Estimation of Soil Salinization by Machine Learning Algorithms in Different Arid Regions of Northwest China
Abstract
:1. Introduction
2. Material and Methods
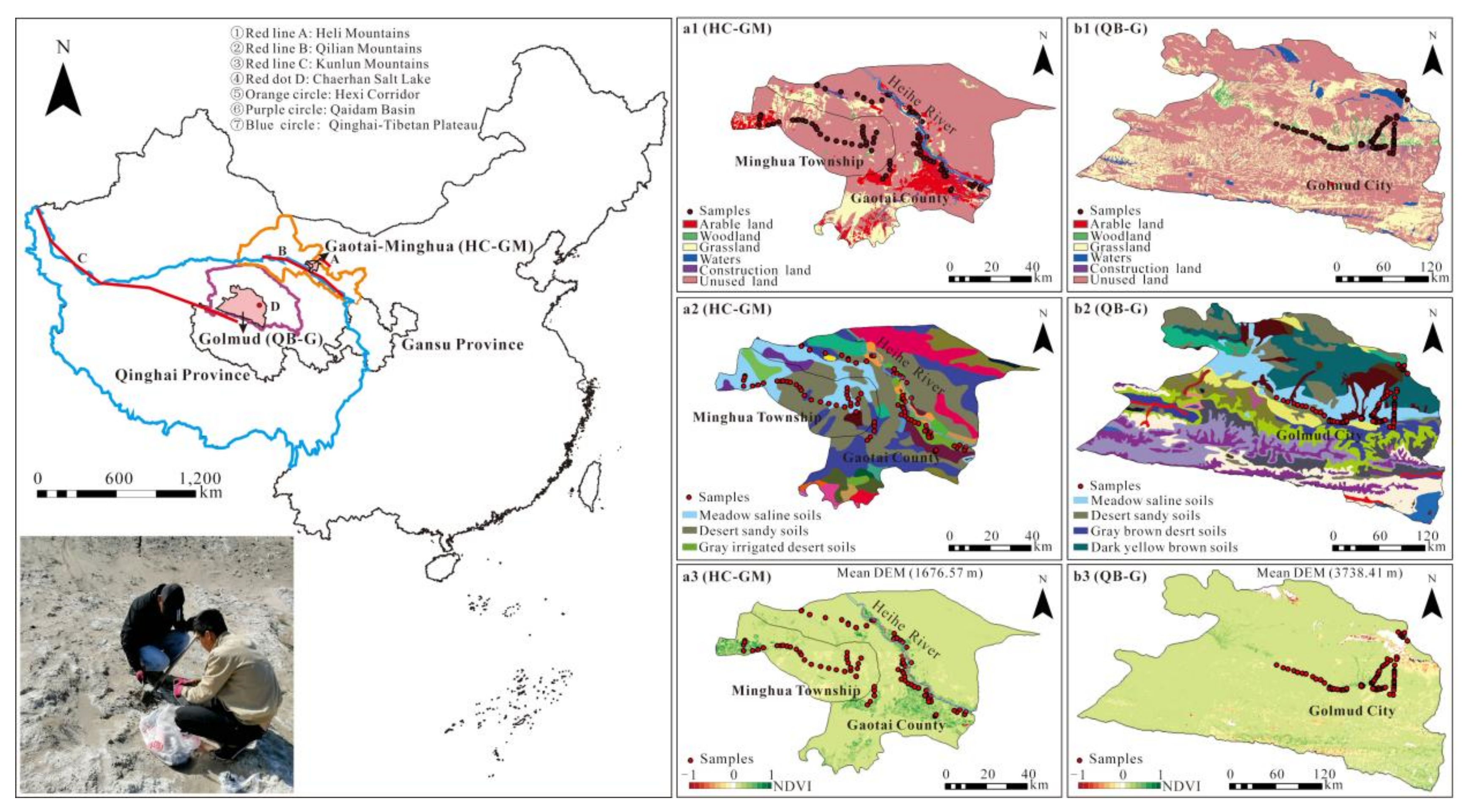
2.1. Study Area
2.2. Datasets
2.2.1. Electrical Conductivity and Eight Major Saline Ions Data
2.2.2. Hyperspectral Reflectance Data
2.2.3. Landsat 8 Remote Sensing Data
2.2.4. Soil and Terrain Influence Factors
2.3. Methods
2.3.1. Hyperspectral Reflectance Data Processing
