Estimating the Horizontal and Vertical Distributions of Pigments in Canopies of Ginkgo Plantation Based on UAV-Borne LiDAR, Hyperspectral Data by Coupling PROSAIL Model
Abstract
:1. Introduction
2. Materials and Methods
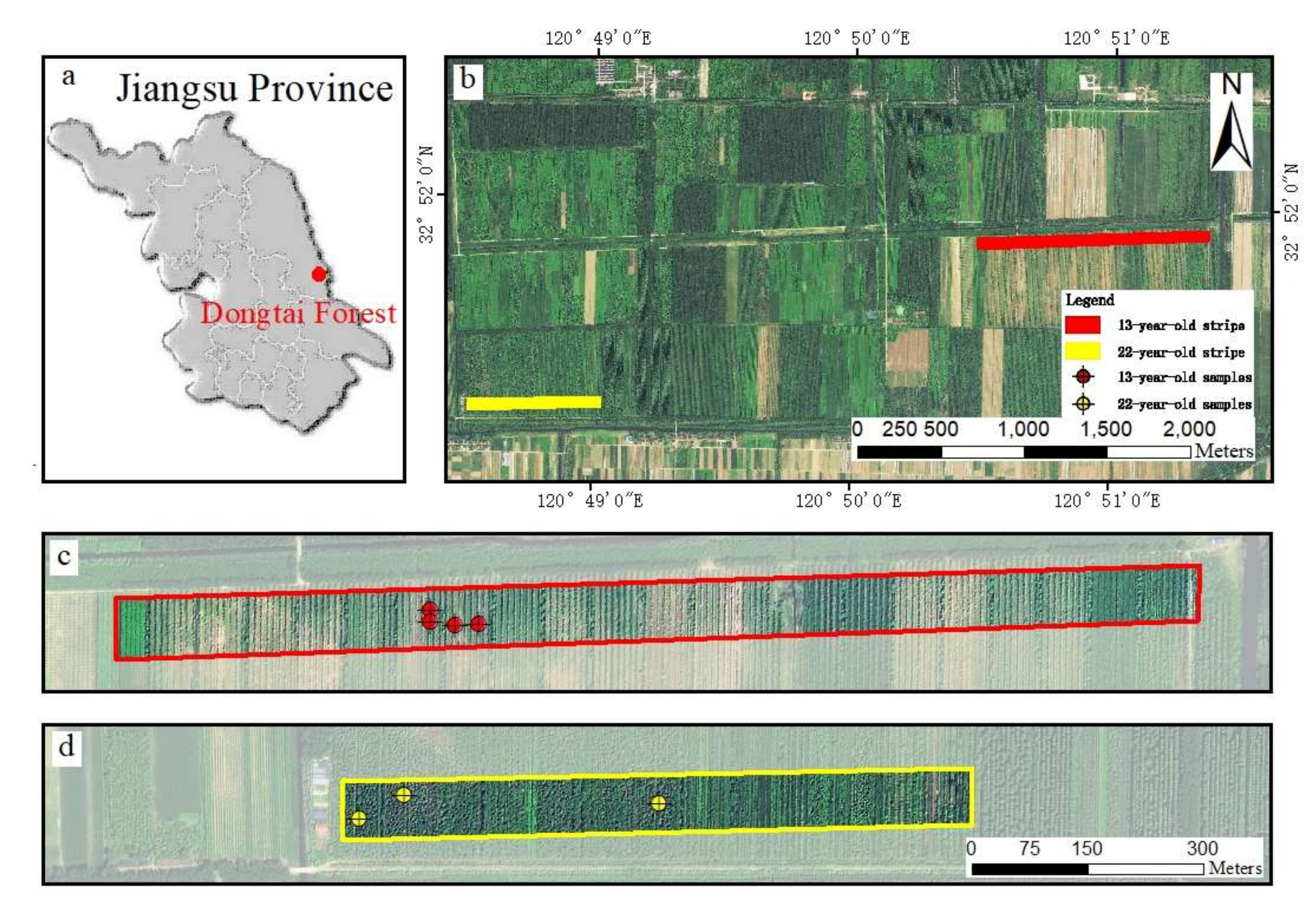
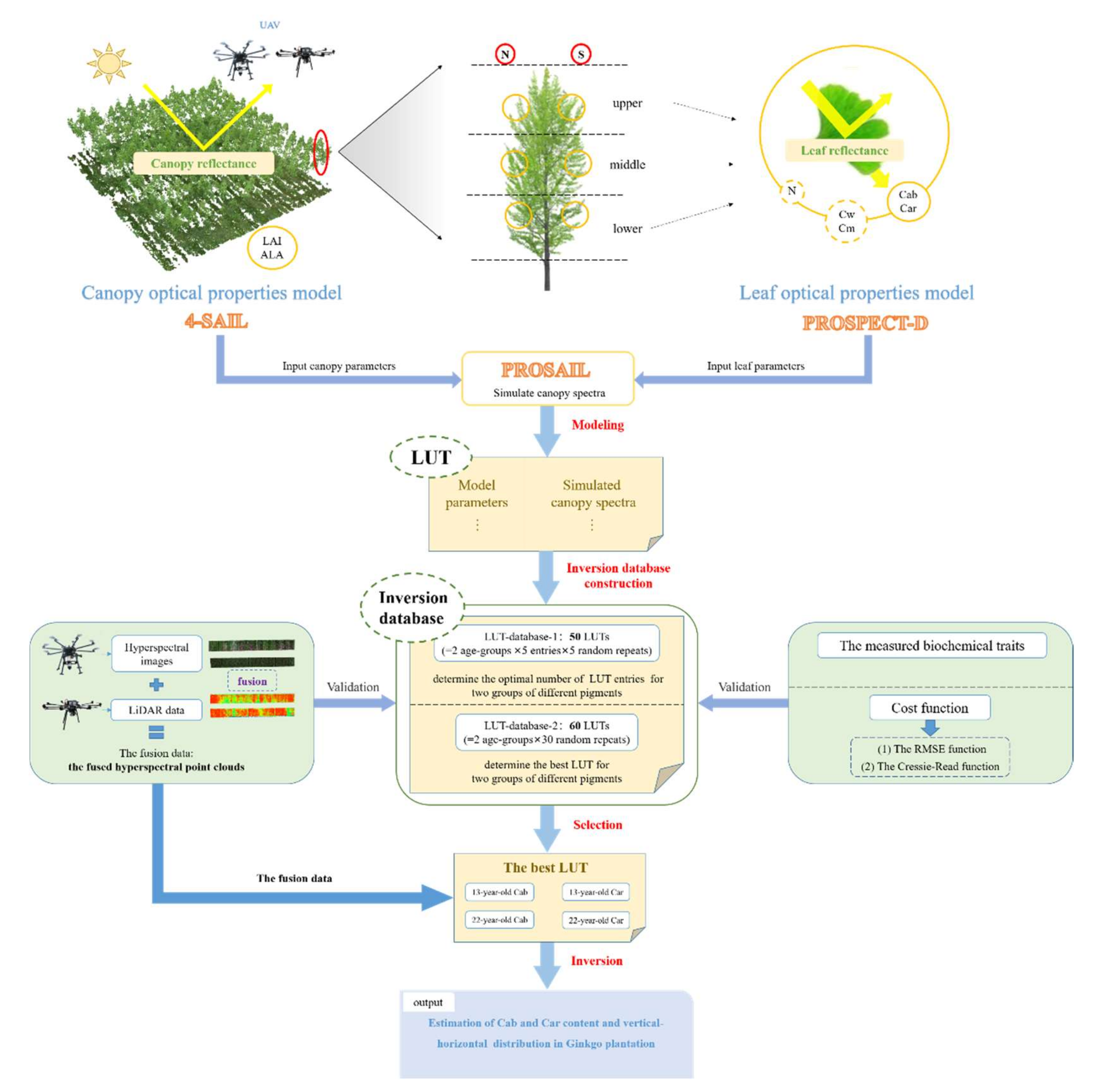
2.1. Study Area and Workflow
2.2. Data Collection and Processing
2.2.1. Field Data Collection and Measurement
2.2.2. Remote Sensing Data Acquisition and Processing
2.2.3. Remote Sensing Data Fusion
2.2.4. Extracting Profile Characteristic Variables
2.3. PROSAIL Model
2.3.1. Local Sensitivity Analysis
2.3.2. Model Parameters Setting
2.3.3. LUT-Based-Inversion and Pigment Estimations
3. Results
3.1. Remote Sensing Data Processing Results
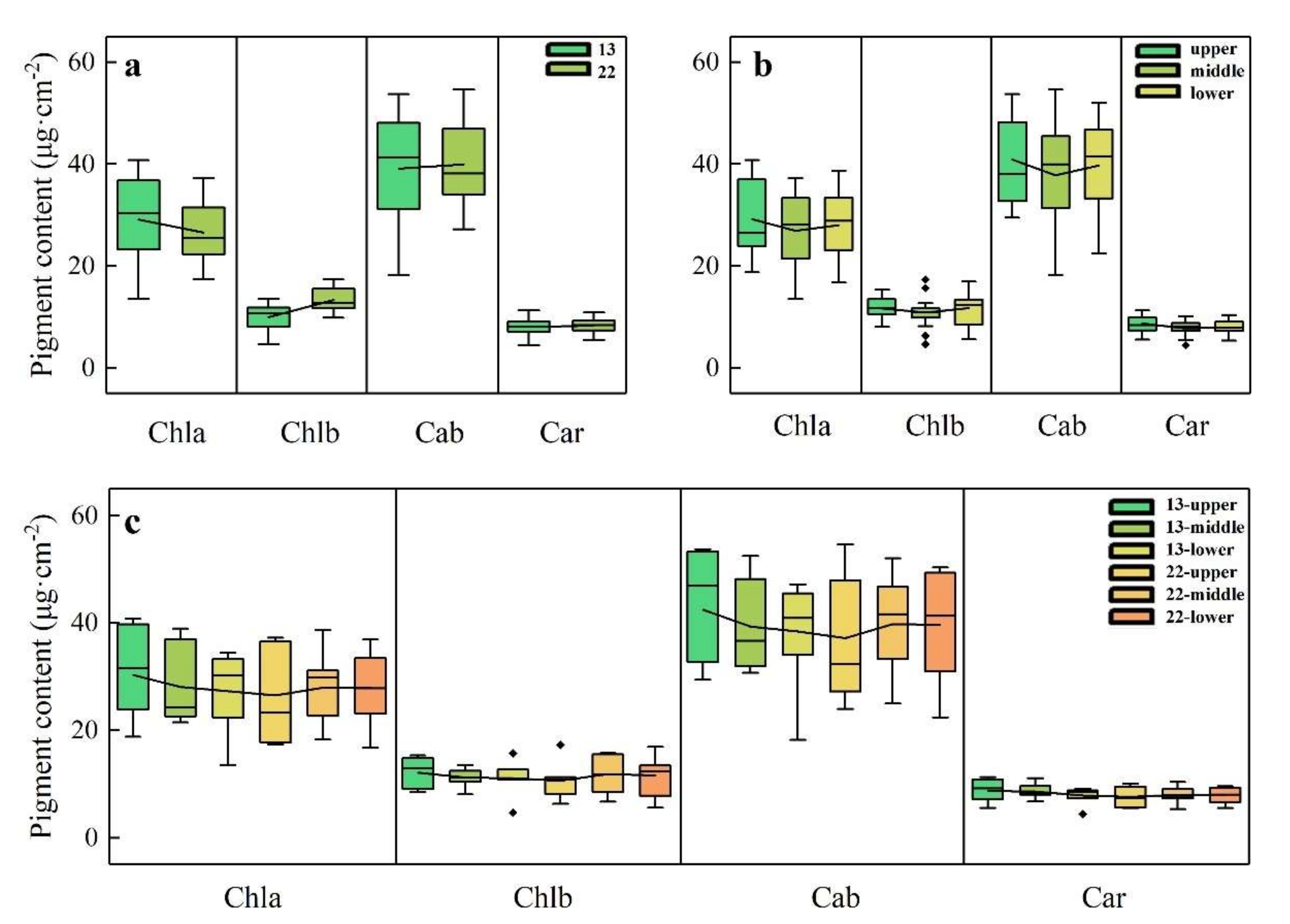
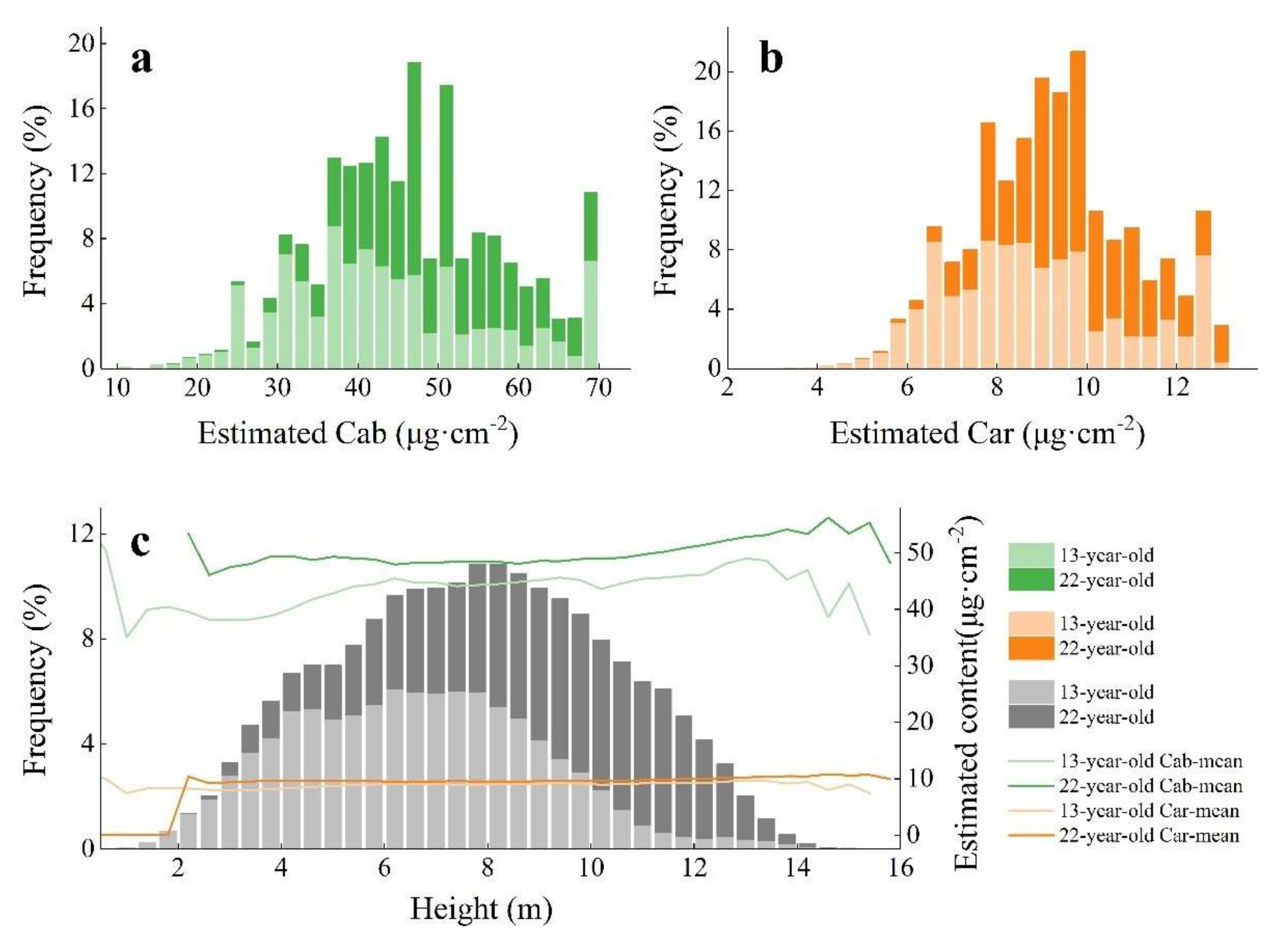
3.2. Statistical Distribution of Biochemical Pigments in Ginkgo Leaves
3.3. Pigment Estimation Performance Using LUT-Based-Inversion Mothod
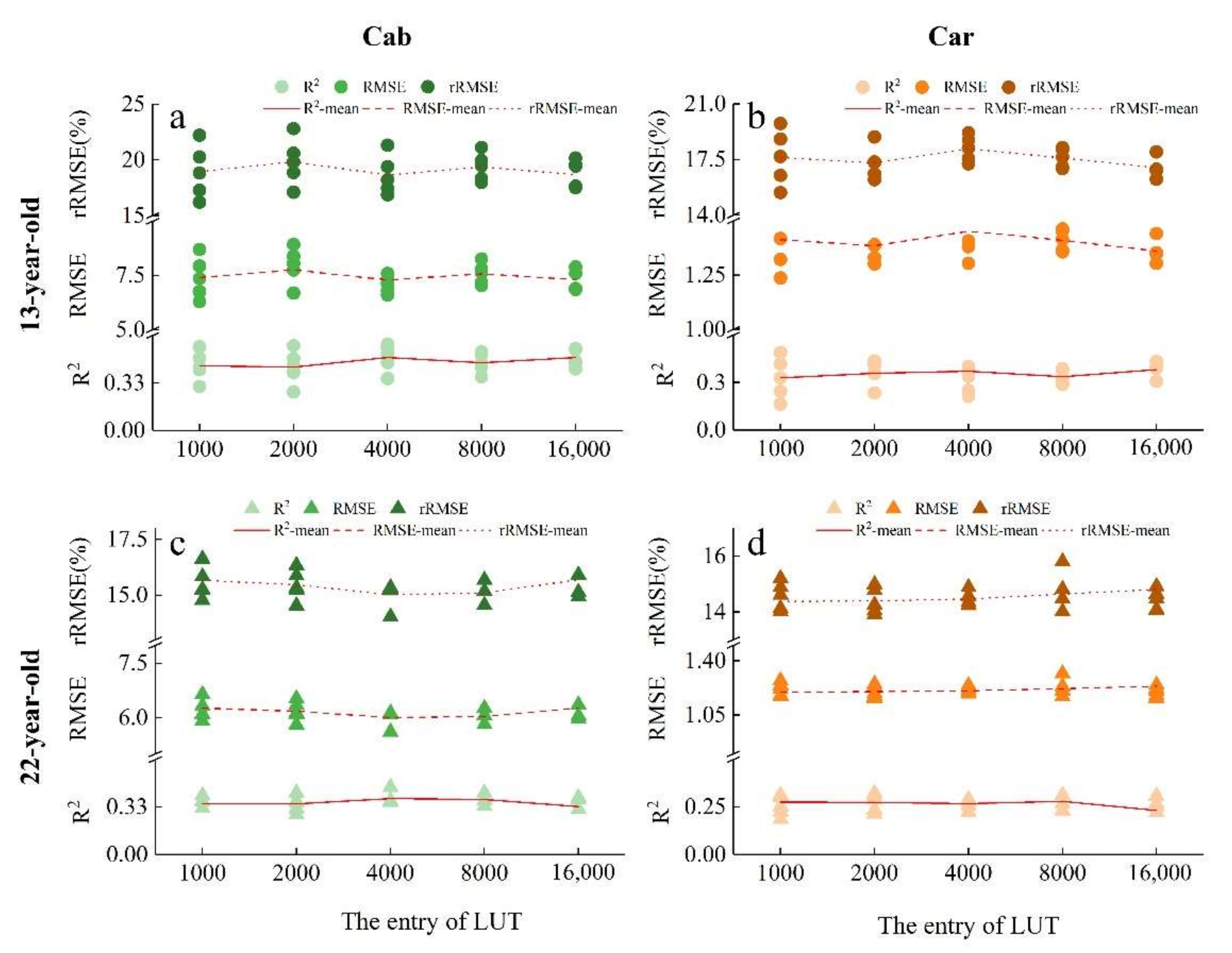
3.3.1. The Selection of Optimal LUT Entries for Estimating Cab and Car
3.3.2. The Selection of Specific Optimal LUTs for Estimating Cab and Car
4. Discussion
4.1. Performance of LUT-Based-Inversion in Estimating Pigment Content
4.2. Potential for Mapping Pigments Content Based on Hyperspectral Fusion Point Cloud Coupling with PROSAIL Model
4.3. Factors Affecting Pigment Distribution on Canopy Surfaces
4.4. Future Research and Application
5. Conclusions
- The special LUTs for Ginkgo plantations were constructed with higher estimation accuracies (R2 = 0.36–0.60, rRMSE = 13.53–16.86% for PROSAIL models, Figure 8 R2 = 0.25–0.46, rRMSE = 16.25–19.37% for VIs models, Supplementary Figure S3). Considering the computation work, we calibrated the parameters of PROSAIL and found the optimal LUT entry (i.e., 4000);
- The pigment content of Ginkgo trees increased from inward to outward in the horizontal direction, but decreased along with increase in height in the vertical direction;
- The hierarchical study in optimizing the inversion model once again emphasized its importance.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brockerhoff, E.G.; Jactel, H.; Parrotta, J.A.; Quine, C.P.; Sayer, J. Plantation forests and biodiversity: Oxymoron or opportunity? Biodivers. Conserv. 2008, 17, 925–951. [Google Scholar] [CrossRef]
- National Forestry and Grassland Administration. China Forest Resources Report 2009–2013; China Forestry Publishing House: Beijing, China, 2014. [Google Scholar]
- Yan-Long, W.; Xiao-Yu, C.H.E.; Yu-Guang, L.I.U.; Ya-Nan, L.I.; Yan-Hui, L.I. Study on the changes of leaf color parameter and pigment content of Ginkgo biloba leaf in autumn. J. Hebei Agric. Univ. 2015, 38, 28–32. [Google Scholar]
