Abstract
Monitoring and assessing vegetation using deep learning approaches has shown promise in forestry applications. Sample labeling to represent forest complexity is the main limitation for deep learning approaches for remote sensing vegetation classification applications, and few studies have focused on the impact of sample labeling methods on model performance and model training efficiency. This study is the first-of-its-kind that uses Mask region-based convolutional neural networks (Mask R-CNN) to evaluate the influence of sample labeling methods (including sample size and sample distribution) on individual tree-crown detection and delineation. A flight was conducted over a plantation with Fokienia hodginsii as the main tree species using a Phantom4-Multispectral (P4M) to obtain UAV imagery, and a total of 2061 manually and accurately delineated tree crowns were used for training and validating (1689) and testing (372). First, the model performance of three pre-trained backbones (ResNet-34, ResNet-50, and ResNet-101) was evaluated. Second, random deleting and clumped deleting methods were used to repeatedly delete 10% from the original sample set to reduce the training and validation set, to simulate two different sample distributions (the random sample set and the clumped sample set). Both RGB image and Multi-band images derived from UAV flights were used to evaluate model performance. Each model’s average per-epoch training time was calculated to evaluate the model training efficiency. The results showed that ResNet-50 yielded a more robust network than ResNet-34 and ResNet-101 when the same parameters were used for Mask R-CNN. The sample size determined the influence of sample labeling methods on the model performance. Random sample labeling had lower requirements for sample size compared to clumped sample labeling, and unlabeled trees in random sample labeling had no impact on model training. Additionally, the model with clumped samples provides a shorter average per-epoch training time than the model with random samples. This study demonstrates that random sample labeling can greatly reduce the requirement of sample size, and it is not necessary to accurately label each sample in the image during the sample labeling process.
1. Introduction
Detecting and delineating individual tree crowns is essential for evaluating forest ecosystems and management [1]. Forestry requires up-to-date tree attribute information for forest management [2]. However, traditional in situ field surveys are extremely time-consuming, labor-intensive, and inefficient [3,4,5]. With the application of remote sensing technology in forestry, many researchers have highlighted the potential of unmanned aerial vehicles (UAVs) data for tree-level surveys [6,7,8]. Many remote sensing methods have been applied to assist with tree crown detection, including watershed segmentation [9,10], template matching [11], edge detection, and region-growing [12,13]. Deep learning techniques have been increasingly used in recent years due to the capability of resolving a wide variety of computer vision problems, such as object recognition and detection [14,15,16,17].
In forestry, the deep learning approach is designed to encode spectral and spatial information from remote sensing images for extracting tree-related information, including individual trees detection or the segmentation of vegetation classes [16,18,19,20]. One of the deep learning capabilities is object detection, which is applied to detect countable things with definable spatial extents, such as single-shot multibox detectors (SSDs) [21] and faster-region convolutional neural networks (Faster R-CNNs) [18,22]. The object detection results are presented in the form of the rectangular bounding boxes but cannot explicitly define vegetation boundaries. Compared to object detection, semantic segmentation is capable of segmenting and classifying the target object in remote sensing data, such as fully convolutional networks (FCN) [23,24], DeepLabv3+ [25], and U-Net [19,26,27]. However, the main problem with semantic segmentation is that it is unable to segment continuous individuals from the same class. Further instance segmentation is developed to perform object detection and semantic segmentation tasks simultaneously and is considered as a combination of object detection and semantic segmentation. Mask region-based convolutional neural networks (Mask R-CNNs) are the most common technique for the instance segmentation approach, which was introduced in 2017 [28]. It is designed to detect individual things (e.g., individual plants) in images to predict a segmentation mask for each object. In a forestry context, instance segmentation helps identify tree number, crown size, and the distance between trees, which can contribute to forest management [29,30,31,32]. However, to our knowledge, although there has been an increase in the application of CNNs in recent years, most researches mainly rely on semantic segmentation for tree detection and counting, and few studies have focused on instance segmentation to extract various vegetation properties from remote sensing images, especially when utilizing high spatial resolution UAV imagery [16] with different sample sizes and sample labeling methods.
It is not practical to fully design and train a new CNN in many applications due to the large amount of required training samples and high computational costs [33,34]. Transfer learning is a common technique to overcome this obstacle because a pre-trained network can be applied to a new target task [27,34,35]. The principle of transfer learning can be regarded as using generic features (learned in earlier layers) to extract features from images that do not necessarily contain the target metric or class. Specifically, it is possible to adjust a pre-trained network for a new task. There are many existing transfer learning approaches that have already been applied in forestry [18,20,25,36]. For example, Fromm et al. [21] reported the size of the seedlings for model training could be reduced to a couple of hundred without obvious loss of accuracy with the application of a pre-trained network. However, in the case of transfer learning, it is not possible to adjust the complete deep architecture of a pre-trained backbone. Therefore, it is important to select the appropriate pre-trained backbone. Comparing three different detectors for conifer seedling identification, Fromm et al. [21] reported that Faster R-CNN and R-FCN benefit from a pre-trained backbone; however, the mean average precision of SSD was decreased by 0.07 when the pre-trained backbone was used. Iqbal et al. [37] compared the Mask R-CNN model with ResNet-50 and ResNet-101 to identify and segment coconut trees and reported that ResNet-101 provided for more accurate detection. Mo et al. [38] reported that the YOLACT model with ResNet-50 is sufficient for the segmentation task and that the deeper backbone of ResNet-101 did not improve the results because the binary classification task is less difficult. In addition, different backbone networks can influence the training speed. For example, Ferreira et al. [31] reported ResNet-18 provided the shortest training time compared to the ResNet-50 and MobileNetV2, although a similar average producer’s accuracy was achieved for Brazil nut tree (Bertholletia excelsa) detection. The input image can also impact model performance during transfer learning. For example, Mahdianpari et al. [39] demonstrated that higher accuracy was achieved by the full-training of pre-existing ConvNets using five bands compared to the full-training and fine-tuning using tree bands.
