Abstract
Miscanthus is one of the most promising perennial crops for bioenergy production, with high yield potential and a low environmental footprint. The increasing interest in this crop requires accelerated selection and the development of new screening techniques. New analytical methods that are more accurate and less labor-intensive are needed to better characterize the effects of genetics and the environment on key traits under field conditions. We used persistent multispectral and photogrammetric UAV time-series imagery collected 10 times over the season, together with ground-truth data for thousands of Miscanthus genotypes, to determine the flowering time, culm length, and biomass yield traits. We compared the performance of convolutional neural network (CNN) architectures that used image data from single dates (2D-spatial) versus the integration of multiple dates by 3D-spatiotemporal architectures. The ability of UAV-based remote sensing to rapidly and non-destructively assess large-scale genetic variation in flowering time, height, and biomass production was improved through the use of 3D-spatiotemporal CNN architectures versus 2D-spatial CNN architectures. The performance gains of the best 3D-spatiotemporal analyses compared to the best 2D-spatial architectures manifested in up to 23% improvements in R2, 17% reductions in RMSE, and 20% reductions in MAE. The integration of photogrammetric and spectral features with 3D architectures was crucial to the improved assessment of all traits. In conclusion, our findings demonstrate that the integration of high-spatiotemporal-resolution UAV imagery with 3D-CNNs enables more accurate monitoring of the dynamics of key phenological and yield-related crop traits. This is especially valuable in highly productive, perennial grass crops such as Miscanthus, where in-field phenotyping is especially challenging and traditionally limits the rate of crop improvement through breeding.
1. Introduction
New and improved bioenergy crops are needed to enhance the domestic production of energy, stimulate rural economies, make agriculture more resilient to climate change, and reduce greenhouse gas emissions by reducing dependence on fossil-fuel-based products [1]. Meeting this challenge requires the rapid development of crops optimized for their ecosystem services, harvestable biomass, and high-value compounds [2]. Miscanthus is a perennial rhizomatous C4 grass originating from East Asia, with high photosynthetic efficiency, high nutrient- and water-use efficiency, and wide adaptability to various climates and soil types [3], i.e., traits that favor high productivity and sustainability on marginal lands [3]. Consequently, it is expected to play a major role in the provision of biomass as a feedstock for energy and bioproducts [4].
However, the need for rapid domestication and improvement of dedicated energy crops presents an unprecedented challenge to plant breeding that can best be addressed with the application of cross-disciplinary approaches and novel technologies [2]. Traditionally, the selection of the best candidate genotypes in breeding programs has mainly relied on in situ visual inspection [5] and laborious destructive sampling throughout critical stages of crop development [6]. Remote sensing has been widely adopted for monitoring physiological and structural traits in annual crops [7,8]. Nevertheless, the adoption of this technology has been significantly slower in perennial feedstock grasses [9]. This partially reflects less research investment relative to existing commodity crops, as well as challenges associated with large, complex, rapidly growing canopies, where signal saturation and occlusion can be significant problems [10]. The challenging nature of Miscanthus propagation also means that early-phase selection trials [11] often require evaluation of traits on individual plants [12], rather than as closed stands in plots, which adds to the technical difficulty of using remote sensing. The development of digital tools that provide precise and dedicated characterization of perennial grasses could therefore substantially reduce the cost of evaluating large populations and commercial fields, but requires technical innovation to do so [13].
Vegetation indices [14] and photogrammetric features [15] have been used to estimate biomass yield in bioenergy crops. More recently, vegetation indices derived from remote sensing imagery and machine learning were integrated to assess the relationship between the senescence rate and moisture content in Miscanthus [13]. Research also suggests that the use of trajectories of photogrammetric or vegetation indices can benefit the predictability of traits of interest [16,17]. There is potential to increase the precision and automation of analyses by acquiring images with a richer feature set and through the use of more advanced analytical approaches that could more fully exploit the spatial and temporal information contained in the images. Additionally, the increasing availability of satellite- [18] and UAV-based sensors [19], producing imagery at unprecedented spatial and temporal resolutions, represents a dramatic increase in the volume of data available. This then requires the development of modular and scalable pipelines for actionable impact in agricultural applications. Convolutional neural networks (CNNs) have proven powerful in extracting information from very-high-spatial-resolution aerial imagery [20,21]. The capacity of CNNs to automatically generate representations of complex features provides an advantage over traditional machine learning, which has mainly relied on manual supervision, e.g., feature generation and selection during modelling implementation [16,21]. However, most existing applications of CNNs evaluate variation in image features over the two dimensions of images collected at a single point in time. Given the dynamic nature of plant growth, there is an increasing interest in adopting models capable of also integrating time as a third dimension explicitly captured by the CNN to better understand and gain novel insights about the plant trait(s) under study [22,23]. Previous studies suggest that multi-date integration of images can perform better than single-date CNNs in a limited set of circumstances. For example, using time-series UAV RGB and weather data collected from nine farm fields including oats, barley, and wheat, spatiotemporal deep learning architectures were able to outperform single-time CNNs in intra-field crop yield estimates [24]. In a more recent publication, a spatiotemporal CNN architecture was superior to a traditional CNN for detecting and determining the severity of lodging in biomass sorghum [25]. However, these are comparatively simple phenotyping challenges. The value of adopting CNN architectures capable of exploiting the temporal information contained in a sequence of images over time may be even greater when presented with the challenge of accurately distinguishing differences in complex traits within a breeding population.
The overall aim of this study was to evaluate how spectral and geometric features in aerial imagery collected throughout the growing season under field conditions could best be exploited in analyses by CNNs for phenotyping time-series-based plant-level predictions of three relevant Miscanthus productivity traits: flowering time, culm length, and biomass yield. We propose to adopt CNNs as a means to ease and semi-automate both feature extraction and the estimation of these three relevant Miscanthus traits. In the context of performing data-based modelling to quantify relevant descriptive traits of Miscanthus, we first utilized a timepoint CNN architecture (i.e., 2D-CNN) to understand the predictive ability of the proposed approach to determine the traits throughout the season. We then focused on determining the value of persistent aerial monitoring by implementing a spatiotemporal CNN architecture (i.e., 3D-CNN) based on sequences of time-steps to understand whether it outperformed the predictive ability of single-time models. The third element of the study was an assessment of how the accuracy of trait prediction depends on the use of the various spectral and geometric features derived from the UAV imagery. Finally, we visualized activation maps to derive insights about the CNN learning and predictive ability [26].
2. Materials and Methods
2.1. Experimental Site
The data collected consisted of three Miscanthus diversity trials located at the University of Illinois Energy Farm, in Urbana (40.06722°N, 88.19583°W) (Figure 1a,b). In one trial we evaluated M. sacchariflorus, and in the other trials we evaluated M. sinensis. The trials were planted in spring 2019; this study focused on the second year (2020)—the first growing season in which we typically phenotyped Miscanthus plants for yield and related traits. The M. sacchariflorus trial (Figure 1a, north field, rectangular yellow area divided into four quadrants) included 2000 entries as single-plant plots in four blocks, each block including 58 genetic backgrounds (half-sib families). The analysis was limited to 46 of these genetic backgrounds from South Japan. The size of the trial was 79 m long × 97 m wide, and each plot (plant) was 1.83 × 1.83 m size. A full set of images of the plots were prepared for this trial, and 1307, 1867, and 1864 images for flowering time, culm length, and biomass yield, respectively, were used in the analysis to allow for comparison with the ground-truth data. Two adjacent M. sinensis trials were studied, with one for the South Japan genetic group (Figure 1a, southern field, two southern tiers in yellow) and the other for the Central Japan genetic group [27] (Figure 1a, the two yellow-highlighted northern tiers in the southern field). Each trial consisted of five blocks (tiers). Between blocks, there were 1.83 m wide alleys (Figure 1a, south field, in pink). Each plot consisted of up to 10 plants spaced 0.91 m within and between plots within each block. The plots contained seedlings from a single half-sib family. Miscanthus is an obligate outcrosser due to self-incompatibility, so each seedling was genetically unique. Thus, families were replicated in these trials. Each block had 130 plots (up to 1300 plants). In the M. sinensis South Japan trial there were 124 families, and in the Central Japan trial there were 117 families; some large families were included in more than one plot per block. Only two middle blocks of each M. sinensis trial were considered in the analysis (Figure 1a, south field, in yellow). Additionally, subsets of 1722, 2519, and 2481 images for flowering time, culm length, and biomass yield, respectively, were used to allow for comparison with the ground-truth data. The size of the field that included both of the M. sinensis trials was 115 m long × 121 m wide.