- Yanling, N.; Zhao, Y. Study on Inversion of Vegetation Biochemical Parameters through Hyperspectral Data; Northeast Normal University: Changchun, China, 2010. [Google Scholar]
- Gara, T.W.; Darvishzadeh, R.; Skidmore, A.K.; Tiejun, W.; Heurich, M. Evaluating the performance of PROSPECT in the retrieval of leaf traits across canopy throughout the growing season. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101919. [Google Scholar] [CrossRef]
- Jagodziński, A.M.; Dyderski, M.K.; Rawlik, K.; Katna, B. Seasonal variability of biomass, total leaf area and specific leaf area of forest understory herbs reflects their life strategies. For. Ecol. Manag. 2016, 374, 71–81. [Google Scholar] [CrossRef]
- Meng, W.; Xidong, W. Effects of Planting Densities and Spatial Distribution Patterns on Canopy Structure and Physiological Characters of Summer Maize; Tianjin Agricultural University: Tianjin, China, 2016. [Google Scholar]
- Sellin, A.; Kupper, P. Effects of light availability versus hydraulic constraints on stomatal responses within a crown of silver birch. Oecologia 2005, 142, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Fu, W.; Yue-Li, H. Study of the Dynamics Models of Forest Growth and Nutrition Ⅷ Diameter Age and Growth Parameter Determination. J. Cent. South For. Univ. 2001, 21, 1–5. [Google Scholar]
- Clevers, J.G.P.W.; Kooistra, L. Using hyperspectral remote sensing data for retrieving canopy chlorophyll and nitrogen content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Paź-Dyderska, S.; Dyderski, M.K.; Nowak, K.; Jagodziński, A.M. On the sunny side of the crown-quantification of intra-canopy SLA variation among 179 taxa. For. Ecol. Manag. 2020, 472, 118254. [Google Scholar] [CrossRef]
- Dong-Yan, X. Application Status and Prospects of Remote Sensing in Forestry. Sci. Technol. Vis. 2014, 21, 309–311. [Google Scholar] [CrossRef]
- Qingwang, L.; Bingxiang, T.; Kailong, H.; Xue, F.; Zengyuan, L.; Yong, P.; Shiming, L. The remote sensing experiment on airborne LiDAR and hyperspectral integrated system for subtropical forest estimation. Chin. High Technol. Lett. 2016, 26, 264–274. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, Z.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Liu, H.; Shen, X.; Wu, X. Assessment of approaches for monitoring forest structure dynamics using bi-temporal digital aerial photogrammetry point clouds. Remote Sens. Environ. 2021, 255, 112300. [Google Scholar] [CrossRef]