A training dataset with extensive training samples is one of the main limitations for CNN utilization (including Mask R-CNN) [18,40]. It is a considerable task to measure tree crown boundaries in the field [3,4], and this information is commonly acquired by using GIS-based visual image interpretation to annotate samples [4,41]. It is time-consuming and costly to delineate large numbers of training samples by manual methods because the delineation of tree crowns needs to be cross-checked by an experienced visual interpreter. Several strategies have been used to manage the high training set requirements. One approach is to apply data augmentation during model training. Data augmentation is the process of generating more samples by manipulating the original data, including shifting, flipping, rotation, translation, shearing, scaling, and brightness changes [18,42,43,44]. By transforming original images into Laplacian pyramid images as multiscale datasets, Zhao and Du [44] reported a significant increase in classification accuracy. Similarly, Fromm et al. [21] reported that data augmentation for SSD improved the performance of smaller seedling detection by 0.27. Another alternative method is to use a weakly- and semi-supervised learning method to compensate for the limited availability of reference data and decrease manual labeling costs [45]. For example, Wu et al. [46] proposed a weakly-supervised learning method using only scribble annotations to supervise deep convolutional neural networks for segmentation. Additionally, researchers recently combined hyperspectral data to reduce reliance on the size of the annotation samples. La Rosa et al. [24] proposed a partial loss function to train an FCN, and achieved good accuracy (average user’s accuracy: 88.63%, average producer’s accuracy: 88.59%) for tree species classification using only 38 sparse and scarce trees as the training samples to detect 14 tree species. Zhang et al. [47] employed a 3D-1D CNN model to classify tree species using airborne hyperspectral data, yielding a 93.14% classification accuracy. In addition, previous studies have reported that CNN models can partially compensate for the errors in training sample delineation [18,48,49] and suggested that exhaustive annotation may not be warranted in some cases [18]. In general, the predictive accuracy of CNNs commonly benefits from many training samples, but it is often challenging to obtain this data. Additionally, having numerous training samples can increase the time required for model training because of computational demand. Conversely, if the number of training samples is too small, it may not be sufficient to avoid overfitting during the model training and can yield poor performance [24], even when data augmentation is used. Therefore, it is an important part of the deep learning approach to determine suitable sample size.
The main objective of this study is to explore the influence of sample labeling and spatial distribution on deep learning models for individual tree-crown detection and delineation. Here, two sample labeling methods were proposed to evaluate model performance. The first approach is to randomly reduce the training samples based on all of the manually delineated tree crowns. The remaining samples for the random sample set are distributed throughout the whole image, with many unlabeled samples remaining. The latter approach is to reduce the same training samples by region. This clumped sample set, which is accurately labeled in some regions of the image, is a commonly used method for sample labeling. The different sample sets that were developed by the two-sample labeling approaches were coupled with UAV-derived RGB imagery and Multi-band imagery for Mask R-CNN model training. The main reason for choosing Mask R-CNN is that transfer learning has a small sample size requirement, so it is possible to carry out field verification of the samples by visual interpretation to ensure label accuracy. Finally, 372 Fokienia hodginsii trees were delineated in the same location as test data to evaluate the Mask R-CNN model performance on the individual tree-crown detection and delineation.
2. Materials and Methods
2.1. Study Site
The study site is located in Anxi county, Fujian, China (coordinate: 25°20′55″N, 118°0′50″E) and covers an approximately 3.26 ha area. The region has a subtropical marine monsoon climate, with a mean annual temperature is 20 °C and a mean annual precipitation of 1600 mm. The site was previously a mid-subtropical evergreen broad-leaved forest. However, due to anthropogenic factors, this area is mainly covered by Fokienia hodginsii, and a small amount of Eucalyptus robusta and other broad-leaved trees (Figure 1). The Fokienia hodginsii was planted in 2003 and thinned in 2017. The elevation in this region is between 450 m and 540 m. The terrain of the study site is high in the west and low in the east, with different slopes and aspects in different locations.
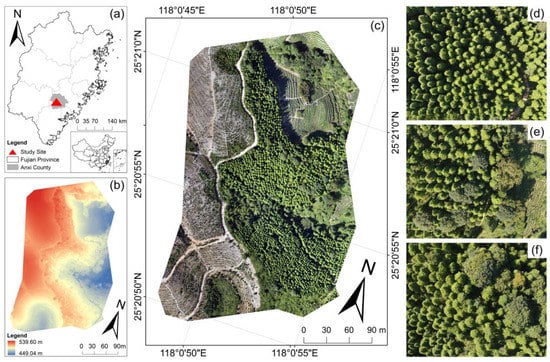
Figure 1.
Location of the study site, Anxi County, Fujian, China. (a) study site in Anxi County, Fujian. (b) the digital surface model (DSM) derived from UAV. (c) UAV image. (d–f) are detailed examples of Fokienia hodginsii with other tree species.
2.2. UAV Image Acquisition and Processing
The UAV imagery was acquired from a camera with six integrated lenses (1600 × 1300 pixels) using a Phantom4-Multispectral (P4M) (https://www.dji.com/p4-multispectral, 6 February 2022) in October 2020. Imagery with blue (450 ± 16 nm), green (560 ± 16 nm), red (650 ± 16 nm), red edge (730 ± 16 nm), near-infrared band (NIR) (840 ± 26 nm), and visible light (RGB) spectrum were captured with the camera. The P4M camera has a focal length of 5.74 mm, a sensor size of 4.87 × 3.96 mm, and a global 2 MP shutter. Similar to other UAV-mounted multispectral cameras (e.g., Parrot Sequoia), the P4M demonstrates good accuracy and consistent data generation in terms of spectral reflectance and vegetation index acquisition [50] and is capable of obtaining the multispectral and RGB image simultaneously.
The flight plan was programmed to acquire the UAV imagery in the study area using the DJI Ground Station Pro application. The flight altitude was 80 m, with an 85% forward overlap rate and 80% side-lap [51]. In addition, the acquired images were directly georeferenced by the global positioning system (GPS) during the flight because the GPS is included in the P4M (https://www.dji.com/p4-multispectral/specs, 6 February 2022). The georeferencing system’s vertical and horizontal location can reach the positioning accuracy of ±0.1 m and ±0.3 m, respectively [52,53].
A total of 3456 images were acquired, with each camera lens taking 576 images for this study. The UAV imagery was processed in the DJI Terra software to generate blue, green, red, red edge, near-infrared-band ortho-mosaics, RGB, and a digital surface model (DSM). Then the ortho-mosaics were created as a Multi-band image (blue, green, red, red edge, and near-infrared bands) in this study. The pixel resolution of RGB image and Multi-band image was 4 cm pixel−1.
2.3. Individual Tree-Crown Sample Collection
Deep learning models require accurately delineated tree crowns for input data and validation data. In this study, it was possible to delineate tree crowns manually due to the Fokienia hodginsii clear and concentrated crown shape and the ability to avoid mixing the pixels from surrounding trees by visual interpretation of the imagery. All Fokienia hodginsii tree crowns were delineated manually based on the UAV imagery and GIS-based visual interpretation. Additionally, the delineated tree crowns were cross-checked by another interpreter to ensure the accuracy of tree crown delineation [19]. In total, 2061 tree crowns were delineated in this study.