Figure 1.
(a) Aerial overview of Miscanthus sacchariflorus (top) and Miscanthus sinensis (bottom) trials and plots analyzed in this study (in yellow) located at the Energy Farm, University of Illinois, Urbana. (b) Close-up images of plots and individual plants under intensive growth during the summer of 2020 (top, M. sacchariflorus; bottom, M. sinensis).
2.2. Traits of Interest and Ground-Truthing
The target traits—flowering time, culm length, and biomass yield—were selected as relevant descriptors of the phenology and aboveground productivity of the crop [5]. Manual screening of Miscanthus trials is a massive repeated effort carried out by researchers at the University of Illinois as part of its breeding program. Regular field activities include visual determination of flowering time (Figure 2a), collection of mature stems for measurement of culm length, and end of season harvest (i.e., destructive sampling) to estimate biomass yield (Figure 2b,c). In these trials, each plant was phenotyped directly for the target traits and other traits of interest.

Figure 2.
Example cases of plant traits collected during field activities and used as ground-truthing for model implementation and validation. Examples of (a) early (back) and late (front) flowering time, (b) culm length measurement, and (c) biomass yield collected in seasons 2020–2021.
In this study, the data collected manually provided the inputs for implementing models based on UAV data. The target traits are described as follows:
Flowering time (50% heading date): Determined as the date on which 50% of the culms that contribute to the plant’s canopy height had inflorescences that emerged ≥1 cm beyond the flag leaf sheath. Each plant was phenotyped weekly between days of the year (DOYs) 238 and 285 (Table 1).
Culm length: Determined as the length in centimeters of the plant’s tallest culm in late autumn, measured from the base of the stem to the tip of the panicle if present, or otherwise to the highest part of the highest leaf. Culms were collected in December 2020 (Table 1) from dormant plants by cutting at ground level, tagged with a barcode label, and transported to a laboratory for measurement.
Biomass yield: After dormancy, the stems of each plant were cut 15–20 cm from the ground. To determine the dry biomass yield, samples were oven-dried at 60 °C until a constant weight was obtained before recording the dry weights. For the M. sinensis trials, whole plants were tied in a bundle, tagged with a barcode label, cut with a handheld bush cutter (Stihl Inc., Virginia Beach, VA, USA), dried, and then weighed. For the M. sacchariflorus trial, plots were harvested using a Wintersteiger Cibus Harvester (Wintersteiger AG, Ried, Austria) with a Kemper Champion C 1200 Forage Harvester attachment (Kemper GmbH & Co. KG, Stadtlohn, Germany). With the Wintersteiger harvester, the plots were chopped, and fresh weights of the main sample were measured using the machine’s weigh-wagon. A 1 kg subsample was taken from the harvester for measuring the fresh and dry weights to estimate the water content and calculate the dry weight of the plot. The yield of dry weight per plot was expressed in g per plant. Biomass harvesting was conducted between February and March, 2021 (Table 1).

Table 1.
Timing of data collection in the field: ground-truthing in the trials and UAV flights throughout the season considered for the CNN modelling setup.
Table 1.
Timing of data collection in the field: ground-truthing in the trials and UAV flights throughout the season considered for the CNN modelling setup.
| Traits | Data Collection | Dates (DOY) |
|---|---|---|
| Flowering time | Emergence of inflorescences |  |
| Ground truth (M. sacchariflorus) |  | |
| Ground truth (M. sinensis) |  | |
| UAV flights |  | |
| UAV (time sequence) 262_279 (2) |  | |
| UAV (time sequence) 247_262_279 (3) |  | |
| UAV (time sequence) 221_247_262_279 (4) |  | |
| UAV (time sequence) 247_262_279_310 (4) |  | |
| UAV (time sequence) 221:310 (5) |  | |
| Culm length or biomass yield | Ground truth (both species) |  |
| UAV flights |  | |
| UAV (time sequence) 247_332 (2) |  | |
| UAV (time sequence) 157_174_190 (3) |  | |
| UAV (time sequence) 205_221_247 (3) |  | |
| UAV (time sequence) 174_247_332 (3) |  | |
| UAV (time sequence) 279_310_332 (3) |  | |
| UAV (time sequence) 247:332 (5) |  | |
| UAV (time sequence) 157:332 (10) |  |
2.3. Aerial Data Collection and Imagery Preprocessing
The aerial platform utilized was a Matrice 600 Pro hexacopter (DJI, Shenzhen, China) equipped with a Gremsy T1 gimbal (Gremsy, Ho Chi Minh, Vietnam), which held a multispectral (MSI) Rededge-M sensor (Micasense, Seattle, WA, USA). The camera included five spectral bands in the blue (465 to 485 nm), green (550 to 570 nm), red (663 to 673 nm), red edge (712 to 722 nm), and near-infrared (820 to 860 nm) regions of the electromagnetic spectrum. Flights were conducted 10 times in the season between DOYs 157 and 332, under clear-sky conditions and ±1 h from solar noon. Once Miscanthus is senesced late in the growing season, the plants stay dormant during winter, so there is no further gain in aboveground biomass. The standard agronomic practice is for this crop to be harvested in the winter. During winter, cold temperatures make UAV operation very unreliable, so image acquisition cannot be continued into that period. The altitude of the flights was set to 20 m above ground level, resulting in a ground sampling distance (GSD) of 0.8 cm/pixel. The flights were planned with 90% forward and 80% side-overlapping during image acquisition to ensure high-quality image stitching and photogrammetric reconstruction. Ten black-and-white square panels (70 cm × 70 cm) were distributed in the trials as ground control points (GCPs). An RTK (real-time kinematic) survey was implemented using a Trimble R8 global navigation satellite system (GNSS) integrated with the CORS-ILUC local station to survey the GCPs for accurate co-registration of orthophotos throughout the season. A standard Micasense calibration panel was imaged on the ground before and after each flight for spectral calibration. The images were further imported into Metashape version 1.7.4 (Agisoft, St. Petersburg, Russia) to generate two types of aerial image features: (1) multispectral, 5-band orthophotos; and (2) geometric orthophotos, resulting from the photogrammetric reconstruction of the canopy, referred to as crop surface models (CSMs). The computation time for processing each orthophoto was 3 h and 10 min for the M. Sacchariflorus trial and 4 h and 20 min for the M. Sinensis trial, using an Intel i7 16GB RAM CPU and 8GB GPU. An early-season flight before plant emergence provided a ground-level reference for the extraction of the absolute height from the CSM files on subsequent sampling dates (n = 10). The resulting multispectral and CSM orthophotos were resampled to a common 1.4 cm/pixel resolution and stacked into one multi-feature and multi-date raster stack object. Image chips for each plot/plant were generated by clipping the stacked orthophoto objects using a polygonal shapefile including each plot polygon of the trials. Three variations of the image chips were generated: (1) spectral features from RGB images (RGB); (2) spectral features from the RGB, red-edge, and NIR bands (MS); and (3) spectral and geometric features from the RGB, red-edge, and NIR bands along with a crop surface model (CSM_MS) (Table S1). A customized pipeline for the production of image chips, data import, and matching between images and labels was implemented in Python 3.7.11.
2.4. CNN Modelling
The architecture of a convolutional neural network consists of a sequence of layers, where every layer transforms the activations or outputs of the previous layer through another differentiable function. The core component of the CNN algorithm is the convolution operation [28], where a set of trainable kernels are applied to an input image (Figure 3a) to generate a set of spatial features that describe the target predictor. The first part of the architecture, known as the backbone feature generator, learns basic features in the first layers and more complex feature representations at deeper layers [28] (Figure 3b). The final output of the feature generator is a set of feature maps that are directly fed into the regressor head, which uses a linear function to deliver the prediction (Figure 3c). The 3D-CNN architecture belongs to the group of spatiotemporal CNN architectures and can be defined as a variation of the traditional 2D-CNN [29]. It includes all of the standard features but adds a new dimension across the time sequence of the image, enabling capture of the temporal dynamics of vegetation that can contribute to better characterizing the trait of interest. The spatiotemporal architecture adopts a 3D kernel that operates in both the spatial and temporal dimensions of the multi-timepoint image.