- Qinghua, G.; Jin, L.; Shengli, T.; Baolin, X.; Le, L.; Guangcai, X.; Wenkai1, L.; Fangfang, W.; Yumei, L.; Linhai, C.; et al. Perspectives and prospects of LiDAR in forest ecosystem monitoring and modeling. Chin. Sci. Bull 2014, 59, 459–478. (in Chinese). [Google Scholar] [CrossRef]
- Kai, Z.; Lin, C. The status and prospects of remote sensing applications in precision silviculture. J. Remote Sens. 2021, 25, 423–438. [Google Scholar] [CrossRef]
- Li-Juan, C.; Lin, S.; Yan-Juan, Y.; Yan, S. Research Developments on Inversion of Vegetation Biochemistry Compositions by Quantitative Remote Sensing. J. Atmos. Environ. Opt. 2011, 6, 163–178. [Google Scholar] [CrossRef]
- Yanfang, X.; Demin, Z.; Wenji, Z. Review of inversing biophysical and biochemical vegetation parameters in various spatial scales using radiative transfer models. J. Remote Sens. 2013, 33, 3291–3297. [Google Scholar] [CrossRef]
- De-Hua, Z.; Jian-Long, L.; Zi-Jian, S. Hyperspectral Remote Sensing for Estimating Biochemical Variables of Canopy. Adv. Earth Sci. 2003, 18, 94–99. [Google Scholar] [CrossRef]
- Changshan, W.; Qingx, T.; Lanfen, Z.; Weidong, L. Correlation Analysis Between Spectral Data and Chlorophyll of Rice and Maize. J. Basic Sci. Eng. 2002, 6, 1–5. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Ustin, S.L.; Verdebout, J.; Schmuck, G.; Andreoli, G.; Hosgood, B. Estimating leaf biochemistry using the PROSPECT leaf optical properties model. Remote Sens. Environ. 1996, 56, 194–202. [Google Scholar] [CrossRef]
- Baret, F.; Fourty, T. Estimation of leaf water content and specific leaf weight from reflectance and transmittance measurements. Agronomie 1997, 17, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Feret, J.B.; François, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.P.R.; Ustin, S.L.; le Maire, G.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Barry, K.M.; Newnham, G.J.; Stone, C. Estimation of chlorophyll content in Eucalyptus globulus foliage with the leaf reflectance model PROSPECT. Agric. For. Meteorol. 2009, 149, 1209–1213. [Google Scholar] [CrossRef]
- Féret, J.-B.; François, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef] [Green Version]
- Féret, J.-B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflectance modeling: The SAIL model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef] [Green Version]
- Kuusk, A.; Nilson, T. A Directional Multispectral Forest Reflectance Model. Remote Sens. Environ. 2000, 72, 244–252. [Google Scholar] [CrossRef]
- Demarez, V.; Gastellu-Etchegorry, J.P. A Modeling Approach for Studying Forest Chlorophyll Content. Remote Sens. Environ. 2000, 71, 226–238. [Google Scholar] [CrossRef]