2.4. Experiment Design and Variable Parameters
Figure 2 presents a framework of the proposed experimental design. First, different backbones coupled with a training and validation set (1689) were used for the Mask R-CNN model training to evaluate the accuracy of individual tree crown detection and delineation. The aim was to select the appropriate backbone for Mask R-CNN to detect Fokienia hodginsii. Second, to evaluate the sample size and sample labeling for the model performance, 1689 manually delineated tree crowns were deleted at a rate of 10% percentage by random and in a clumped manner, respectively. Moreover, the RGB image and the Multi-band image were selected as the input image to evaluate the influence of the input image on the model performance. Third, to evaluate the model training efficiency, the average pre-epoch training time of each model was compared. A total of 42 models were designed and tested in this study. The variable parameters are as follows in the following sections.
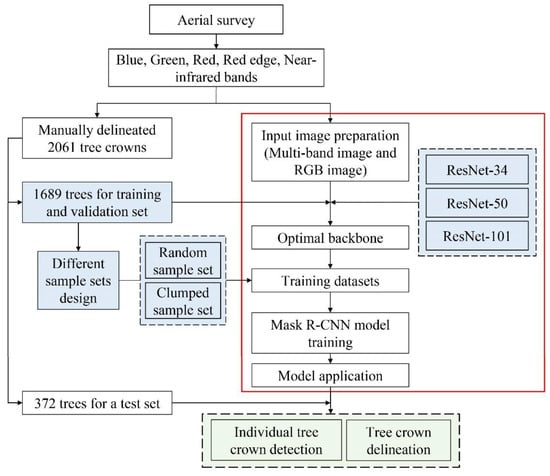
Figure 2.
The flowchart of individual tree crown detection and delineation. The light blue color represents the variables; the light green color represents the results from the Mask R-CNN model. The white color objects inside the red box represent the processing steps of Mask R-CNN.
2.4.1. Input Image
To test the optimal image type for Mask R-CNN, the RGB image from the RGB sensor and the Multi-band image from the UAV multi-band sensor were acquired for the input image.
2.4.2. Backbone
Residual Neural Network (ResNet) architecture proposed by He et al. [54] is a simple and efficient method to develop a deeper network. In this study, several pre-trained ResNet versions (ResNet-34, ResNet-50, and ResNet-101) were employed as the backbone network. The ResNet networks are similar, which consist of several residual blocks that activate the feature maps of a given layer to a deep layer. The ResNet-34, ResNet-50, and ResNet-101 differ by the number of layers, training speed, and intermediate features.
2.4.3. Training Samples
The Fokienia hodginsii trees from the study site were divided into two parts (Figure 3). A total of 1689 manually delineated trees in part 1 were used for model training, and 372 manually delineated trees in part 2 were used as test data. First, 1689 individual tree crowns were used as a training and validation set to choose the optimal backbone between ResNet-34, ResNet-50, and ResNet-101. Subsequently, the training and validation set (1689) was repeatedly reduced by 10%, that is, 1520 (90%), 1351 (80%), 1182 (70%), 1013 (60%), 844 (50%), 675 (40%), 506 (30%), 337 (20%), and 168 (10%) of the training and validation sets for model training, to evaluate the influence of sample size on model performance. Here, two methods for reducing the training and validation set were used: random deleting and clumped deleting, in order to simulate the label accuracy and labeling pattern during the delineated tree-crown acquisition in the sample preparation of the Mask R-CNN model to evaluate the influence of sample labeling on model performance. The training and validation set obtained by random deleting and clumped deleting are subsequently referred to as the random sample set and clumped sample set. The random sample set aims to imitate the situation of labeled samples that were widely distributed, but where not all the samples were labeled. The random sample sets were collected by randomly deleting 10% (169 trees) from the training and validation set (initially 1689), and then repeatedly deleting 169 samples from the former random sample set. A total of 9 random sample sets were developed for this study (Figure 4a–i). The clumped sample set imitates the case when the labeled samples are concentrated in some region of the image, and each tree is labeled. The clumped sample sets were deleted according to the distance from the test set in the image. The trees close to the test set were deleted first, and then 169 trees were deleted repeatedly (Figure 4j–r). A total of 9 clumped sample sets were developed. Overall, a total of 19 groups of training samples were created, and the distribution and location of the training samples are shown in Figure 3 and Figure 4.
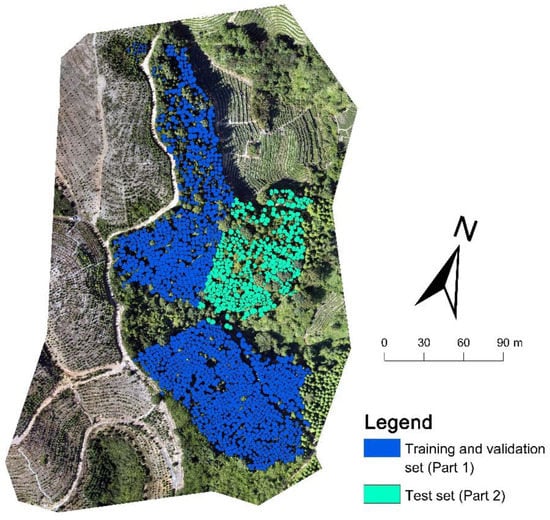
Figure 3.
The distribution of the manually delineated trees. The manually delineated trees in part 1 are shown with blue boundaries, and the manually delineated trees in part 2 are shown with green boundaries.
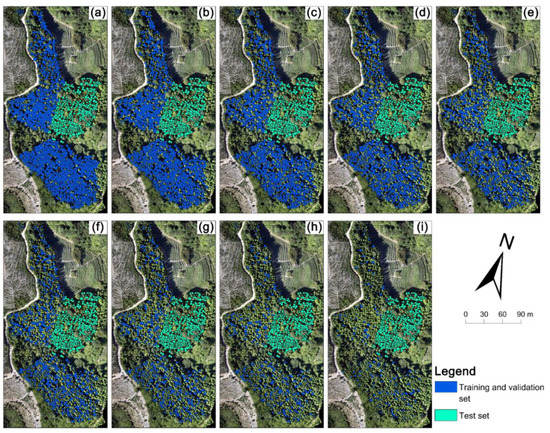
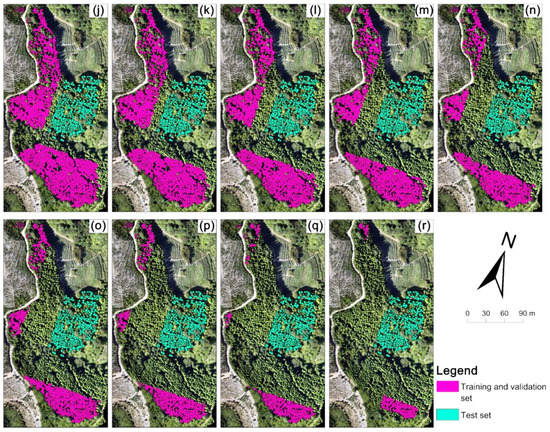
Figure 4.