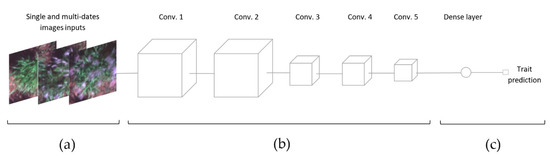
Figure 3.
Diagram of the custom CNN architecture utilized to assess flowering time, culm length, and biomass yield in Miscanthus. (a) Input single- or multi-date image chips with varying numbers of features, as described in Table S1. (b) Backbone feature generator and (c) regressor head for trait prediction.
2.5. CNN Implementation and Metrics
A customized CNN architecture was utilized for all traits under study. The backbone feature generator includes five convolution layers, maximum-pooling operations following the convolution operation in layers 2 and 4, and 50% feature dropout and global average pooling at layer 5, (Figure 3b). After preliminary experimentation, the number of features in each layer is set to 64, 64, 32, 32, and 32, respectively (Figure 3b). Zero padding, stride equal to one with no overlapping, and the rectified linear unit activation function are also considered in the architecture design. The convolution layers are two- and three-dimensional for 2D-CNNs and 3D-CNNs, respectively. In contrast with the 2D-CNN kernel, the 3D-CNN kernel moves in three instead of two dimensions. The 2D-CNN’s kernel filter size was set to (3 pixels × 3 pixels × n features), while the 3D- kernel’s size was set to (3 pixels × 3 pixels × n features × n-time sequence lengths), operating at the sequence of each feature over time. The 2D-max-pooling was set to two following each convolution, with 3D-max-pooling equal to two over the spatial dimensions and equal to one over the time/depth dimension. The feature generator was linked to a fully connected layer or regressor head, which generated the prediction values for each trait.
A 2D-CNN analysis was conducted on data from individual dates for each trait as a means to test the sensitivity between ground-truthing, timing of aerial data collection, and modelling of the CNN architectures. In addition, various configurations of input data from sequences of dates were analyzed using the 3D-CNN architecture to test the sensitivity of the predictions to variations in the length and timepoint selected in the time sequence to the final prediction power of the traits. Table 1 summarizes the dates of ground-truth data collection and aerial data collection, as well as the different timepoints and time ranges selected for analysis in the 3D-CNN architecture. We tailored the time series tested to reflect that flowering time is a discrete event that occurs in the mid-to-late season, while end-of-season culm length and biomass are the product of growth processes that span the whole field season. Flowering occurred from DOY 238 to 283. Therefore, we focused the 3D-CNN analysis around this period, but with different time sequences that varied in the number of dates, their timing with respect to when the crop flowered, and the total duration of the sampling period. Analysis of culm length and biomass yield used the same set of time series. In their case, we considered a wider window of time for aerial coverage, up to and including all phases of the growing season. Periods of early intensive vegetative growth, the transition between the vegetative and reproductive stages, and the senescence period were also selected for isolated analysis.
The modelling step was implemented in the Python Keras API with a TensorFlow-GPU version 2.8.0 backend using a NVIDIA GeForce RTX 3070 (8 GB) GPU unit. The combination of different types of features, CNN architectures, and time sequence lengths of days of flights resulted in the implementation of 138 models (Table S2). In total, across all traits, 81 models were implemented using the 2D-CNN architecture and 57 models were implemented using the 3D-CNN architecture. Each model fitting was iterated 5 times using a random training and testing partition to ease the convergence of the models’ prediction metrics. The dataset used for prediction of the traits varied for each trait according to the ground-truth data available. The total number of image chips for the analysis of flowering time, culm length, and biomass yield on each date for the 2D-CNN and on all dates stacked for the 3D-CNN was 3029, 4296, and 3937, respectively. The input datasets were split (70:30) into training and testing datasets. The training datasets were split further (80:20) into training and validation datasets. The validation datasets were used to optimize the models’ performance and prevent overfitting during training. Mean squared error was used as the loss function. The models were trained up to 500 epochs using an early stop with a lambda value of 0.01 to ensure efficient training time. The learning rate and decay parameters were initialized with values of 0.01 and 0.001, respectively. The test datasets were utilized to expose the models to unseen data to evaluate the generalization ability of the models.
The mean absolute error (MAE), the relative root-mean-square error (RMSE), and the coefficient of determination (R2) were estimated (Equations (1)–(3)) to determine the performance of the CNNs for each of the traits.
In Equations (1)–(3), y, , and are the observed, predicted, and observed mean trait values of the ith plot, respectively, and n is the total number of samples within the study area.
Activation mapping visualization was implemented to improve the interpretation of the CNN learning process [26]. The visualization technique highlighted the importance of different regions of the image in the output prediction by projecting back the weights of the output layer onto the convolutional feature maps [30]. The following steps, as described in [31], were used to generate the activation maps: First, the CNN model mapped the input image to the activations of the last convolution layer as well as the output predictions. The gradient of the predicted value for the input image with respect to the activation of the last convolution layer was computed. Each image channel in the feature map array was weighted by how important the channel was with regard to the predicted value, and then all of the channels were summed to generate the corresponding activation map array. The activation map provided a measure of how strongly portions of the image contributed to the predictions made by the CNN, visualized in a 0–255-scale map array.
3. Results
3.1. Flowering Time
For the task of estimating flowering time, 2D-CNN analyses (Figure 4a) of imagery from any single date throughout the growing season were outperformed by all of the 3D-CNN analyses tested (Figure 4b) in terms of MAE, RMSE, and R2. This was the case regardless of the number or timing of dates from which imagery was used as inputs for the 3D-CNNs. The ability of the 3D-CNN to model variation between dates as well as among images from different genotypes led to the best 3D-CNN model (3D_262_279(2)) performing significantly better (Figure 4d; MAE = 4.61 days, RMSE = 6.05 days, R2 = 0.70) (Figure 4d) than the best 2D-CNN (Figure 4e; 2D_262(1) and 2D_279(1); MAE = 5.20 days, RMSE = 6.69 days, R2 = 0.63) (Figure 4c).
Unsurprisingly, among the 2D-CNN analyses, the model accuracy was substantially greater midway through the flowering period (DOYs 262 and 279) than when using images of plants prior to the onset of flowering for many genotypes (DOY < 262) or after all genotypes had completed flowering and were senescing (DOY > 279) (Figure 4a). Meanwhile, while the results from the 3D-CNNs were generally robust, a slight loss of predictive power from the 3D-CNNs was observed as additional dates of imagery were added beyond the two dates (DOYs 262 and 279) in the middle of the flowering period (Figure 4b).
Compared to using multispectral features or geometric + multispectral features, the effect of analyzing only RGB spectral features on the estimation of flowering time was that the predictions were slightly weaker and inconsistent across the scenarios tested by both 2D-CNNs and 3D-CNNs (Figure 4a,b).
When interpreting the CNN learning process via activation maps, it was notable that the models tended to modulate their activation levels depending on the flowering time and were driven by the presence of inflorescences (silver-white areas) in the imagery. For early flowering time (i.e., lower numerical values), higher activation regions (red) were visibly located over the green areas of the plant, but minimized over the inflorescence (Figure 4e). In contrast, for plants with a later flowering time that had still not flowered (i.e., higher numerical values) at the time of the UAV flight, higher (red) activation was notable in the imagery due to the dominant greenness (i.e., absence of inflorescences) on the plant (Figure 4f).
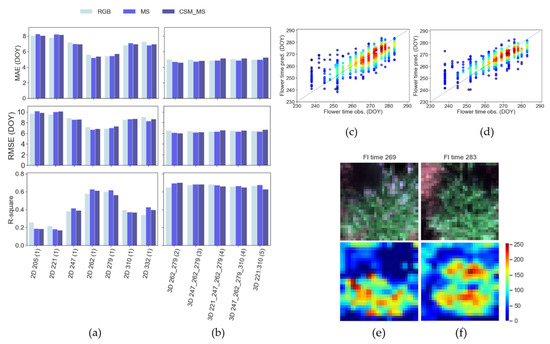
Figure 4.