- Goel, N.S.; Thompson, R.L. A snapshot of canopy reflectance models and a universal model for the radiation regime. Remote Sens. Rev. 2000, 18, 197–225. [Google Scholar] [CrossRef]
- Verhoef, W.; Bach, H. Simulation of hyperspectral and directional radiance images using coupled biophysical and atmospheric radiative transfer models. Remote Sens. Environ. 2003, 87, 23–41. [Google Scholar] [CrossRef]
- Baret, F.; Jacquemoud, S.; Guyot, G.; Leprieur, C. Modeled analysis of the biophysical nature of spectral shifts and comparison with information content of broad bands. Remote Sens. Environ. 1992, 41, 133–142. [Google Scholar] [CrossRef]
- Dawson, T.P.; Curran, P.J.; North, P.R.J.; Plummer, S.E. The Propagation of Foliar Biochemical Absorption Features in Forest Canopy Reflectance: A Theoretical Analysis. Remote Sens. Environ. 1999, 67, 147–159. [Google Scholar] [CrossRef]
- Verhoef, W.; Bach, H. Remote sensing data assimilation using coupled radiative transfer models. Phys. Chem. Earth Parts A/B/C 2003, 28, 3–13. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Kötz, B.; Schaepman, M.; Morsdorf, F.; Bowyer, P.; Itten, K.; Allgöwer, B. Radiative transfer modeling within a heterogeneous canopy for estimation of forest fire fuel properties. Remote Sens. Environ. 2004, 92, 332–344. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Harron, J.; Hu, B.; Noland, T.L.; Goel, N.; Mohammed, G.H.; Sampson, P. Needle chlorophyll content estimation through model inversion using hyperspectral data from boreal conifer forest canopies. Remote Sens. Environ. 2004, 89, 189–199. [Google Scholar] [CrossRef]
- Verhoef, W.; Jia, L.; Xiao, Q.; Su, Z. Unified Optical-Thermal Four-Stream Radiative Transfer Theory for Homogeneous Vegetation Canopies. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1808–1822. [Google Scholar] [CrossRef]
- Dong, L.; Tao, C.; Min, J.; Kai, Z.; Ning, L.; Xia, Y.; Yongchao, T.; Yan, Z.; Weixing, C. PROCWT: Coupling PROSPECT with continuous wavelet transform to improve the retrieval of foliar chemistry from leaf bidirectional reflectance spectra. Remote Sens. Environ. 2018, 206, 1–14. [Google Scholar] [CrossRef]
- Berger, K.; Wang, Z.; Danner, M.; Wocher, M.; Mauser, W.; Hank, T. Simulation of Spaceborne Hyperspectral Remote Sensing to Assist Crop Nitrogen Content Monitoring in Agricultural Crops. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; Volume 2018, pp. 3801–3804. [Google Scholar]