2.3.2. Variable Selection and Inversion of EC
- (1)
- LightGBM
- (2)
- DELM
- (3)
- SCA–Elman
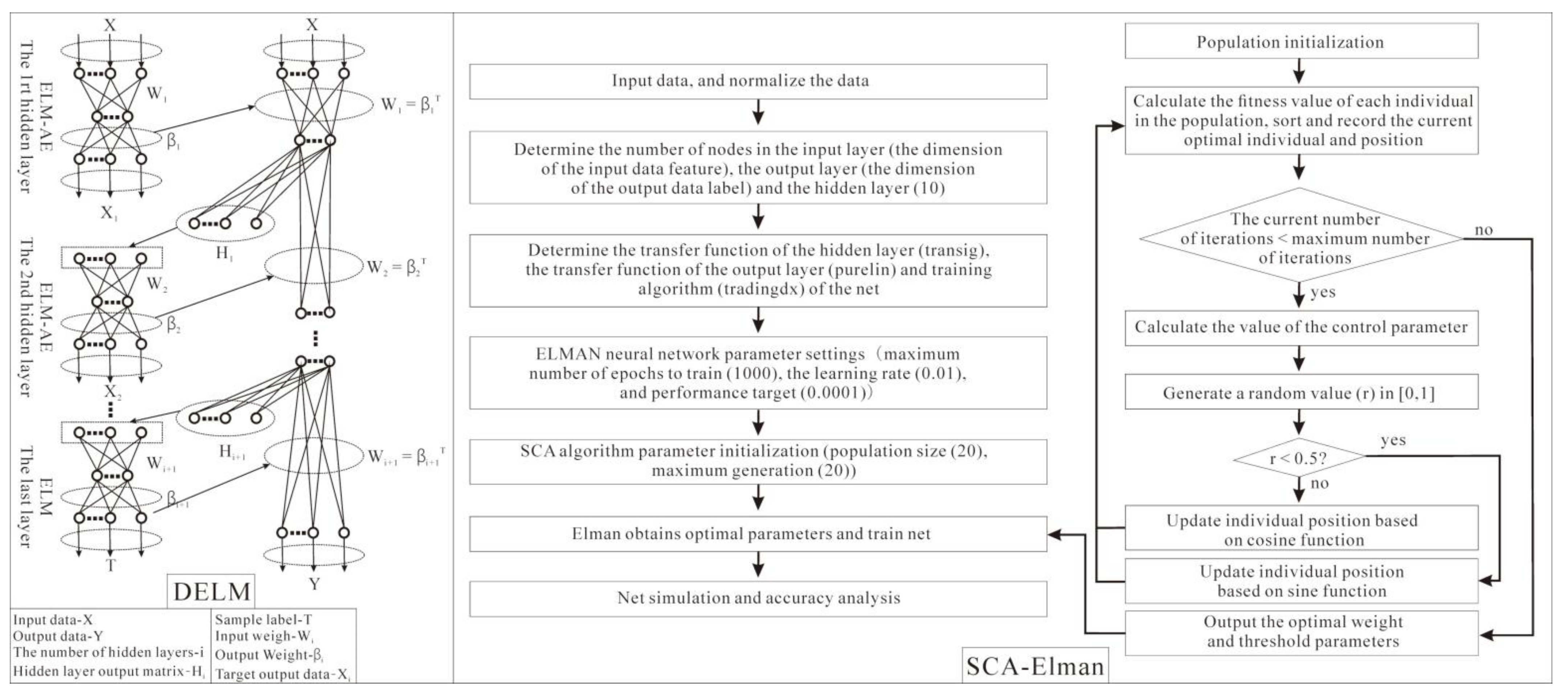
2.3.3. Model Verification
3. Results
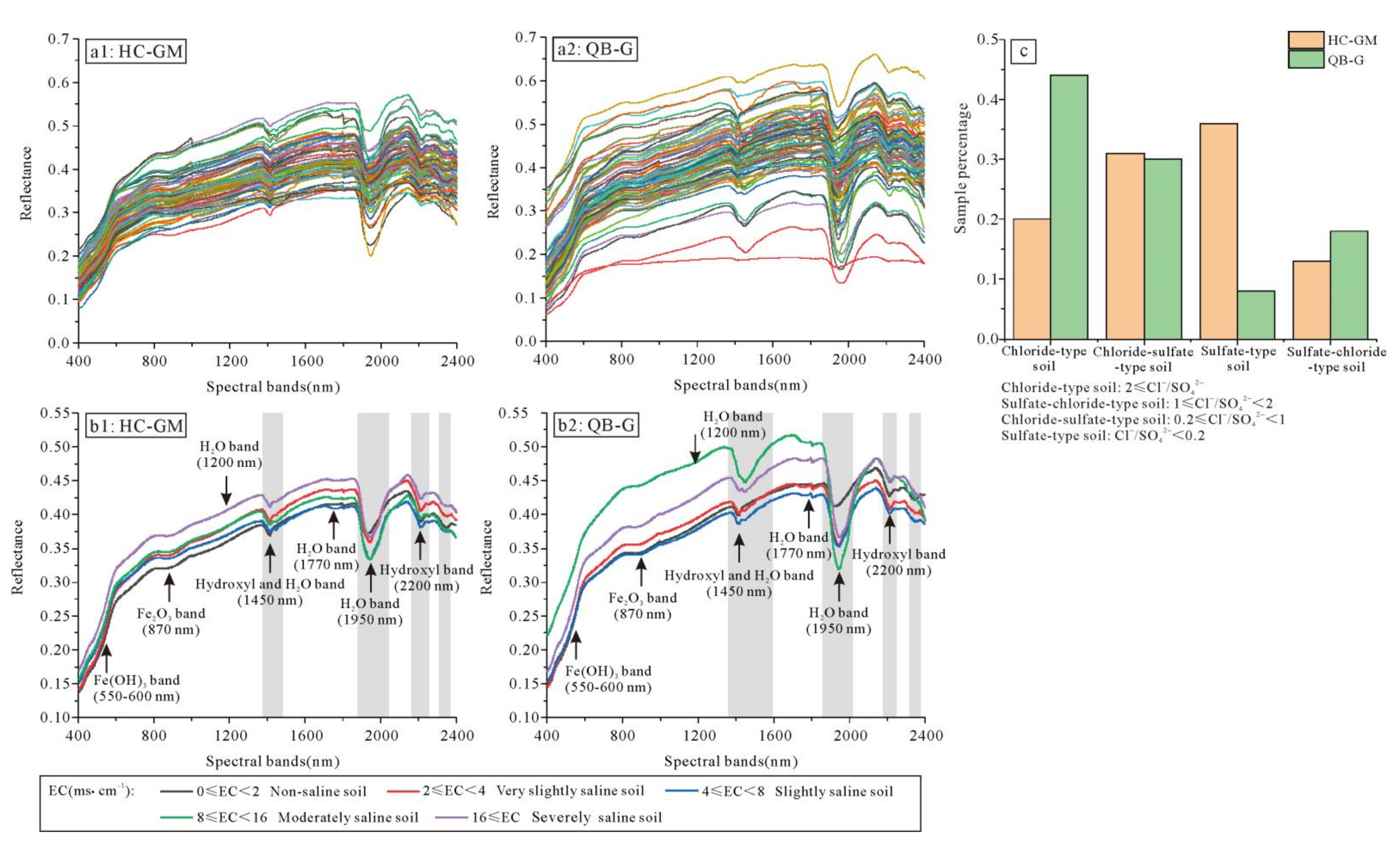
3.1. Descriptive Statistics of the EC and Chemistry Types of the Soil Samples
3.2. Hyperspectral Reflectance Curve of Soil Samples
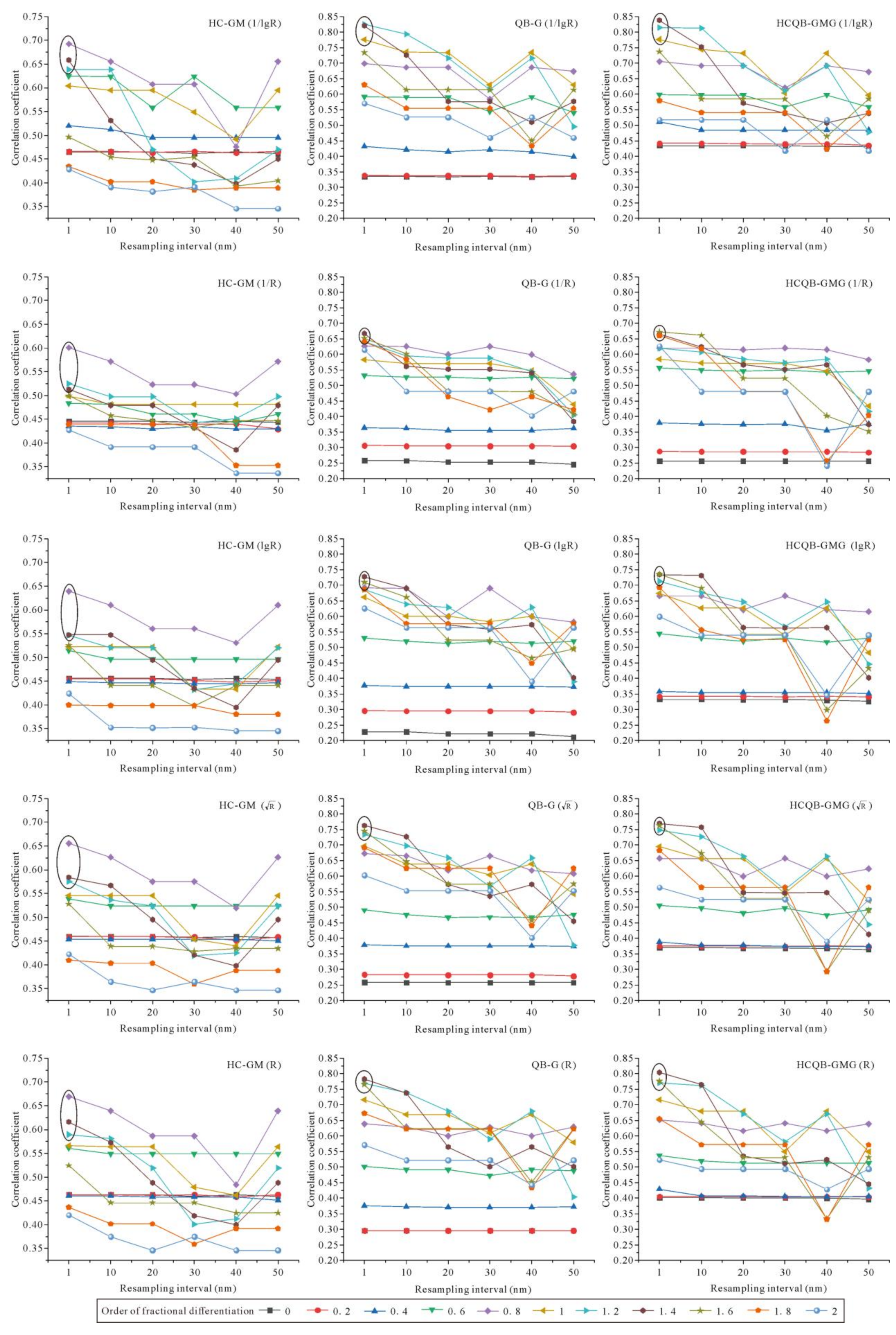
3.3. Correlation between EC and Different Forms of Hyperspectral Reflectance Data