Evaluation of flowering time prediction in testing data for (a) 2D models by DAP and (b) 3D models by number of time-frame sequences via the MAE, RMSE, and R2 metrics. Observed and predicted values for the best (c) 2D models and (d) 3D models for testing datasets with low- (blue) to high-density (red) sample distribution. Example RGB image chips and activation maps for (e) early- and (f) late-flowering-time plants from the 2D-based model of aerial data collected on DOY 279. The level of activation of the deepest CNN layers in the images is represented by low (blue), intermediate (yellow), and high (red) values on a 0–255 scale.
3.2. Culm Length
For the task of estimating culm length, the performance of both 2D-CNN analyses (Figure 5a) and 3D-CNN analyses (Figure 5b) was strongly dependent on the dates in the field season from which imagery was used as an input. Thus, the comparative performance of 2D-CNNs versus 3D-CNNs must be discussed in that context. Nevertheless, a key high-level conclusion is that the best 3D-CNN model (3D_157:332(10)) performed significantly better (Figure 5c; MAE = 19.06 cm, RMSE = 24.93 cm, R2 = 0.66) than the best 2D-CNN (Figure 5d; 2D_262(1) and 2D_279(1); MAE = 20.95 cm, RMSE = 27.08 cm, R2 = 0.58).
For both 2D-CNNs and 3D-CNNs, the best results in terms of MAE, RMSE, and R2 came from using images collected late in the growing season. The performance of the 2D-CNNs was moderate when using imagery from dates early in the growing season, worsened slightly with mid-season imagery, and improved again with late-season imagery, before dropping again on the final date (Figure 5a). The performance of the 3D-CNNs was moderate when relying on only early-season (3D_157_174_190(3)) or mid-season imagery (3D_205_221_247(3)), and improved significantly when using late-season data, especially as the number of dates of imagery analyzed increased from 3 (3D_279_310_332(3)) to 5 (3D_247:332(5)) or 10 (3D_157:332(10)) (Figure 5b). Intermediate predictive power was achieved when imagery from fewer dates, which were spread at lower frequency across the growing season, were used as inputs to the 3D-CNN (3D_174_247_332(3)).
On most individual dates, analysis of geometric plus multispectral features (CSM_MS) led to the best predictions of culm length, followed by multispectral features (MS) and then RGB spectral features, with those differences being most pronounced when analyzing imagery from late-season dates (Figure 5a). Using multispectral + geometric features delivered the most benefit when the 3D-CNNs analyzed data from three or fewer dates. In contrast, in the best-performing 3D-CNN models—which exploited 5 or 10 dates of imagery—adding additional spectral bands or geometric features had modest effects on predictive power.
When evaluating how CNNs exploit the information contained in the imagery, the models assigned high (red) activation levels to the dense greenness of the plant in the image, while low (blue) activation levels were focused on the background (e.g., soil and plant shade) in the images, as shown in Figure 5e,f.
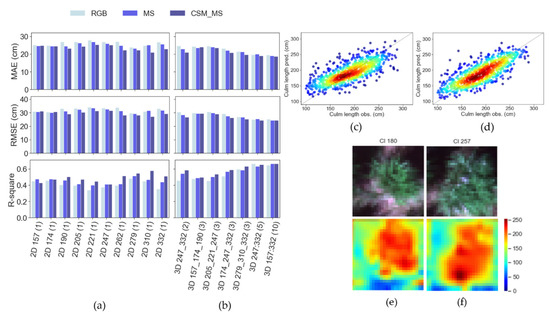
Figure 5.
Evaluation of culm length prediction in testing data for (a) 2D models by DAP and (b) 3D models by number of time-frame sequences via the MAE, RMSE, and R2 metrics. Observed and predicted values for the best (c) 2D models and (d) 3D models for testing datasets with low- (blue) to high-density (red) sample distribution. Example RGB image chips and activation maps for (e) low and (f) high-culm-length plants from the 2D-based model of aerial data collected on DOY 279. The level of activation of the deepest CNN layers in the images is represented by low (blue), intermediate (yellow), and high (red) values on a 0–255 scale.
3.3. Biomass Yield
When determining the biomass yield, 3D analysis generally tended to perform better than 2D analysis, with the best 3D-CNN model (3D_157:332(10)) performing significantly better (Figure 6c; MAE = 110.61 g, RMSE = 149.31 g, R2 = 0.69) than the best 2D-CNN (Figure 6d; 2D_247(1); MAE = 139 g, RMSE = 180 g, R2 = 0.56). The best 3D-CNN predictions (Figure 6c) and the best 2D-CNN predictions (Figure 6d) both had a tendency to underestimate the biomass of the most productive genotypes, but this was more marked for the 2D-CNN models.
The performance of 2D analyses was generally higher in the early–mid growing season (DOYs 157 to 247) than in the late season (DOYs 262 to 332). The best 3D-CNN models were those that integrated early-, mid-, and late-season imagery (Figure 6b), although imagery from 3 dates over these periods of time (3D_174_247_332(3)) resulted in model predictions (MAE = 114.46 g, RMSE = 153.90 g, R2 = 0.68) almost as good as when data from all 10 dates were used (3D_157:332 (10); MAE = 110.61g, RMSE = 149.31 g, R2 = 0.69). The 3D-CNNs’ predictive power was progressively lower when the imagery used was only from dates in the early–mid season (3D_57_174_190(3), 3D_205_221_247(3)) or the late season (Figure 6b; 3D_247:332(5), 3D_279_310_332(3)).
Adding additional spectral features (i.e., MS versus RGB) along with geometric features (i.e., CSM_MS versus MS) as inputs to the analysis progressively improved the predictions of biomass by both 2D- and 3D-CNNs (Figure 6a,b). When using CSM_MS data, 2D-CNN analyses of imagery from any single date throughout the growing season (Figure 6a) were outperformed by all of the 3D-CNN analyses tested (Figure 6b) in terms of MAE, RMSE, and R2.
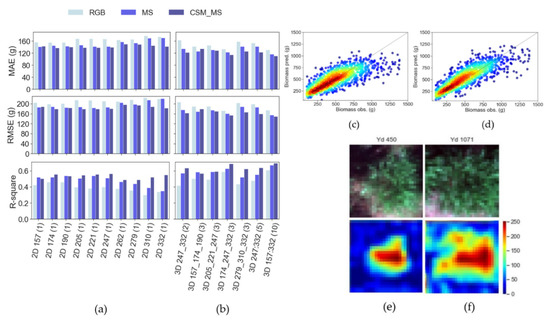
Figure 6.
Evaluation of biomass yield prediction in testing data for (a) 2D models by DAP and (b) 3D models by number of time-frame sequences via the MAE, RMSE, and R2 metrics. Observed and predicted values for the best (c) 2D models and (d) 3D models for testing datasets with low- (blue) to high-density (red) sample distribution. Example RGB image chips and activation maps for (e) low- and (f) high-biomass plants from the 2D-based model of aerial data collected on DOY 279. The level of activation of the deepest CNN layers in the images is represented by low (blue), intermediate (yellow), and high (red) values on a 0–255 scale.
When considering how CNNs exploit the imagery to maximize biomass yield prediction, higher activation regions (red) were located over dense green regions of the plant in the image. Correspondingly, low (blue) activation levels were assigned to the background, e.g., soil and shaded regions of the image. It was notable that the models tended to assign larger areas of high (red) activation levels to plants with larger biomass (Figure 6f) and smaller areas of high activation to plants with a lower biomass yield (Figure 6e).
4. Discussion
This study of large, diverse populations of Miscanthus successfully demonstrated the following: (1) 3D-CNNs, which analyzed the time-courses of remote sensing imagery to assess flowering time, culm length, and aboveground biomass, outperformed 2D-CNNs, which analyzed imagery from single dates; (2) the three traits differed in how the models’ performance depended on the combination of dates from which imagery was selected for input to the 3D-CNN analysis; and (3) combining more spectral features along with geometric features in the imagery led to better trait predictions. Overall, this highlights how high-spatial- and -temporal-resolution UAV time-series imagery can be analyzed by 3D-CNNs to more fully exploit the information contained in imagery than commonly used 2D-CNN methods, while also simplifying workflows by semi-automating feature extraction. Making these advances in a highly productive perennial biomass crop, such as Miscanthus, is particularly challenging and important because such crops are less studied and more difficult to phenotype than the annual grain crops that currently dominate agricultural commodity production [32]. In addition to advancing remote sensing science, this work could accelerate efforts to select Miscanthus for improved productivity and adaptation to regional growth environments by providing a rapid, efficient approach to phenotyping key traits across breeding populations [33]. For example, these analyses could optimize sampling strategies, including whether or not to inspect or collect samples in certain areas with early support from UAV imagery. Otherwise, breeders and farmers are often advised to inspect the entire area or to use a random strategy to sample commercial fields, with the risk of missing relevant areas or expending unnecessary labor [34]. Moreover, the improved efficiency of UAV phenotyping can enable plant breeders to grow larger populations for a given level of research funding, thereby improving the probability of identifying rare individuals that are truly outstanding. This enables efforts to transition to more sustainable and resilient cropping systems that can mitigate greenhouse gas emissions and climate change [35].