- Féret, J.-B.; le Maire, G.; Jay, S.; Berveiller, D.; Bendoula, R.; Hmimina, G.; Cheraiet, A.; Oliveira, J.C.; Ponzoni, F.J.; Solanki, T.; et al. Estimating leaf mass per area and equivalent water thickness based on leaf optical properties: Potential and limitations of physical modeling and machine learning. Remote Sens. Environ. 2019, 231, 110959. [Google Scholar] [CrossRef]
- Duan, S.-B.; Li, Z.-L.; Wu, H.; Tang, B.-H.; Ma, L.; Zhao, E.; Li, C. Inversion of the PROSAIL model to estimate leaf area index of maize, potato, and sunflower fields from unmanned aerial vehicle hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 12–20. [Google Scholar] [CrossRef]
- Zhiqing, C.; Jinsong, Z. Estimation Model of Poplar Plantation Productivity with Hyperspectral Information and Remote Sensing; Chinese Academy of Forestry: Beijing, China, 2015. [Google Scholar]
- Xin, S.; Lin, C.; Coops, N.C.; Hongchao, F.; Xiangqian, W.; Hao, L.; Guibin, W.; Fuliang, C. Quantifying vertical profiles of biochemical traits for forest plantation species using advanced remote sensing approaches. Remote Sens. Environ. 2020, 250, 112041. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Wen-Ting, Q.; Hui, Z.; Hong-Mei, L.; Wan-Juan, Y.; Hua-Long, L. Remote Recognition and Growth Monitoring of Winter Wheat in Key Stages Based on S-G Filter in Guanzhong Region. Chin. J. Agrometeorol. 2015, 36, 93–99. [Google Scholar]
- Morsdorf, F.; Kötz, B.; Meier, E.; Itten, K.I.; Allgöwer, B. Estimation of LAI and fractional cover from small footprint airborne laser scanning data based on gap fraction. Remote Sens. Environ. 2006, 104, 50–61. [Google Scholar] [CrossRef]
- Coops, N.C.; Hilker, T.; Wulder, M.A.; St-Onge, B.; Newnham, G.; Siggins, A.; Trofymow, J.A. (Tony) Estimating canopy structure of Douglas-fir forest stands from discrete-return LiDAR. Trees 2007, 21, 295–310. [Google Scholar] [CrossRef] [Green Version]
- Næsset, E.; Bjerknes, K.-O. Estimating tree heights and number of stems in young forest stands using airborne laser scanner data. Remote Sens. Environ. 2001, 78, 328–340. [Google Scholar] [CrossRef]
- Lovell, J.L.; Jupp, D.L.B.; Culvenor, D.S.; Coops, N.C. Using airborne and ground-based ranging lidar to measure canopy structure in Australian forests. Can. J. Remote Sens. 2003, 29, 607–622. [Google Scholar] [CrossRef]
- Xi-Guang, Y.; Wen-Yi, F.; Ying, Y. Estimation of Forest Canopy Chlorophyll Content Based on PROSPECT and SAIL models. Spectrosc. Spectr. Anal. 2010, 30, 3022–3026. [Google Scholar] [CrossRef]
- Bowyer, P.; Danson, F.M. Sensitivity of spectral reflectance to variation in live fuel moisture content at leaf and canopy level. Remote Sens. Environ. 2004, 92, 297–308. [Google Scholar] [CrossRef]