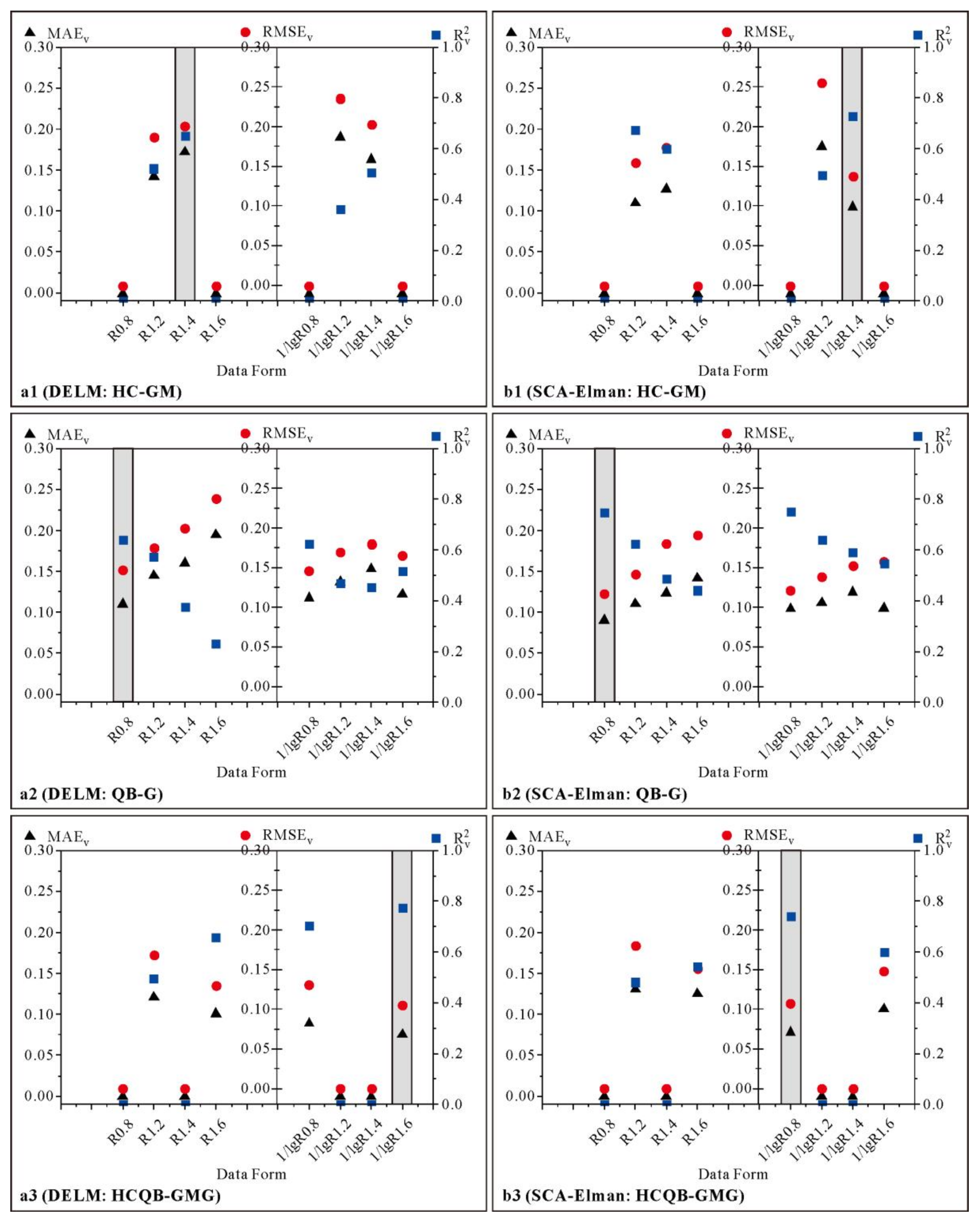
3.4. Simulation of Soil EC Using DELM and SCA–Elman
3.4.1. Modeling Results of DELM and SCA–Elman
3.4.2. Modeling Results for Different Data Forms and Different Regions
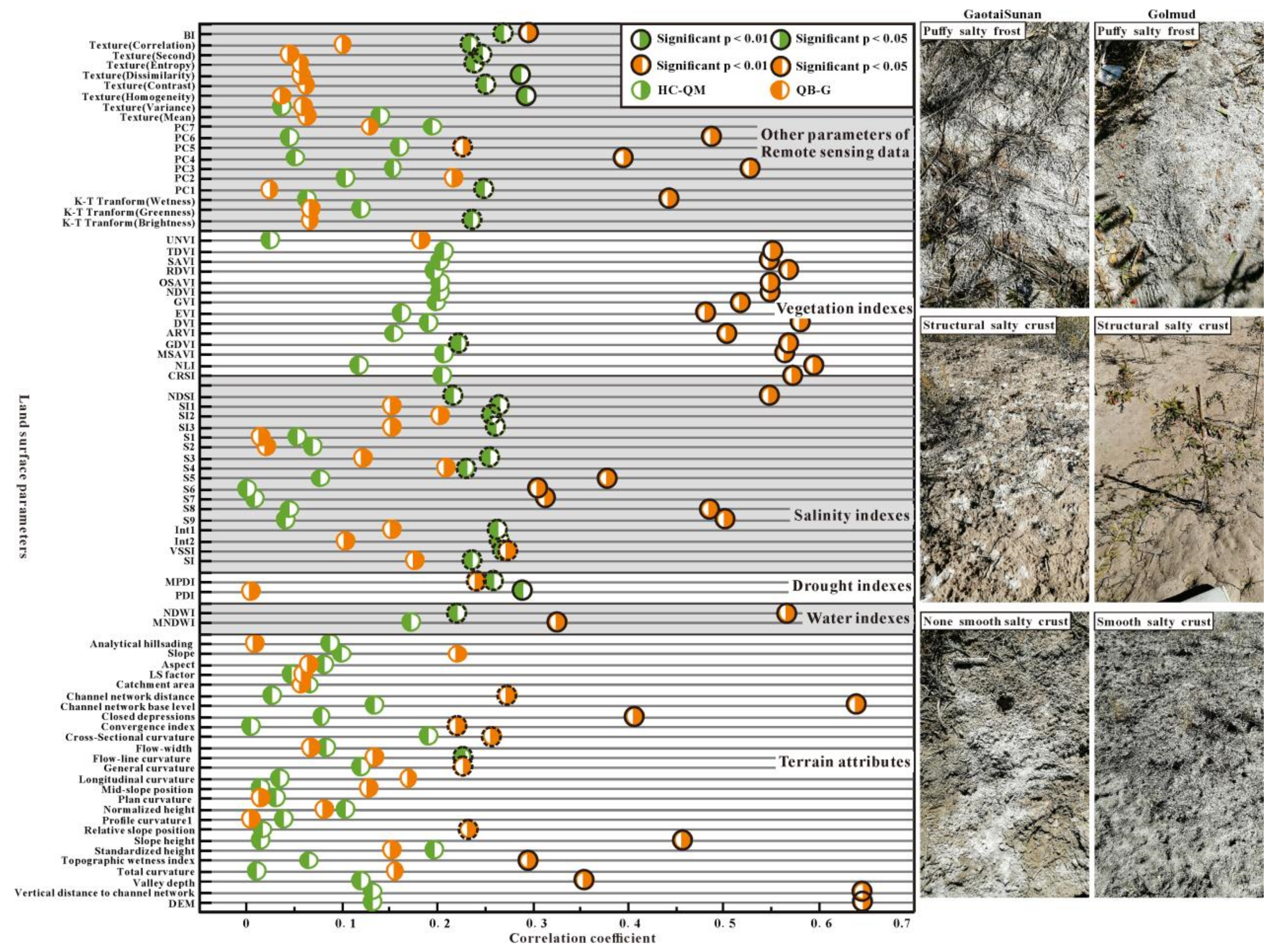
3.5. Correlation between Different Surface Parameters and EC
4. Discussion
4.1. Analysis of Correlation between Different Surface Parameters and EC