4.1. Flowering Time
Previous studies have reported the use of imagery to monitor the vegetative–reproductive transition in annual crops [36,37]. Traditional approaches have mainly been based on using the temporal trajectories of vegetation indices derived from UAV [16,38] and satellite [39] imagery. While these studies have demonstrated significant advances for the rapid assessment of phenological transitions, they mainly relied on manual interventions—for example, to determine the optimal parameters in curve-smoothing algorithms or threshold values to match the phenological transitions, which are important challenges for successful scalability [40]. Success has also been achieved in computer vision applications, reporting significant advances in exploiting very-high-spatial-resolution imagers mounted on close-range [41] or aerial [42] platforms for detecting reproductive organs in plants. While these represent valuable advances, most of these studies have focused on delivering binary outcomes (i.e., flowering versus not-flowering status) [43,44], without accounting for the dynamic nature of the underlying biological process. Implementing scalable remote sensing methodologies that can identify differences in the phenological dynamics of a population with diverse origins and genetic backgrounds can be of significant interest for breeders, because they are otherwise very challenging to assess by traditional manual inspection over large areas [32].
The 3D-CNN analysis of imagery from multiple dates generally outperformed 2D-CNN analysis of imagery from any single date (Figure 4). The performance enhancement produced a 12% greater R2 and 10% lower RMSE for the best 3D-CNN analysis (3D_262_279(2)) versus the best 2D-CNN analysis (2D_262(1)). By definition, flowering time is a trait that involves a single, specific event in the development of the crop over the growing season. When a genotype rapidly transitions between the vegetative and reproductive stages, the appearance of inflorescences produces strong spectral and spatial changes in the images of the plants. The 2D-CNN analysis of single-date data is able to differentiate plots that have or have not passed through this transition, but 3D-CNN analysis can additionally exploit information about the changes in spectral or geometric features that occur between two or more dates [25]. Notably, the potential improvement in the accuracy of phenotyping flowering time with 3D-CNN analysis may be underestimated here. In line with standard practice by breeders and the laborious nature of a person visiting thousands of field plots on foot, ground-truth data were collected on a weekly basis. This set an upper limit on the precision of flowering time estimates. Hence, in the future, higher-frequency ground-truth data could potentially allow the power of 3D-CNNs to be fully exploited.
Since the signal from the canopy is strongly influenced by the presence of inflorescences, the variance between individuals of the populations declined after most individuals flowered. Additionally, autumn senescence began in November, during which the canopy underwent a sequence of profound biochemical and biophysical changes that affected its reflectance. Therefore, it is not surprising that in this case—and others [43]—late-season imagery was not effective for estimating flowering time by 2D-CNN analyses (Figure 4). While performance also declined when 3D-CNNs were provided with images collected from a larger number of dates outside the main period of flowering, the results were more robust. Operationally, this would aid in the adoption of these methods by breeders, because they could rely on acquiring high-quality phenotypic data from a set of regular UAV flights without having to stratify the data before analysis.
While the use of simpler RGB features proved to be effective throughout the season and was consistent with a previous report on maize for tassel detection [37], the inclusion of multispectral or multispectral + geometric features resulted in slightly superior performance. The high sensitivity of near-infrared [45] and geometric [46] features to the fast-emerging new objects (i.e., inflorescences) (Figure 2a) can explain the superior ability of these features in both timepoint and time-series analysis. It was also possible to better understand the learning process of the CNNs via activation maps. The models coherently modulated the levels of activation by assigning high and low values to the greenness and inflorescence components of the plants, respectively. It was notable that the area of high activation tended to be smaller during early flowering, given the presence of inflorescences, while it was relatively larger in late-flowering plants, given the absence of inflorescences or dominance of greenness. This indicates that both the level and the area of high activation played a relevant role in maximizing the predictive ability of the neural networks. Understanding that the models adjusted their learning process according to the ratio of greenness versus inflorescences in the images was a valuable insight when implementing neural-network-based inference processes.
4.2. Culm Length
Sensors mounted on close-range [47] and aerial [15] platforms have recently been adopted to determine culm length in perennial grasses. Traditional approaches to inferring culm length from remote sensing data mainly rely on using point clouds derived from LIDAR or photogrammetry features, which are extracted and summarized over an area of interest via statistical descriptors [17] (e.g., mean, median, quantile) and then fed into a regression step to deliver the final prediction [22]. Even though this methodology has enabled the rapid deployment of data analytics, it relies on a high level of manual supervision, e.g., selecting an adequate vegetation index and statistical descriptor that represent the case under study [48]. Another limiting factor of these approaches is the need for handcrafted feature engineering when using multiple sensors or features [49]. Additionally, while these procedures are feasible at small sample sizes (<200) [14], they can be challenging to implement at larger sample sizes (>1000). Moreover, when considering integrating timepoint features as inputs into time-series analysis, this uncertainty is passed forward into the curve-fitting step [22,50], which can lead to biased inference and limited repeatability. The use of CNNs contributes to a more automated data extraction and data-driven inference procedure to determine culm length from high-resolution aerial imagery.
In this study, the accuracy with which culm length could be phenotyped was improved in several ways by using a 3D-CNN architecture rather than a 2D-CNN architecture (Figure 5). For example, the best 3D-CNN analysis (3D 157:332(10)) achieved a 16% greater R2 and 8% lower RMSE than the best 2D-CNN analysis (2D 310). End-of-season culm length is the consequence of gradual growth over most of the growing season. This distinguishes it from flowering time, which is the product of a discrete event in time. Therefore, it is logical that the 3D-CNN analyses, which could directly exploit information from the changing nature of image features across a series of sampling dates, became more powerful as they were provided with imagery from a larger number of sampling dates (Figure 5). This outcome was consistent with the superior ability of long time-series sequences in 3D-CNNs to predict yield in annual crops compared to both shorter 3D-CNNs and timepoint analysis [24]. It is also intuitive that prediction accuracy increased when the 3D-CNNs were provided with series of images from later in the growing season i.e., when plants were nearer to their final height. In fact, the 3D-CNNs’ performance only dropped to match that of 2D-CNNs when relying solely on imagery from the early season.
The 3D-CNN and 2D-CNN architectures were also distinguished by their dependency on the types of image features used by the model (Figure 5). The 2D-CNN analysis of any sampling date, other than those in the first month of the growing season, was significantly improved when using geometric features in addition to spectral features (i.e., CSM_MS versus RGB or MS). This reflects the fact that the geometric features directly correspond to canopy height. The effect became stronger in the second part of the season, when the spectral features would be saturated [51] due to the large volume of vegetation accumulated at this stage. Exploiting additional image features also improved the 3D-CNN models’ performance when imagery from only two or three dates was analyzed. However, unexpectedly, this effect diminished when analyzing sequences of 5 or 10 dates of imagery with the 3D-CNN, such that the model performance using RGB was essentially equal to that of CSM_MS (Figure 5). This indicates that these models were also able to learn meaningful spectral representations from the temporal dimension of these long time sequences [52], in stark contrast to 2D-CNN analysis of single timepoints.
According to the activation maps (Figure 5e,f), the learning process focused over the area of the plant by assigning high levels of activation to regions characterized by high CSM, greater absorption of green, and high reflectance of the near-infrared light. This confirmed that the models coherently exploited the information that represented meaningful areas of an image to determine the culm length without the need for external supervision. This resulted in a significant reduction in expert supervision needed during modelling implementation. It is important to note that this is likely to be a particularly valuable methodological advance for perennial crops with many stems, such as Miscanthus, because heterogeneity in the height and ground cover both within and between plants of different genotypes is much greater than for annual row crops with simpler plant architectures (e.g., maize).