The spatial distribution of training samples in the study site: (a–i) random sample sets with 1520, 1351, 1182, 1013, 844, 675, 506, 337, and 168 samples, which were the random labeling pattern. (j–r) clumped sample sets, which were the clumped labeling pattern. Green boundaries represent the test set, blue and pink boundaries represent the training and validation sets.
2.5. Mask R-CNN Model Training and Application
The Mask R-CNN mainly contains the following steps:
- (1)
- Preparation for the training dataset
To match the input constraints of Mask R-CNN architecture, the input image (RGB image and Multi-band image) (Section 2.4.1) was used to convert the training samples (Section 2.4.3) into the training dataset [18]. Moreover, the input image was split as image tiles (256 × 256 pixels), and a 50% overlap (128 × 128 pixels) was set for processing [55]. Then, the original orientation of the training samples was rotated (90°, 180°, and 270°) to increase the size of the training samples [20]. Finally, 38 training datasets were created, and each training dataset contained the generated tiles and features presented in Table 1.

Table 1.
The number of image tiles and features on each training dataset.
- (2)
- Model training
The Mask R-CNN model was trained in ArcGIS API for Python. In order to complete the task, the backbone can be adjusted to learn new features during the transfer learning process [56,57]. In addition, the epoch was set to 100 to train the networks, with a batch size of 4. Early stopping was used to reduce overfitting. If the validation loss did not improve in 5 epochs, the training would be stopped [58]. In this study, each training dataset was randomly divided into 90% training data and 10% validation data. The different training datasets and backbones were mentioned above (Section 2.4), and each training dataset implemented the same processing. A total of 2 training datasets (RGB image and Multi-band image) with a 1689 sample size were used to test the three backbone networks. The optimal backbone was then chosen for model training with the other 36 training datasets.
- (3)
- Model application
The trained Mask R-CNN model was applied to the corresponding input image to detect individual trees in the study site. The model output is a vector file containing the boundary of each identified tree. Overlapping and redundant tree crowns were removed using the non-maximum suppression algorithm [59]. For the tree crown identification, the confidence score >0.2 was acceptable, and the maximum overlap was 0.2 [58].
All models were run on a laptop with an Nvidia GeForce RTX 2060 GPU and 16 GB of memory.
2.6. Estimation of Individual Tree Detection and Delineation
Model performance was assessed by the accuracy of individual tree-crown detection and delineation. The assessment of each model was carried out by comparing model output with a test set of 372 manually delineated tree crowns. The individual tree-crown detection was evaluated using recall, precision, and F1 score [60,61]. Recall is the ratio of correctly identified trees of the test set. Precision is the ratio of correctly identified trees of the model. F1 score is the overall accuracy considering the recall and precision.
The precision of tree crown delineation was evaluated using the Intersection over Union (IoU). The definition of IoU is the ratio of the union and intersection between the area of the test set and the predicted tree-crown polygons [62]. The predicted tree-crown polygons were considered acceptable if the IoU was higher than 50% [20,63].
where TP (true positive) represents the correctly predicted trees by the model, FP (false positive) represents the erroneously detected trees by the model, such as other tree species, FN (false negative) represents the actual trees that were not identified by the model, Bactual is the tree crown boundaries of the test set, Bpredicted is the predicted tree crown boundaries from the model with the confidence score higher than 0.2.
3. Results
3.1. The Impact of Backbone for Model Performance
The accuracy assessment of individual tree-crown detection and delineation for the Mask R-CNN model with different backbones is shown in Figure 5. For the Multi-band image, the F1 score was 88.44% and the IoU was 79.04% for the model with ResNet-50, followed by ResNet-101 (F1 score: 86.25%, IoU: 77.92%) and ResNet-34 (F1 score: 72.89%, IoU: 74.81%). For the RGB image, the F1 score was 89.01% and the IoU was 79.91% for the model with ResNet-50, followed by ResNet-101 (F1 score: 84.52%, IoU: 77.20%) and ResNet-34 (F1 score: 62.83%, IoU: 75.15%). Overall, despite two input images (RGB image and Multi-band image) being used for the model, the ResNet-50 showed higher accuracy than ResNet-34 and ResNet-101.
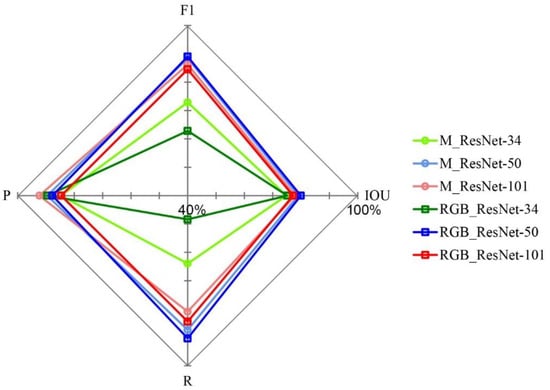
Figure 5.
The detection accuracy of Fokienia hodginsii using Mask R-CNN with three different backbones (ResNet-34, ResNet-50, and ResNet-101). The same number of training samples (1689 tree crowns) and two input images (RGB image and Multi-band image) were used as input into the model, and other parameters were kept the same. The accuracy was achieved using ResNet-34 with the Multi-band image, which is highlighted in light green, ResNet-34 with the RGB image is in dark green, ResNet-50 with the Multi-band image is in light blue, ResNet-50 with the RGB image is in dark blue, ResNet-101 with the Multi-band image is in light red, and ResNet-101 with the RGB image is in dark red. R, P, F1, and IoU represent recall, precision, F1 score, and Intersection over Union, respectively.
3.2. The Impact of the Number and Sample Labeling Method on the Model Performance
When different training datasets were used for model training, the performance of the Mask R-CNN models with ResNet-50 is shown in Table 2. It was found that with increasing sample size, the model’s accuracy first increased and then tended to stabilize. According to the F1 score and IoU values, it can be seen that the random sample set had lower sample size requirements. When the sample size reached 337 trees (337/1689), the model’s accuracy yielded an F1 score > 85% and IoU > 75%. While the clumped sample set needed 675 trees (675/1689) to reach a similar accuracy. From the perspective of the input image, Mask R-CNN with an RGB image achieved poorer accuracy than Mask R-CNN with a Multi-band image. From the sample labeling pattern, Mask R-CNN with a random sample set achieved superior accuracy when compared with Mask R-CNN with a clumped sample set. The results indicate that a model with the combination of a Multi-band image and the random sample set had the lowest requirements for sample size. For example, for the same number of training samples (337), the accuracy of the model using a random sample set with a Multi-band image yielded an F1 score of 87.92% and an IoU of 77.98%, followed by the model using a random sample set and RGB image (F1 score: 85.71%, IOU: 77.16%). The model using a clumped sample set and an RGB image had the lowest accuracy (F1 score: 78.04%, IoU: 70.66%).