- Keenan, T.F.; Niinemets, Ü. Global leaf trait estimates biased due to plasticity in the shade. Nat. Plants 2016, 3, 16201. [Google Scholar] [CrossRef] [Green Version]
- Junhua, Z.; Jiabao, Z. Response of the Spectral Reflectance to Different Pigments of Summer Maize. Acta Agric. Boreali-Occident. Sin. 2010, 19, 70–76. [Google Scholar]
- La, Q.; Chun-Jiang, Z.; Wen-Jiang, H.; Han-Hai, L. Sensitivity Analysis of Canopy Spectra to Canopy Structural Parameters Based on Multi-temporal Data. Geogr. Geo-Inf. Sci. 2009, 25, 17–25. [Google Scholar]
- Zongjian, Z.; Yuyan, L.; Mingchun, G. Study on the Canopy Structure and Photosynthetic Characteristics of Ginkgo Biloba L.Saplings; Hebei Normal University of Science and Technology: Qin Huang Dao Shi, China, 2014. [Google Scholar]
- Xiangqian, W.; Lin, C.; Xin, S.; Guibin, W.; Fuliang, C. Estimation of Effective Leaf Area Index Using UAV-Based LiDAR in Ginkgo Plantations. For. Resour. Manag. 2020, 56, 74–86. [Google Scholar]
- Dong, L.; Jing, M.C.; Xiao, Z.; Yan, Y.; Jie, Z.; Hengbiao, Z.; Kai, Z.; Xia, Y.; Yongchao, T.; Yan, Z.; et al. Improved estimation of leaf chlorophyll content of row crops from canopy reflectance spectra through minimizing canopy structural effects and optimizing off-noon observation time. Remote Sens. Environ. 2020, 248, 111985. [Google Scholar] [CrossRef]
- Cheng-Yan, G.; Hua-Qiang, D.; Guo-Mo, Z.; Ning, H.; Xiao-Jun, X.; Xiao, Z.; Xiao-Yan, S. Retrieval of leaf area index of Moso bamboo forest with Landsat Thematic Mapper image based on PR OSAIL canopy radiative transfer model. Chin. J. Appl. Ecol. 2013, 24, 2248–2256. [Google Scholar]
- Sinha, S.K.; Padalia, H.; Dasgupta, A.; Verrelst, J.; Rivera, J.P. Estimation of leaf area index using PROSAIL based LUT inversion, MLRA-GPR and empirical models: Case study of tropical deciduous forest plantation, North India. Int. J. Appl. Earth Obs. Geoinf. 2020, 86, 102027. [Google Scholar] [CrossRef]
- Zhenhai, L.; Zhenhong, L.; Fairbairn, D.; Na, L.; Bo, X.; Haikuan, F.; Guijun, Y. Multi-LUTs method for canopy nitrogen density estimation in winter wheat by field and UAV hyperspectral. Comput. Electron. Agric. 2019, 162, 174–182. [Google Scholar] [CrossRef]
- Verrelst, J.; Romijn, E.; Kooistra, L. Mapping Vegetation Density in a Heterogeneous River Floodplain Ecosystem Using Pointable CHRIS/PROBA Data. Remote Sens. 2012, 4, 2866–2889. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Muñoz, J.; Alonso, L.; Delegido, J.; Rivera, J.P.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Chun-Xia, H.; Ji-Yue, L.; Yan-Xiang, Z.; Quan-Shui, Z.; Xie, B.; Yi-Ting, D. Differences in leaf mass per area, photosynthetic pigments and δ13C by orientation and crown position in five greening tree species. Chin. J. Plant Ecol. 2010, 34, 134–143. [Google Scholar] [CrossRef]