4.3. Biomass Yield
Remote sensing has been extensively adopted in agriculture to determine biomass in annual crops [53], and more recently in perennial grasses [54]. Traditional approaches to determine biomass have been mainly based on using multiple vegetation indices summarized over an area of interest via statistical descriptors, which are then fed as tabular features into a regression step for predictive inference [55]. This approach, together with handcrafted feature engineering [56] (e.g., select optimal vegetation index, determine feature importance), has significantly advanced the adoption of remote sensing for modelling biomass [57]. However, the early dimension reduction of the original image (i.e., two- or three-dimensional matrices) into an unique tabular value and the manual supervision needed (i.e., selecting optimal statistical descriptor) during the process [58] can limit the scalability and repeatability of the approach. A similar challenge is faced when determining biomass using time-series analysis via traditional curve-fitting procedures [59]. The sequential convolutional design of CNNs, which directly operate in the original image to automatically learn features that maximize the predictive ability of a certain trait, can significantly reduce the need for expert knowledge and the uncertainty [60] related to the manual supervision when determining biomass from aerial sensors.
Biomass yield is the most complex trait evaluated in this study, but also the most important trait that Miscanthus breeders are trying to improve. The complexity stems from trying to accurately evaluate the biomass of all aboveground tissues in plants across a genetically diverse population that vary considerably in canopy architecture (i.e., height, stem numbers, stem widths, leaf size, leaf angles). Perhaps as a result of this complexity, the benefits of using 3D-CNN analysis over 2D-CNN analysis were even greater for phenotyping of biomass than for flowering time or culm height. The accuracy of biomass predictions from all of the 3D-CNN analyses performed exceeded that of any 2D-CNN analysis (Figure 6). For example, the best 3D-CNN analysis (3D 157:332(10)) achieved a 23% greater R2 and 17% lower RMSE than the best 2D-CNN analysis (2D 247). While the 3D-CNN analysis using imagery from 10 dates across the whole growing season proved to be the best-performing (3D_157:332(10)), a model with fewer timepoints that still spanned all phases of the growing season (i.e., early, mid, and late season) performed almost as well (3D_174_247_332(3)). This was consistent with the rate of early-season growth, as well as the rate and timing of canopy closure, providing valuable information from which the CNN could learn beyond exploiting measures of final size late in the growing season. This highlighted that time-series analysis was not dependent on high-temporal-resolution imagery to produce accurate estimates of biomass.
Throughout the season, estimates of biomass from multispectral and multispectral + geometric features were significantly better than estimates from RGB features alone. This gap in performance increased further at the end of the season, when the very best estimates could be generated. Spectral saturation is a limiting factor when determining biomass using passive optical sensors mounted on aerial platforms [61]. The signal becomes more rapidly saturated in visible wavelengths than in the longer wavelengths of the electromagnetic spectrum as the amount of vegetation accumulated on the ground increases. In contrast, there is no saturation of geometric features with increasing crop biomass. In fact, as crop stature increases, the power to detect equivalent relative differences between genotypes is likely to grow. The multiple elements of plant growth dynamics—i.e., increasing leaf expansion, ground cover (perimeter), and height—produce considerable complexity throughout the season, which needs to be considered in order to understand the seasonal patterns of the different features (Figure 6a).
Regarding the learning process of the neural networks to determine biomass, the models mostly focused over regions of the plant related to dense green coverage, which represented meaningful regions of the images in terms of biomass accumulation. It was notable that the level and the area of high activation tended to be positively correlated with the level of biomass (Figure 6e,f). This was consistent with the higher predictive ability in the first part of the season and with the capacity of the models to exploit non-vertical descriptors—e.g., horizontal green coverage—to maximize the information gain from images.
4.4. Limitations
The process of cropping individual plots’ plants from the orthophotos into individual plant image chips can add noise in the analysis. Given the massive growth of these plants, some contamination from neighboring plants may also have occurred. Future studies should investigate the integration of plant detection techniques as an early step in the analysis pipeline.
The multispectral sensor used here was able to generate both photogrammetric and multispectral features from a single flight. However, the limited spatial resolution of the sensor compared to other widely available RGB sensors may have penalized the learning ability of the neural networks. Future research should investigate this, e.g., whether more detailed identification of small inflorescences in a higher-resolution image can lead to a more accurate determination of flowering time.
Although photogrammetry provided a low-cost alternative to generate geometric descriptors of the plants, future studies should also explore the benefits of integrating more descriptors of the internal and external structure of the canopy (e.g., via LIDAR technology) for the performance of the neural networks, given the massive vertical growth and complex plant architecture of this crop.
5. Conclusions
Our analysis demonstrates that Miscanthus can be effectively characterized using high-spatial- and -temporal-resolution aerial imagery integrated with convolutional neural networks. To the best of our knowledge, this is the first effort to characterize individual plant traits in large, genetically diverse populations of Miscanthus in large-scale field trials using remote sensing. Flowering time, culm length, and biomass yield were chosen as target traits due to their importance for Miscanthus breeding and the sustainable production of feedstock for bioenergy and bioproducts. The different natures of these traits present a range of challenges and opportunities for interpreting the outcomes of this work. While flowering time can be considered to be a short–medium-term process, culm length and biomass yield are the result of growth processes that progress throughout the entire growing season. This work indicates that the estimation of phenological transitions and yield-related traits is feasible and can produce results accurate enough to monitor Miscanthus using very-high-spatial-resolution remote sensing imagery. The study shows that feature-rich (i.e., multispectral + geometric) data produce meaningful gains in the prediction accuracy of flowering time, culm length, and biomass yield, regardless of the CNN architecture considered. Additionally, (2D-spatial) timepoint CNN analysis was outperformed by 3D-spatiotemporal CNN analysis. Predictions of flowering time were highly sensitive to the period of time over which the imagery was collected. Meanwhile, estimates of culm length and biomass improved as sequences of images from more dates, which were also spread out over more of the growing season, were used. These outcomes are valuable for informing future strategies by aligning the biological processes of perennial grasses and the timing of aerial data collection. In practice, this can result in large benefits for breeding selection, e.g., by adopting the proposed methodology for within-season yield prediction to make selections prior to flowering, thereby saving a whole year in the breeding cycle. This work partially addresses the need for advanced modelling techniques that can take advantage of recently improved temporal and spatial resolution sensors and aerial platforms, the ease of collecting data, cloud storage, and scalable modelling for UAV remote sensing imagery.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/rs14215333/s1, Table S1: Description of image features, number of flights, number of image features, and dimensions of image chips for the three imaging approaches tested. Table S2: Description of the 138 CNN models implemented according to combinations of UAV-based image features, CNN architectures, and target traits.
Author Contributions
S.V. and A.D.B.L. conceived the study, interpreted the data, and wrote the manuscript; E.J.S., X.Z. and J.N.N. established, maintained, and collected the ground-truth data in the field trials; J.R. and D.P.A. collected the aerial data; S.V. implemented the data pipeline in the study. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the DOE Center for Advanced Bioenergy and Bioproducts Innovation (U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018420). Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Energy.
Data Availability Statement
The datasets used and analyzed during this study are available from the corresponding author via the Illinois Databank at https://doi.org/10.13012/B2IDB-5689586_V1 (accessed on 19 October 2022).
