---|---|---|
| Maximum Number of Change Points | Models | Temperature | Rainfall | Radiation | Windspeed |
| 1 | Multi-linear | 0.58 | −0.39 | −0.34 | 0.39 |
| GP | 0.26 | 0.14 | 0.53 | 0.05 | |
| RF | 0.31 | −0.84 | −0.21 | −0.41 | |
| BRT | 0.50 | −0.65 | 0.28 | - | |
| BP | −0.02 | 0.42 | - | 0.35 | |
| GRNN | 0.58 | −0.21 | −0.28 | 0.38 | |
| LSTM | 0.03 | −0.67 | −0.13 | 0.02 | |
| Mean deviation | 0.23 | 0.43 | 0.32 | 0.29 | |
| Mean relative variation | 11.5% | 21.4% | 15.9% | 14.3% | |
| 2 | Multi-linear | [−0.25, 0.61] | [−0.61, 0.05] | −0.34 | 0.39 |
| GP | [−0.47, 0.58] | [0.02, 0.90] | [−0.20, 0.46] | [−0.11, 0.25] | |
| RF | [−0.32, 0.56] | [−0.93, −0.83] | [−0.60, −0.22] | [−0.39, 0.07] | |
| BRT | [−0.21, 0.56] | [−0.91, −0.65] | [−0.42, 0.28] | [−0.32, −0.04] | |
| BP | [−0.26, 0.37] | [−0.64, 0.90] | [−0.28, 0.75] | [−0.01, 0.52] | |
| GRNN | [−0.25, 0.61] | [−0.59, 0.09] | [−0.44, 0.02] | [0.18, 0.53] | |
| LSTM | [−0.21, 0.69] | [−0.59, −0.58] | −0.13 | [−0.08, 0.07] | |
| Mean deviation | 0.09 | 0.48 | 0.24 | 0.21 | |
| Mean relative deviation | 4.4% | 23.8% | 11.9% | 10.3% | |
| 3 | Multi-linear | [−0.25, 0.61] | [−0.63, −0.10, 0.90] | [−0.44, 0.02, 0.75] | [−0.68, 0.13, 0.52] |
| GP | [−0.46, −0.02, 0.61] | [−0.88, 0.02, 0.90] | [−0.25, 0.40, 0.75] | [−0.23, 0.09, 0.42] | |
| RF | [−0.26, 0.29, 0.63] | [−0.93, −0.83, −0.65] | [−0.60, −0.22, 0.52] | [−0.45, −0.32, 0.07] | |
| BRT | [−0.54, −0.16, 0.56] | [−0.91, −0.65, −0.39] | [−0.42, 0.20 ,0.42] | [−0.32, −0.05, 0.47] | |
| BP | [−0.39, −0.02, 0.47] | [−0.72, 0.05, 0.90] | [−0.44, 0.06, 0.75] | [−0.19, 0.21, 0.54] | |
| GRNN | [−0.42, −0.02, 0.63] | [−0.63, −0.07, 0.90] | [−0.44, 0.02, 0.75] | [−0.68, 0.13, 0.52] | |
| LSTM | [−0.21, 0.69] | [−0.97, −0.59, −0.58] | [−0.22, 0.07, 0.16] | [−0.11, −0.08, 0.07] | |
| Mean deviation | 0.10 | 0.40 | 0.17 | 0.19 | |
| Mean relative variation | 5.0% | 19.8% | 8.5% | 9.6% | |
| Sum_sq | df | F | p | |
|---|---|---|---|---|
| C(Factor, Sum) | 51.57 | 3 | 0.24 | 0.87 |
| C(Model, Sum) | 186,817.21 | 6 | 427.85 | 0.00 |
| Residual | 1309.93 | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, B.; Liu, H.; Cressey, E.L.; Xu, C.; Shi, L.; Wang, L.; Dai, J.; Wang, Z.; Wang, J. Uncertainty of Partial Dependence Relationship between Climate and Vegetation Growth Calculated by Machine Learning Models. Remote Sens. 2023, 15, 2920. https://doi.org/10.3390/rs15112920
Liang B, Liu H, Cressey EL, Xu C, Shi L, Wang L, Dai J, Wang Z, Wang J. Uncertainty of Partial Dependence Relationship between Climate and Vegetation Growth Calculated by Machine Learning Models. Remote Sensing. 2023; 15(11):2920. https://doi.org/10.3390/rs15112920
Chicago/Turabian StyleLiang, Boyi, Hongyan Liu, Elizabeth L. Cressey, Chongyang Xu, Liang Shi, Lu Wang, Jingyu Dai, Zong Wang, and Jia Wang. 2023. "Uncertainty of Partial Dependence Relationship between Climate and Vegetation Growth Calculated by Machine Learning Models" Remote Sensing 15, no. 11: 2920. https://doi.org/10.3390/rs15112920
APA StyleLiang, B., Liu, H., Cressey, E. L., Xu, C., Shi, L., Wang, L., Dai, J., Wang, Z., & Wang, J. (2023). Uncertainty of Partial Dependence Relationship between Climate and Vegetation Growth Calculated by Machine Learning Models. Remote Sensing, 15(11), 2920. https://doi.org/10.3390/rs15112920