- Feng-Li, H.; Fei, W.; Qin-Ping, W.; Xiao-Wei, W.; Qiang, Z. Relationship Between Distribution of Relative Light Intensity in Canopy and Yield and Quality of Peach Fruit. Sci. Agric. Sin. 2008, 41, 502–507. [Google Scholar]
- Shaoxuan, L.; Fuliang, C. Study of Crown Structure Feacture in the Timber Ginkgo; Nanjing Forestry University: Nanjing, China, 2014. [Google Scholar]
- Yong, L.; Hairong, H.; Fengfeng, K.; Xiaoqin, C.; Ke, L.; Bin, Z.; Yali, S. Spatial Heterogeneity of Photosynthetic Characterisitics of Pinus tabulaeformis Canopy. J. Northeast For. Univ. 2013, 41, 32–35. [Google Scholar]
- Xue-Hong, Z.; Qing-Jiu, T.; Run-Ping, S. Analysis of Directional Characteristics of Winter Wheat Canopy Spectra. Spectrosc. Spectr. Anal. 2010, 30, 1600–1605. [Google Scholar] [CrossRef]
- Ghosh, A.P.; Dass, A.; Krishnan, P.; Kaur, D.; Rana, K. Assessment of photosynthetically active radiation (PAR), photosynthetic rate (NPR), biomass and yield of two maize varieties under varied planting dates and nitrogen application. J. Environ. Biol. 2017, 38, 683–688. [Google Scholar] [CrossRef]
- Wei, L.; Kun-Fang, K. Effects of Drought Stress on Photosynthetic Characteristics and Chlorophyll Fluorescence Parameters in Seedlings of Terminthia paniculata Grown under Different Light level. Acta Bot. Boreali-Occident. Sin. 2006, 26, 266–275. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
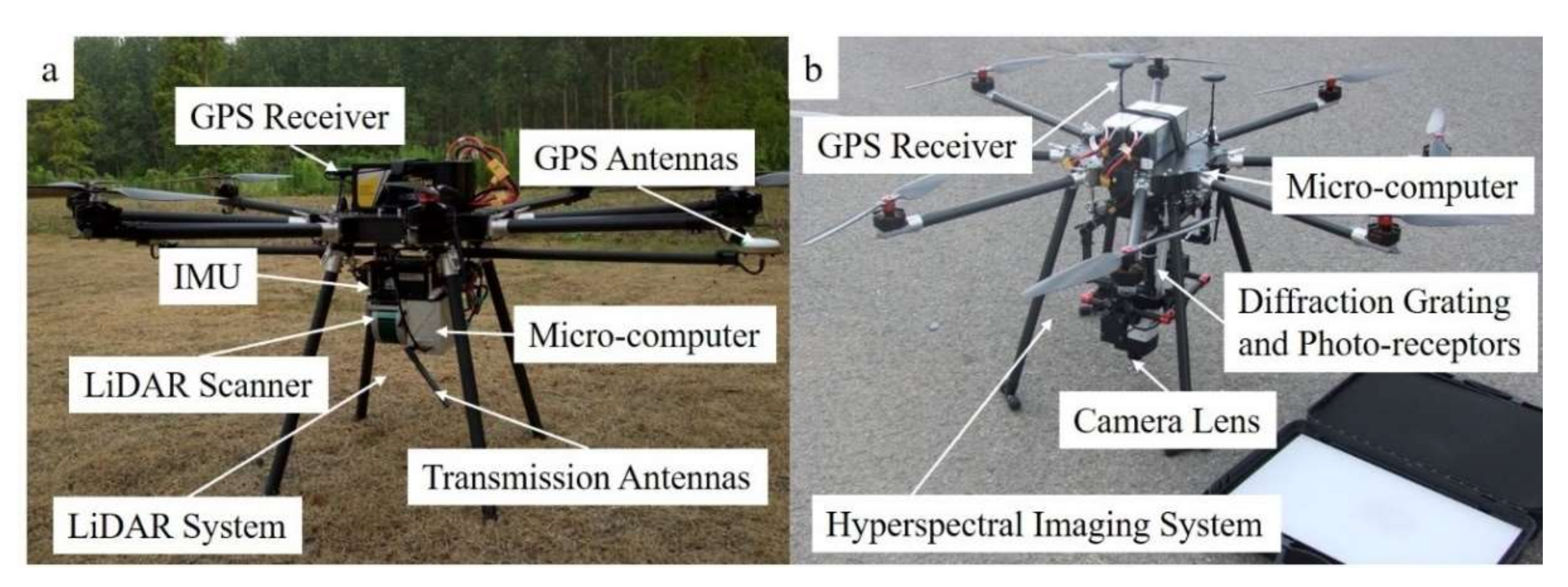



| Parameters | UAS-Hyperspectral System | UAS-LiDAR System |
|---|---|---|
| Sensors | ZK-VNIR-FPG480 | Velodyne Puck VLP-16 |
| Data of acquisition | 17 Aug. 2019 | 18 Aug. 2019 |
| Flight height (m) | 80 | 80 |
| Flight speed (m·s−1) | 4.8 | 6 |
| Side overlap (%) | >20 | 100 |
| Focal length (mm) | 16 | - |
| IFOV/Beam divergence (mrad) | 0.9 | 3 |
| Spatial resolution/Footprint (cm) | 18.75 | 17 |
| FOV/Maximum scan angle (°) | 26 | ± 30 |
| Wavelength (nm) | 403–929 | 903 |
| Spectral sampling (nm) | 2.3 | - |
| Number of bands | 226 | 1 |
| Average point density (pts·m−2) | - | 169 |
| Model Parameters | Variable (Unit) | Range | Step |
|---|---|---|---|
| Structural coefficient | N (-) | 1.1–1.7 | 0.1 |
| Total chlorophyll content | Cab (μg·cm−2) | 10–70 | 10 |
| Carotenoid content | Car (μg·cm−2) | 0–15 | 2.5 |
| Leaf area index | LAI (m2·m−2) | 0–3 | 0.5 |
| Average leaf angle | ALA (°) | 0–90 | 15 |
| Hotspot | hspot | 0.1–0.4 | 0.05 |
| Fraction of dry soil | psoil | 0.4–1 | 0.1 |
| Fraction of diffuse radiation | skyl (%) | 10–70 | 10 |