Acknowledgments
We thank Tim Mies at the Energy Farm at University of Illinois for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Whitaker, J.; Field, J.L.; Bernacchi, C.J.; Cerri, C.E.P.; Ceulemans, R.; Davies, C.A.; DeLucia, E.H.; Donnison, I.S.; McCalmont, J.P.; Paustian, K.; et al. Consensus, Uncertainties and Challenges for Perennial Bioenergy Crops and Land Use. GCB Bioenergy 2018, 10, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Robson, P.; Jensen, E.; Hawkins, S.; White, S.R.; Kenobi, K.; Clifton-Brown, J.; Donnison, I.; Farrar, K. Accelerating the Domestication of a Bioenergy Crop: Identifying and Modelling Morphological Targets for Sustainable Yield Increase in Miscanthus. J. Exp. Bot. 2013, 64, 4143–4155. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Kong, Y.; Hu, R.; Zhou, G. Miscanthus: A Fast-Growing Crop for Environmental Remediation and Biofuel Production. GCB Bioenergy 2021, 13, 58–69. [Google Scholar] [CrossRef]
- Clifton-Brown, J.; Hastings, A.; Mos, M.; McCalmont, J.P.; Ashman, C.; Awty-Carroll, D.; Cerazy, J.; Chiang, Y.-C.; Cosentino, S.; Cracroft-Eley, W.; et al. Progress in Upscaling Miscanthus Biomass Production for the European Bio-Economy with Seed-Based Hybrids. GCB Bioenergy 2017, 9, 6–17. [Google Scholar] [CrossRef]
- Clark, L.V.; Dwiyanti, M.S.; Anzoua, K.G.; Brummer, J.E.; Ghimire, B.K.; Głowacka, K.; Hall, M.; Heo, K.; Jin, X.; Lipka, A.E.; et al. Biomass Yield in a Genetically Diverse Miscanthus Sinensis Germplasm Panel Evaluated at Five Locations Revealed Individuals with Exceptional Potential. GCB Bioenergy 2019, 11, 1125–1145. [Google Scholar] [CrossRef]
- Knörzer, H.; Hartung, K.; Piepho, H.-P.; Lewandowski, I. Assessment of Variability in Biomass Yield and Quality: What Is an Adequate Size of Sampling Area for Miscanthus? GCB Bioenergy 2013, 5, 572–579. [Google Scholar] [CrossRef]
- Sishodia, R.P.; Ray, R.L.; Singh, S.K. Applications of Remote Sensing in Precision Agriculture: A Review. Remote Sens. 2020, 12, 3136. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote Sensing for Agricultural Applications: A Meta-Review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Ahamed, T.; Tian, L.; Zhang, Y.; Ting, K.C. A Review of Remote Sensing Methods for Biomass Feedstock Production. Biomass Bioenergy 2011, 35, 2455–2469. [Google Scholar] [CrossRef]
- Molijn, R.A.; Iannini, L.; Vieira Rocha, J.; Hanssen, R.F. Sugarcane Productivity Mapping through C-Band and L-Band SAR and Optical Satellite Imagery. Remote Sens. 2019, 11, 1109. [Google Scholar] [CrossRef]
- Krause, M.R.; Mondal, S.; Crossa, J.; Singh, R.P.; Pinto, F.; Haghighattalab, A.; Shrestha, S.; Rutkoski, J.; Gore, M.A.; Sorrells, M.E.; et al. Aerial High-Throughput Phenotyping Enables Indirect Selection for Grain Yield at the Early Generation, Seed-Limited Stages in Breeding Programs. Crop Sci. 2020, 60, 3096–3114. [Google Scholar] [CrossRef]
- Crain, J.; Wang, X.; Evers, B.; Poland, J. Evaluation of Field-Based Single Plant Phenotyping for Wheat Breeding. Plant Phenome J. 2022, 5, e20045. [Google Scholar] [CrossRef]
- Kubiak, K.; Rotchimmel, K.; Stereńczak, K.; Damszel, M.; Sierota, Z. Remote Sensing Semi-Automatic Measurements Approach for Monitoring Bioenergetic Crops of Miscanthus Spp. Pomiary Autom. Robot. 2019, 23, 77–85. [Google Scholar] [CrossRef]
- Li, F.; Piasecki, C.; Millwood, R.J.; Wolfe, B.; Mazarei, M.; Stewart, C.N. High-Throughput Switchgrass Phenotyping and Biomass Modeling by UAV. Front. Plant Sci. 2020, 11, 574073. [Google Scholar] [CrossRef]
- Miura, N.; Yamada, S.; Niwa, Y. Estimation of Canopy Height and Biomass of Miscanthus Sinensis in Semi-Natural Grassland Using Time-Series Uav Data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, V-3–2020, 497–503. [Google Scholar] [CrossRef]
- Adak, A.; Murray, S.C.; Božinović, S.; Lindsey, R.; Nakasagga, S.; Chatterjee, S.; Anderson, S.L.; Wilde, S. Temporal Vegetation Indices and Plant Height from Remotely Sensed Imagery Can Predict Grain Yield and Flowering Time Breeding Value in Maize via Machine Learning Regression. Remote Sens. 2021, 13, 2141. [Google Scholar] [CrossRef]
- Anderson, S.L.; Murray, S.C.; Malambo, L.; Ratcliff, C.; Popescu, S.; Cope, D.; Chang, A.; Jung, J.; Thomasson, J.A. Prediction of Maize Grain Yield before Maturity Using Improved Temporal Height Estimates of Unmanned Aerial Systems. Plant Phenome J. 2019, 2, 1–15. [Google Scholar] [CrossRef]
- The Rise of Cubesats: Opportunities and Challenges|Wilson Center. Available online: https://www.wilsoncenter.org/blog-post/rise-cubesats-opportunities-and-challenges (accessed on 9 April 2022).
- UAV in the Advent of the Twenties: Where We Stand and What Is next|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0924271621003282?token=6713E23A68E83727FE3D141161298A4D7AF0FFEFACDEF7162016781D00A1A5CB1461CFFCB2C9158B51D9CFF2AE76E8F9&originRegion=us-east-1&originCreation=20220410025754 (accessed on 9 April 2022).
- Koh, J.C.O.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C. Convolutional Neural Networks for Image-Based High-Throughput Plant Phenotyping: A Review. Plant Phenomics 2020, 2020, 4152816. [Google Scholar] [CrossRef]
- Varela, S.; Pederson, T.; Bernacchi, C.J.; Leakey, A.D.B. Understanding Growth Dynamics and Yield Prediction of Sorghum Using High Temporal Resolution UAV Imagery Time Series and Machine Learning. Remote Sens. 2021, 13, 1763. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D Convolutional Neural Networks for Crop Classification with Multi-Temporal Remote Sensing Images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Linna, P.; Lipping, T. Crop Yield Prediction Using Multitemporal UAV Data and Spatio-Temporal Deep Learning Models. Remote Sens. 2020, 12, 4000. [Google Scholar] [CrossRef]
- Varela, S.; Pederson, T.L.; Leakey, A.D.B. Implementing Spatio-Temporal 3D-Convolution Neural Networks and UAV Time Series Imagery to Better Predict Lodging Damage in Sorghum. Remote Sens. 2022, 14, 733. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In European Conference on Computer Vision, Proceedings of the Computer Vision—ECCV 2014, Zurich, Switzerland, 6–12 September 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Clark, L.V.; Stewart, J.R.; Nishiwaki, A.; Toma, Y.; Kjeldsen, J.B.; Jørgensen, U.; Zhao, H.; Peng, J.; Yoo, J.H.; Heo, K.; et al. Genetic Structure of Miscanthus Sinensis and Miscanthus Sacchariflorus in Japan Indicates a Gradient of Bidirectional but Asymmetric Introgression. J. Exp. Bot. 2015, 66, 4213–4225. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional Neural Networks: An Overview and Application in Radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Tran, D.; Bourdev, L.; Fergus, R.; Torresani, L.; Paluri, M. Learning Spatiotemporal Features with 3D Convolutional Networks. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 4489–4497. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
- Team, K. Keras Documentation: Grad-CAM Class Activation Visualization. Available online: https://keras.io/examples/vision/grad_cam/ (accessed on 1 December 2021).