Table 2.
Comparison of model performance with different sample sizes and sampling labeling. Sample sizes are sorted in descending order by a percentage of 10%. R, P, F1, and IoU represent recall, precision, F1 score, and Intersection over Union, respectively.
3.3. The Impact of the Different Sample Labeling Methods on Model Training Efficiency
Table 3 presents the average per-epoch time required for model training using different sample sizes, sample labeling methods, and input images. The average per-epoch time was calculated by the average training time for all epochs in each model. It can be seen that the training time was increased with increasing sample size and that random sample labeling provided a longer training time than clumped sample labeling. The Multi-band image provided a slightly longer training time than the RGB image. For the same sample labeling method, the average epochs time were similar for the Mask R-CNN models using the RGB image and the Multi-band image, with only a small difference that ranged between 0.2–0.9 min. For example, when the random sample set decreased from 1520 to 168, the average epoch time was reduced from 21.3 min to 11.1 min for the model using the Multi-band image, and the average epoch time was reduced from 21.1 min to 10.2 min for the model using the RGB image. It can be seen that the time is approximately reduced by half. However, the average epoch time decreased to 2.6 min for the Multi-band image and 2.0 min for the RGB image (168 training samples) when the clumped sample set was used, and the time efficiency was increased by ten times.

Table 3.
The average per-epoch time was obtained with the random and clumped labeling approaches used to train the Mask R-CNN architecture with the RGB and Multi-band images. Sample sizes are sorted in descending order by a percentage of 10%.
4. Discussion
4.1. Study Contribution
The use of Mask R-CNN for tree detection is still in development. To our knowledge, this is the first study that showed a model with different input variables to predict individual tree crowns. The variables, such as sample labeling, sample size, backbone, and input image type, should be considered when training the Mask R-CNN model. This study explores the accuracy of the models with these variables and the efficiency of model training for individual tree crown detection and delineation and explores the influence of the sample labeling method on model performance in detail. It can help improve the understanding of the Mask R-CNN model and leads to more accurate detection and delineation of individual trees.
4.2. Transfer Learning
In this study, a fine-tuning Mask R-CNN architecture was used to detect Fokienia hodginsii from a multi-tree species plantation. The results showed that fine-tuning the parameters for the model architecture could reduce sample size requirements. When the random sample set and the clumped sample set reached 337 trees and 675 trees, respectively, the model performance can achieve promising accuracy (F1 score > 85%). Transfer learning is the most effective method when it is difficult or challenging to obtain a large number of training samples to train a new model [33,35]. The core of the transfer learning approach is using the pre-trained network to another site and then fine-tuning the parameters to adapt the current object detection. Weinstein et al. demonstrated that conducting transfer learning experiments to detect trees based on an existing model with a small amount of local training data can achieve a similar accuracy when compared to a fully-trained model [13]. Previous studies also proved the value of transfer learning for forestry remote sensing applications using a small training sample size [18,20,64].
However, transfer learning has several drawbacks. The effectiveness of transfer learning depends on the relationship between the target task and the source task [33]. In the case of a weak relationship, it could cause negative transfer and result in poor performance for the target task. Mahdianpari et al. [39] stated that the full-training strategy is more accurate than the fine-tuning for classification. This conclusion may be because their dataset had five spectral bands, which differed from the ImageNet dataset [39]. Similarly, Nogueira et al. [33] reported that a difference between datasets could impact the accuracy of the transfer learning network, and higher accuracy of fine-tuning is achieved when the original and current datasets are similar. Fine-tuning is only able to adjust the parameters of the network, rather than the complete deep architecture (e.g., the number and types of the layers, layers organization) during the learning step. If the selected model architecture is not suitable, it will achieve the opposite result. For instance, Fromm et al. [21] found that pre-training cannot always improve the accuracy of tree seedling detection significantly, which may be attributed to the use of shallow architectures that are less likely to benefit from pre-training. Thus, it is suggested that an appropriate backbone is required for successful transfer learning.
4.3. Sample Size
In this study, the sample sizes between 168 and 1689 were compared to evaluate Fokienia hodginsii detection accuracy. It was found that sample size is the main factor impacting the accuracy of tree-crown detection and delineation by Mask R-CNN. According to our results, the model’s accuracy was increased with an increasing number of samples when the sample size was small, and the predicted accuracy of the model tended to be stable when the sample size was higher than 337 for random sampling and 675 for clumped sampling. Previous studies have also mentioned the influence of sample size on model performance [21,42]. Weinstein et al. showed that the accuracy first increased and then became stable with increasing the number of training samples [13]. Hartling et al. reported that the detection accuracy only decreased 3.02% when decreasing the quantity of training samples from 70% to 30% for tree species classification using the DenseNet classifier, but that the detection accuracy decreased 8.79% when lowering the number of training samples from 30% to 10% [65]. In practice, it is suggested that selecting a minimum number of training samples for CNN models that still ensure accurate classification could reduce the workload of training sample collection and training time and improve the working efficiency.
4.4. Sample Labeling
In this study, the strategy for testing different sample labeling patterns and sample sizes were accomplished by deleting samples from the original 1689 manually delineated training samples by repeatedly reducing the number by a percentage of 10% using both random and clumped sample labeling. This study found that the model with the random sample set achieved better accuracy than the clumped sample set at smaller sample sizes. This result can be explained by the study site being located in a mountain area with trees under different illumination conditions that cause the spectrum characteristic to vary, which can affect the model training accuracy. For different sample labeling methods, random sample sets are more representative because the sampling could cover the different locations and conditions of the study site. While clumped sample sets are only able to represent some locations, which indicated that the corresponding information of trees at the given location could reduce the sample representativeness and cause bias. Thus, it is the main reason that random and clumped samples delivered different accuracy under the same sample size. In addition, although the model with random sampling has lower requirements for sample size, it had a higher model training time cost than clumped sampling. The main reason for the difference in time utilization is that the number of image tiles for random sampling was larger than that for clumped sampling, and unlabeled trees on each tile were still read during the model training for random sampling, even though unlabeled trees were not used for actual model training.