| Parameters of PROSPECT-D Model | ||||
|---|---|---|---|---|
| Parameters | Variable (Unit) | Rang (13-year-old) | Rang (22-year-old) | Reference |
| Structural coefficient | N (-) | Fixed at 1.4 | Fixed at 1.4 | [21] |
| Total chlorophyll content | Cab (μg·cm−2) | 10–70 | 10–70 | The measured data |
| Carotenoid content | Car (μg·cm−2) | 0–15 | 0–15 | 0.1512 × Cab + 2.1864 |
| Water content | Cw (g·cm−2) | Fixed at 0.017 | Fixed at 0.017 | The mean of measured data |
| Dry matter content | Cm (g·cm−2) | Fixed at 0.009 | Fixed at 0.009 | The mean of measured data |
| Anthocyanin content | Canth (μg·cm−2) | Fixed at 0.0 | Fixed at 0.0 | [60] |
| Brown pigment content | Cbrown (-) | Fixed at 0.0 | Fixed at 0.0 | [60] |
| Parameters of SAIL model | ||||
| Parameters | Variable (Unit) | Rang (13-year-old) | Rang (22-year-old) | Reference |
| Leaf area index | LAI (m2·m−2) | 0.3–3 | 0.3–3 | [59] |
| Average leaf angle | ALA (°) | Fixed at 45 | Fixed at 45 | [58] |
| Hotspot | hspot | Fixed at 0.15 | Fixed at 0.15 | [61] |
| Fraction of dry soil | psoil | Fixed at 0.8 | Fixed at 0.8 | [62] |
| Fraction of diffuse radiation | skyl (%) | Fixed at 10 | Fixed at 10 | The LSA result |
| Viewing zenith angle | VZA(°) | Fixed at 0 | Fixed at 0 | The flight time |
| Solar zenith angle | SZA (°) | Fixed at 35.5 | Fixed at 67.5 | The flight time |
| Relative azimuth angle | RAA(°) | Fixed at 115.2 | Fixed at 88.1 | The flight time |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, S.; Zhou, K.; Cao, L.; Shen, X. Estimating the Horizontal and Vertical Distributions of Pigments in Canopies of Ginkgo Plantation Based on UAV-Borne LiDAR, Hyperspectral Data by Coupling PROSAIL Model. Remote Sens. 2022, 14, 715. https://doi.org/10.3390/rs14030715
Yin S, Zhou K, Cao L, Shen X. Estimating the Horizontal and Vertical Distributions of Pigments in Canopies of Ginkgo Plantation Based on UAV-Borne LiDAR, Hyperspectral Data by Coupling PROSAIL Model. Remote Sensing. 2022; 14(3):715. https://doi.org/10.3390/rs14030715
Chicago/Turabian StyleYin, Shiyun, Kai Zhou, Lin Cao, and Xin Shen. 2022. "Estimating the Horizontal and Vertical Distributions of Pigments in Canopies of Ginkgo Plantation Based on UAV-Borne LiDAR, Hyperspectral Data by Coupling PROSAIL Model" Remote Sensing 14, no. 3: 715. https://doi.org/10.3390/rs14030715
APA StyleYin, S., Zhou, K., Cao, L., & Shen, X. (2022). Estimating the Horizontal and Vertical Distributions of Pigments in Canopies of Ginkgo Plantation Based on UAV-Borne LiDAR, Hyperspectral Data by Coupling PROSAIL Model. Remote Sensing, 14(3), 715. https://doi.org/10.3390/rs14030715