- Huang, L.S.; Flavell, R.; Donnison, I.S.; Chiang, Y.-C.; Hastings, A.; Hayes, C.; Heidt, C.; Hong, H.; Hsu, T.-W.; Humphreys, M.; et al. Collecting Wild Miscanthus Germplasm in Asia for Crop Improvement and Conservation in Europe Whilst Adhering to the Guidelines of the United Nations’ Convention on Biological Diversity. Ann. Bot. 2019, 124, 591–604. [Google Scholar] [CrossRef]
- Lewandowski, I.; Clifton-Brown, J.; Trindade, L.M.; van der Linden, G.C.; Schwarz, K.-U.; Müller-Sämann, K.; Anisimov, A.; Chen, C.-L.; Dolstra, O.; Donnison, I.S.; et al. Progress on Optimizing Miscanthus Biomass Production for the European Bioeconomy: Results of the EU FP7 Project OPTIMISC. Front. Plant Sci. 2016, 7, 1620. [Google Scholar] [CrossRef]
- Loures, L.; Chamizo, A.; Ferreira, P.; Loures, A.; Castanho, R.; Panagopoulos, T. Assessing the Effectiveness of Precision Agriculture Management Systems in Mediterranean Small Farms. Sustainability 2020, 12, 3765. [Google Scholar] [CrossRef]
- Martinez-Feria, R.A.; Basso, B.; Kim, S. Boosting Climate Change Mitigation Potential of Perennial Lignocellulosic Crops Grown on Marginal Lands. Environ. Res. Lett. 2022, 17, 044004. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, X. Mapping Crop Phenology in Near Real-Time Using Satellite Remote Sensing: Challenges and Opportunities. J. Remote Sens. 2021, 2021, 8379391. [Google Scholar] [CrossRef]
- Guo, Y.; Fu, Y.H.; Chen, S.; Robin Bryant, C.; Li, X.; Senthilnath, J.; Sun, H.; Wang, S.; Wu, Z.; de Beurs, K. Integrating Spectral and Textural Information for Identifying the Tasseling Date of Summer Maize Using UAV Based RGB Images. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102435. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Z.; Shang, J.; Liu, J.; Dong, T.; Tang, M.; Feng, S.; Cai, H. Detecting Winter Canola (Brassica Napus) Phenological Stages Using an Improved Shape-Model Method Based on Time-Series UAV Spectral Data. Crop J. 2022, 10, 1353–1362. [Google Scholar] [CrossRef]
- Ji, Z.; Pan, Y.; Zhu, X.; Zhang, D.; Wang, J. A Generalized Model to Predict Large-Scale Crop Yields Integrating Satellite-Based Vegetation Index Time Series and Phenology Metrics. Ecol. Indic. 2022, 137, 108759. [Google Scholar] [CrossRef]
- Hu, P.; Sharifi, A.; Tahir, M.N.; Tariq, A.; Zhang, L.; Mumtaz, F.; Shah, S.H.I.A. Evaluation of Vegetation Indices and Phenological Metrics Using Time-Series MODIS Data for Monitoring Vegetation Change in Punjab, Pakistan. Water 2021, 13, 2550. [Google Scholar] [CrossRef]
- Li, G.; Suo, R.; Zhao, G.; Gao, C.; Fu, L.; Shi, F.; Dhupia, J.; Li, R.; Cui, Y. Real-Time Detection of Kiwifruit Flower and Bud Simultaneously in Orchard Using YOLOv4 for Robotic Pollination. Comput. Electron. Agric. 2022, 193, 106641. [Google Scholar] [CrossRef]
- Liu, Y.; Cen, C.; Che, Y.; Ke, R.; Ma, Y.; Ma, Y. Detection of Maize Tassels from UAV RGB Imagery with Faster R-CNN. Remote Sens. 2020, 12, 338. [Google Scholar] [CrossRef]
- Alzadjali, A.; Alali, M.H.; Veeranampalayam Sivakumar, A.N.; Deogun, J.S.; Scott, S.; Schnable, J.C.; Shi, Y. Maize Tassel Detection From UAV Imagery Using Deep Learning. Front. Robot. AI 2021, 8, 600410. [Google Scholar] [CrossRef]
- Ghosal, S.; Zheng, B.; Chapman, S.C.; Potgieter, A.B.; Jordan, D.R.; Wang, X.; Singh, A.K.; Singh, A.; Hirafuji, M.; Ninomiya, S.; et al. A Weakly Supervised Deep Learning Framework for Sorghum Head Detection and Counting. Plant Phenomics 2019, 2019, 1525874. [Google Scholar] [CrossRef]
- Kumar, A.; Rajalakshmi, P.; Guo, W.; Naik, B.B.; Marathi, B.; Desai, U.B. Detection and Counting of Tassels for Maize Crop Monitoring Using Multispectral Images. In Proceedings of the 2020 IEEE International Conference on Computing, Power and Communication Technologies (GUCON), Greater Noida, India, 2–4 October 2020; pp. 789–793. [Google Scholar]
- Niu, Y.; Zhang, L.; Zhang, H.; Han, W.; Peng, X. Estimating Above-Ground Biomass of Maize Using Features Derived from UAV-Based RGB Imagery. Remote Sens. 2019, 11, 1261. [Google Scholar] [CrossRef]
- Zhang, L.; Grift, T.E. A LIDAR-Based Crop Height Measurement System for Miscanthus Giganteus. Comput. Electron. Agric. 2012, 85, 70–76. [Google Scholar] [CrossRef]
- Li, J.; Veeranampalayam-Sivakumar, A.-N.; Bhatta, M.; Garst, N.D.; Stoll, H.; Stephen Baenziger, P.; Belamkar, V.; Howard, R.; Ge, Y.; Shi, Y. Principal Variable Selection to Explain Grain Yield Variation in Winter Wheat from Features Extracted from UAV Imagery. Plant Methods 2019, 15, 123. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Schachtman, D.P.; Creech, C.F.; Wang, L.; Ge, Y.; Shi, Y. Evaluation of UAV-Derived Multimodal Remote Sensing Data for Biomass Prediction and Drought Tolerance Assessment in Bioenergy Sorghum. Crop J. 2022, 10, 1363–1375. [Google Scholar] [CrossRef]
- Impollonia, G.; Croci, M.; Ferrarini, A.; Brook, J.; Martani, E.; Blandinières, H.; Marcone, A.; Awty-Carroll, D.; Ashman, C.; Kam, J.; et al. UAV Remote Sensing for High-Throughput Phenotyping and for Yield Prediction of Miscanthus by Machine Learning Techniques. Remote Sens. 2022, 14, 2927. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide Dynamic Range Vegetation Index for Remote Quantification of Biophysical Characteristics of Vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Impollonia, G.; Croci, M.; Martani, E.; Ferrarini, A.; Kam, J.; Trindade, L.M.; Clifton-Brown, J.; Amaducci, S. Moisture Content Estimation and Senescence Phenotyping of Novel Miscanthus Hybrids Combining UAV-Based Remote Sensing and Machine Learning. GCB Bioenergy 2022, 14, 639–656. [Google Scholar] [CrossRef]
- Habyarimana, E.; Baloch, F.S. Machine Learning Models Based on Remote and Proximal Sensing as Potential Methods for In-Season Biomass Yields Prediction in Commercial Sorghum Fields. PLoS ONE 2021, 16, e0249136. [Google Scholar] [CrossRef]
- Hamada, Y.; Zumpf, C.R.; Cacho, J.F.; Lee, D.; Lin, C.-H.; Boe, A.; Heaton, E.; Mitchell, R.; Negri, M.C. Remote Sensing-Based Estimation of Advanced Perennial Grass Biomass Yields for Bioenergy. Land 2021, 10, 1221. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of Biomass in Wheat Using Random Forest Regression Algorithm and Remote Sensing Data. Crop J. 2016, 4, 212–219. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High Density Biomass Estimation for Wetland Vegetation Using WorldView-2 Imagery and Random Forest Regression Algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Cen, H.; Wan, L.; Zhu, J.; Li, Y.; Li, X.; Zhu, Y.; Weng, H.; Wu, W.; Yin, W.; Xu, C.; et al. Dynamic Monitoring of Biomass of Rice under Different Nitrogen Treatments Using a Lightweight UAV with Dual Image-Frame Snapshot Cameras. Plant Methods 2019, 15, 32. [Google Scholar] [CrossRef]
- Li, J.; Shi, Y.; Veeranampalayam-Sivakumar, A.-N.; Schachtman, D.P. Elucidating Sorghum Biomass, Nitrogen and Chlorophyll Contents With Spectral and Morphological Traits Derived From Unmanned Aircraft System. Front. Plant Sci. 2018, 9, 1406. [Google Scholar] [CrossRef]
- Perry, E.; Sheffield, K.; Crawford, D.; Akpa, S.; Clancy, A.; Clark, R. Spatial and Temporal Biomass and Growth for Grain Crops Using NDVI Time Series. Remote Sens. 2022, 14, 3071. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Chen, Y.; Du, K.; Zheng, F.; Zhang, L.; Sun, Z. Estimating above Ground Biomass of Winter Wheat at Early Growth Stages Using Digital Images and Deep Convolutional Neural Network. Eur. J. Agron. 2019, 103, 117–129. [Google Scholar] [CrossRef]
- Chen, P.; Wang, F. New Textural Indicators for Assessing Above-Ground Cotton Biomass Extracted from Optical Imagery Obtained via Unmanned Aerial Vehicle. Remote Sens. 2020, 12, 4170. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).