4.5. Label Accuracy
Although it is common to delineate tree crowns from remote sensing images using visual interpretation for training sets, the target tree species must be clearly visible in the image [16]. However, this technique can inevitably cause inaccuracies during the delineation process due to differences in tree morphology and overlapping between trees [49]. Previous studies have reported that CNN models can partially compensate for the errors in training sample delineation [18,48,49]. Pearse et al. and Kattenborn et al. showed the model was capable of detecting the target objects that were ignored during the manual labeling process [18,49]. In our study, the results further showed that the model could achieve reasonable accuracy (F1 score > 85%) even with only 20% of tree crowns being annotated in the study site. The results indicated that even when there are only one or two labeled trees are in an individual tile, that is little impact on the model’s ability to detect other trees (Figure 6). This study shows that it is not necessary to annotate every tree in an image as long as the labeled trees are sufficiently representative. This is consistent with the speculation in a previous study using CNN to detect tree seedlings [18].
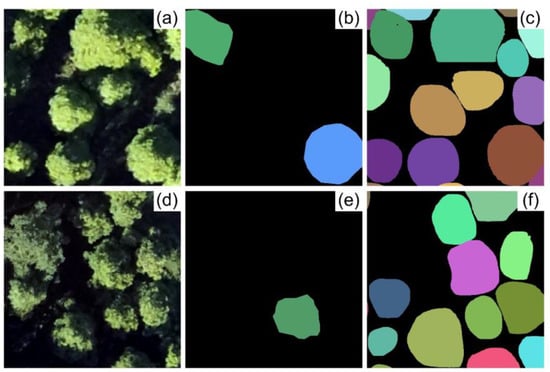
Figure 6.
Example of the detected results of Mask R-CNN with a combination of Multi-band image and a random sample set (using 337 manually delineated tree crowns). (a,d) are input images; (b,e) are the labels by random sampling; (c,f) are predicted results. Different colors represent the different trees.
At the same time, this finding can help solve the problem of extremely imbalanced samples. In a given region or a forest with unevenly distributed tree species, it may be dominated by one tree species, with others being rare. In that case, it would result in an imbalanced training set if all the tree species were annotated in that image [66]. Therefore, a Class-Balanced Cross-Entropy Loss (CBCEL) and a Class-Balanced Smooth L1 Loss (CBSLL) have been proposed by Zheng et al. for multi-class oil palm detection to solve the problem of imbalanced samples [22]. Our results provide a new idea to overcome this problem, which is to annotate the same number of samples for different tree species, with no need to annotate an additional amount of the dominant tree species.
4.6. Input Image
The generated Multi-band image and RGB image for this study were obtained from a single flight, which makes the Multi-band image and RGB image comparable. This study found that there is no apparent difference in model accuracy using Multi-band or RGB images when the sample size is sufficient. Our result is consistent with previous studies in this respect. For instance, Osco et al. reported that the identification of citrus trees achieved excellent accuracy (R2 between 0.92 and 0.96) using different combined input images (two bands, three bands, and four bands combinations) with 37,353 manually delineated trees [67]. Nezami et al. proposed that no matter if hyperspectral or RGB channels were used, the accuracy of 3D-CNN models with 3039 manually labeled trees was similar [68].
It is important to note that the accuracy of the model using a Multi-band image is superior to the model using RGB image when the sample size is small (<675/1689) in this study. This can be explained by the Multi-band image often providing more predictors. In the case of a small sample size (e.g., only several training samples), using hyperspectral data or multiple combined data may be the preferred input image type because it can provide more features for model training. For example, La Rose et al. reported that 23 individual tree crowns (maximum two individual tree crowns per species) could perform with reasonable accuracy for 14 tree species detections using the hyperspectral data with 25 spectral bands (OA: 72.55%) [24]. However, although the model training benefits from the increase in the image dimension, it increases the computational load and may cause a high correlation between bands. Therefore, it may be necessary to design a special network, such as a 3D network [47], a partial loss function to train an FCN [24] to deal with this problem, which may outweigh the convenience mentioned above.
With the increased availability of UAVs, RGB imagery derived from UAVs is relatively accessible and low-cost compared with Multi-band imagery and hyperspectral data. In addition, the acquisition of high-resolution Multi-band image and hyperspectral data is more difficult than the RGB image. This study showed the input image had little impact on the model performance compared to the sample size and sample distribution. In general, it is suggested to consider the RGB image and large training samples for model training.
5. Conclusions
This is the first study to use Mask R-CNN as a model to evaluate the influence of sample labeling, sample size, and input image type for deep learning approaches to detect and delineate individual tree crowns. It was found that the best model performance used the ResNet-50 as a network architecture for tree detection and delineation. The sample size is a crucial parameter that can affect model performance, followed by sample labeling and the input image type. Random sample labeling is able to greatly reduce the sample size requirements for model training. The model with a random sample set achieved higher accuracy than the model with a clumped sample set, even if many of the trees were unlabeled in each tile during the process of random sample labeling. The accuracy of the model with a Multi-band image is higher than the model with an RGB image. In addition, the average per-epoch training time for the model with a clumped sample set was shorter than the model with a random sample set. Considering the difficulty of image acquisition, sample labeling, and training time, it is suggested to use RGB imagery from UAVs with random sample labeling for model training. This study contributes to a better understanding of the influence of sample labeling and provides the reference for sample labeling on vegetation remote sensing.
Author Contributions
Conceptualization, Z.H. and L.L.; methodology, Z.H.; software, Z.H.; validation, Z.H.; formal analysis, Z.H.; investigation, Z.H. and L.L.; resources, K.Y.; data curation, Z.H.; writing—original draft preparation, Z.H. and L.L.; writing—review and editing, C.J.P. and E.A.M.; visualization, Z.H. and L.L.; supervision, L.L. and J.L.; project administration, J.L.; funding acquisition, K.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fujian Provincial Department of Science and Technology (grant number 2019N5012 and 2020N5003).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We wish to thank the anonymous reviewers and editors for their detailed comments and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chopping, M.; Moisen, G.G.; Su, L.; Laliberte, A.; Rango, A.; Martonchik, J.V.; Peters, D.P. Large area mapping of southwestern forest crown cover, canopy height, and biomass using the NASA Multiangle Imaging Spectro-Radiometer. Remote Sens. Environ. 2008, 112, 2051–2063. [Google Scholar] [CrossRef] [Green Version]
- Holopainen, M.; Vastaranta, M.; Hyyppä, J. Outlook for the Next Generation’s Precision Forestry in Finland. Forest 2014, 5, 1682–1694. [Google Scholar] [CrossRef] [Green Version]
- Gardner, T.A.; Barlow, J.; Araujo, I.S.; Ávila-Pires, T.C.; Bonaldo, A.B.; Costa, J.E.; Esposito, M.C.; Ferreira, L.V.; Hawes, J.; Hernandez, M.I.M.; et al. The cost-effectiveness of biodiversity surveys in tropical forests. Ecol. Lett. 2008, 11, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Mohan, M.; Silva, C.A.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.T.; Dia, M. Individual Tree Detection from Unmanned Aerial Vehicle (UAV) Derived Canopy Height Model in an Open Canopy Mixed Conifer Forest. Forest 2017, 8, 340. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Shao, G. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry Remote Sensing from Unmanned Aerial Vehicles: A Review Focusing on the Data, Processing and Potentialities. Remote Sens. 2020, 12, 1046. [Google Scholar] [CrossRef] [Green Version]
- Jurjević, L.; Liang, X.; Gašparović, M.; Balenović, I. Is field-measured tree height as reliable as believed—Part II, A comparison study of tree height estimates from conventional field measurement and low-cost close-range remote sensing in a deciduous forest. ISPRS J. Photogramm. Remote Sens. 2020, 169, 227–241. [Google Scholar] [CrossRef]
- Swayze, N.C.; Tinkham, W.T.; Vogeler, J.C.; Hudak, A.T. Influence of flight parameters on UAS-based monitoring of tree height, diameter, and density. Remote Sens. Environ. 2021, 263, 112540. [Google Scholar] [CrossRef]
- Imangholiloo, M.; Saarinen, N.; Markelin, L.; Rosnell, T.; Näsi, R.; Hakala, T.; Honkavaara, E.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Characterizing Seedling Stands Using Leaf-Off and Leaf-On Photogrammetric Point Clouds and Hyperspectral Imagery Acquired from Unmanned Aerial Vehicle. Forest 2019, 10, 415. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.; Sun, Q.C.; Hally, B.; Hillman, S.; Both, A.; Hurley, J.; Martin Saldias, D.S. Linking urban tree in-ventories to remote sensing data for individual tree mapping. Urban For. Urban Green. 2021, 61, 127106. [Google Scholar] [CrossRef]
- Larsen, M.; Eriksson, M.; Descombes, X.; Perrin, G.; Brandtberg, T.; Gougeon, F.A. Comparison of six individual tree crown detection algorithms evaluated under varying forest conditions. Int. J. Remote Sens. 2011, 32, 5827–5852. [Google Scholar] [CrossRef]
- Pouliot, D.A.; King, D.J.; Bell, F.W.; Pitt, D.G. Automated tree crown detection and delineation in high-resolution digital camera imagery of coniferous forest regeneration. Remote Sens. Environ. 2002, 82, 322–334. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; White, E.P. Cross-site learning in deep learning RGB tree crown detection. Ecol. Inform. 2020, 56, 101061. [Google Scholar] [CrossRef]
- Cheng, G.; Han, J.; Lu, X. Remote Sensing Image Scene Classification: Benchmark and State of the Art. Proc. IEEE 2017, 105, 1865–1883. [Google Scholar] [CrossRef] [Green Version]
- Diez, Y.; Kentsch, S.; Fukuda, M.; Caceres, M.; Moritake, K.; Cabezas, M. Deep Learning in Forestry Using UAV-Acquired RGB Data: A Practical Review. Remote Sens. 2021, 13, 2837. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Pearse, G.D.; Tan, A.Y.; Watt, M.S.; Franz, M.O.; Dash, J.P. Detecting and mapping tree seedlings in UAV imagery using convolutional neural networks and field-verified data. ISPRS J. Photogramm. Remote Sens. 2020, 168, 156–169. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Braga, J.R.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragão, L.E.O.C.; De Campos Velho, H.F.; Shiguemori, E.H.; Wagner, F.H. Tree crown delineation algorithm based on a convolutional neural network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef] [Green Version]
- Fromm, M.; Schubert, M.; Castilla, G.; Linke, J.; McDermid, G. Automated Detection of Conifer Seedlings in Drone Imagery Using Convolutional Neural Networks. Remote Sens. 2019, 11, 2585. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Yu, L.; Yuan, S.; Tao, W.Y.W.; Pang, T.K.; Kanniah, K.D. Growing status ob-servation for oil palm trees using Unmanned Aerial Vehicle (UAV) images. ISPRS J. Photogramm. 2021, 173, 95–121. [Google Scholar] [CrossRef]
- Xiao, C.; Qin, R.; Huang, X. Treetop detection using convolutional neural networks trained through automatically generated pseudo labels. Int. J. Remote Sens. 2019, 41, 3010–3030. [Google Scholar] [CrossRef]
- La Rosa, L.E.C.; Sothe, C.; Feitosa, R.Q.; de Almeida, C.M.; Schimalski, M.B.; Oliveira, D.A.B. Multi-task fully convolutional network for tree species mapping in dense forests using small training hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2021, 179, 35–49. [Google Scholar] [CrossRef]
- Ferreira, M.P.; de Almeida, D.R.A.; Papa, D.D.A.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Anagnostis, A.; Tagarakis, A.; Kateris, D.; Moysiadis, V.; Sørensen, C.; Pearson, S.; Bochtis, D. Orchard Mapping with Deep Learning Semantic Segmentation. Sensors 2021, 21, 3813. [Google Scholar] [CrossRef]
- Brandt, M.; Tucker, C.J.; Kariryaa, A.; Rasmussen, K.; Abel, C.; Small, J.; Chave, J.; Rasmussen, L.V.; Hiernaux, P.; Diouf, A.A.; et al. An unexpectedly large count of trees in the West African Sahara and Sahel. Nature 2020, 587, 78–82. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Wagner, F.H.; Ferreira, M.P.; Sanchez, A.; Hirye, M.C.M.; Zortea, M.; Gloor, E.; Phillips, O.L.; de Souza Filho, C.R.; Shimabukuro, Y.E.; Aragão, L.E.O.C. Individual tree crown delineation in a highly diverse tropical forest using very high resolution satellite images. ISPRS J. Photogramm. Remote Sens. 2018, 145, 362–377. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Lotte, R.G.; D’Elia, F.V.; Stamatopoulos, C.; Kim, D.-H.; Benjamin, A.R. Accurate mapping of Brazil nut trees (Bertholletia excelsa) in Amazonian forests using WorldView-3 satellite images and convolutional neural networks. Ecol. Inform. 2021, 63, 101302. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Nogueira, K.; Penatti, O.A.; dos Santos, J.A. Towards better exploiting convolutional neural networks for remote sensing scene classification. Pattern Recognit. 2017, 61, 539–556. [Google Scholar] [CrossRef] [Green Version]
- Rezaee, M.; Mahdianpari, M.; Zhang, Y.; Salehi, B. Deep Convolutional Neural Network for Complex Wetland Classification Using Optical Remote Sensing Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3030–3039. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.; Hu, J.; Zhang, L. Transferring Deep Convolutional Neural Networks for the Scene Classifica-tion of High-Resolution Remote Sensing Imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Yang, G.; Yang, H.; Zhu, Y.; Li, Z.; Lei, L.; Zhao, C. Extracting apple tree crown information from remote imagery using deep learning. Comput. Electron. Agric. 2020, 174, 105504. [Google Scholar] [CrossRef]
- Iqbal, M.S.; Ali, H.; Tran, S.N.; Iqbal, T. Coconut trees detection and segmentation in aerial imagery using mask region-based convolution neural network. IET Comput. Vis. 2021, 15, 428–439. [Google Scholar] [CrossRef]
- Mo, J.; Lan, Y.; Yang, D.; Wen, F.; Qiu, H.; Chen, X.; Deng, X. Deep Learning-Based Instance Segmentation Method of Litchi Canopy from UAV-Acquired Images. Remote Sens. 2021, 13, 3919. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Rezaee, M.; Mohammadimanesh, F.; Zhang, Y. Very Deep Convolutional Neural Networks for Complex Land Cover Mapping Using Multispectral Remote Sensing Imagery. Remote Sens. 2018, 10, 1119. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. In Proceedings of the 16th IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Kattenborn, T.; Eichel, J.; Fassnacht, F.E. Convolutional Neural Networks enable efficient, accurate and fine-grained segmentation of plant species and communities from high-resolution UAV imagery. Sci. Rep. 2019, 9, 17656. [Google Scholar] [CrossRef]
- Liu, T.; Abd-Elrahman, A. Deep convolutional neural network training enrichment using multi-view object-based analysis of Unmanned Aerial systems imagery for wetlands classification. ISPRS J. Photogramm. 2018, 139, 154–170. [Google Scholar] [CrossRef]
- Yu, X.; Wu, X.; Luo, C.; Ren, P. Deep learning in remote sensing scene classification: A data augmentation enhanced convolutional neural network framework. GISci. Remote Sens. 2017, 54, 741–758. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Du, S. Learning multiscale and deep representations for classifying remotely sensed imagery. ISPRS J. Photogramm. Remote Sens. 2016, 113, 155–165. [Google Scholar] [CrossRef]
- Lu, Z.; Fu, Z.; Xiang, T.; Han, P.; Wang, L.; Gao, X. Learning from Weak and Noisy Labels for Semantic Segmentation. IEEE T. Pattern Anal. 2017, 39, 486–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Qi, H.; Rong, Z.; Liu, L.; Su, H. Scribble-Supervised Segmentation of Aerial Building Footprints Using Adversarial Learning. IEEE Access 2018, 6, 58898–58911. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, L.; Zhang, X. Three-dimensional convolutional neural network model for tree species classification using airborne hyperspectral images. Remote Sens. Environ. 2020, 247, 111938. [Google Scholar] [CrossRef]
- Hamdi, Z.M.; Brandmeier, M.; Straub, C. Forest Damage Assessment Using Deep Learning on High Resolution Remote Sensing Data. Remote Sens. 2019, 11, 1976. [Google Scholar] [CrossRef] [Green Version]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Net-works accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sens. Ecol. Conserv. 2020, 6, 472–486. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Fan, T.; Ghimire, P.; Deng, L. Experimental Evaluation and Consistency Comparison of UAV Multi-spectral Minisensors. Remote Sens. 2020, 12, 2542. [Google Scholar] [CrossRef]
- Hao, Z.; Lin, L.; Post, C.J.; Jiang, Y.; Li, M.; Wei, N.; Yu, K.; Liu, J. Assessing tree height and density of a young forest using a consumer unmanned aerial vehicle (UAV). New For. 2021, 52, 843–862. [Google Scholar] [CrossRef]
- Gallardo-Salazar, J.L.; Pompa-García, M. Detecting Individual Tree Attributes and Multispectral Indices Using Unmanned Aerial Vehicles: Applications in a Pine Clonal Orchard. Remote Sens. 2020, 12, 4144. [Google Scholar] [CrossRef]
- Syetiawan, A.; Gularso, H.; Kusnadi, G.I.; Pramudita, G.N. Precise topographic mapping using direct georef-erencing in UAV. IOP conference series. Earth Environ. Sci. 2020, 500, 12029. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hao, Z.; Lin, L.; Post, C.J.; Mikhailova, E.A.; Li, M.; Chen, Y.; Yu, K.; Liu, J. Automated tree-crown and height detection in a young forest plantation using mask region-based convolutional neural network (Mask R-CNN). ISPRS J. Photogramm. Remote Sens. 2021, 178, 112–123. [Google Scholar] [CrossRef]
- Chadwick, A.; Goodbody, T.; Coops, N.; Hervieux, A.; Bater, C.; Martens, L.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-Off Conditions Using UAV Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, F.; Zhang, H.; Liang, Z. Convolutional neural network based heterogeneous transfer learning for remote-sensing scene classification. Int. J. Remote Sens. 2019, 40, 8506–8527. [Google Scholar] [CrossRef]
- Pleșoianu, A.; Stupariu, M.; Șandric, I.; Pătru-Stupariu, I.; Drăguț, L. Individual tree-crown detection and species classification in very high-resolution remote sensing imagery using a deep learning ensemble model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Wu, G.; Li, Y. Non-maximum suppression for object detection based on the chaotic whale optimization algorithm. J. Vis. Commun. Image Represent. 2021, 74, 102985. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation. In Proceedings of the 27th European Conference on Information Retrieval, Santiago de Compostela, Spain, 21–23 March 2005; pp. 345–359. [Google Scholar]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In Proceedings of the 19th Australian Joint Conference on Artificial Intelligence, Hobart, Australia, 4–8 December 2006; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. [Google Scholar]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection Over Union: A Metric and a Loss for Bounding Box Regression. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2019, Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Sahoo, D.; Hoi, S.C.H. Recent advances in deep learning for object detection. Neurocomputing 2020, 396, 39–64. [Google Scholar] [CrossRef] [Green Version]
- Mehdipour Ghazi, M.; Yanikoglu, B.; Aptoula, E. Plant identification using deep neural networks via optimization of transfer learning parameters. Neurocomputing 2017, 235, 228–235. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban Tree Species Classification Using a WorldView-2/3 and LiDAR Data Fusion Approach and Deep Learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [Green Version]
- Mellor, A.; Boukir, S.; Haywood, A.; Jones, S. Exploring issues of training data imbalance and mislabelling on random forest performance for large area land cover classification using the ensemble margin. ISPRS J. Photogramm. Remote Sens. 2015, 105, 155–168. [Google Scholar] [CrossRef]
- Osco, L.P.; Arruda, M.D.S.D.; Marcato Junior, J.; Da Silva, N.B.; Ramos, A.P.M.; Moryia, É.A.S.; Imai, N.N.; Pe-reira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. ISPRS J. Photogramm. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).