4.2. Advantages of Hyperspectral Data and Fractional Differential Transformation
4.3. Analysis of the Different Machine Learning Algorithms
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- FAO. Status of the world’s soil resources. Agric. Compr. Dev. China 2021, 10, 64. [Google Scholar]
- Csillag, F.; Pasztor, L.; Biehl, L.L. Spectral band selection for the characterization of salinity status of soils. Remote Sens. Environ. 1993, 43, 231–242. [Google Scholar] [CrossRef]
- Dwivedi, R.S.; Sreenivas, K. Image transforms as a tool for the study of soil salinity and alkalinity dynamics. Int. J. Remote Sens. 1998, 19, 605–619. [Google Scholar] [CrossRef]
- Muller, S.J.; van Niekerk, A. Identification of WorldView-2 spectral and spatial factors in detecting salt accumulation in cultivated fields. Geoderma 2016, 273, 1–11. [Google Scholar] [CrossRef]
- Fathizad, H.; Ardakani, M.A.H.; Sodaiezadeh, H.; Kerry, R.; Taghizadeh-Mehrjardi, R. Investigation of the spatial and temporal variation of soil salinity using random forests in the central desert of Iran. Geoderma 2020, 365, 114233. [Google Scholar] [CrossRef]
- Mirjalili, S.; Saremi, S.; Mirjalili, S.M.; Coelho, L.D.S. Multi-objective grey wolf optimizer: A novel algorithm for multi-criterion optimization. Expert Syst. Appl. 2016, 47, 106–119. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Mirjalili, S. SCA: A sine cosine algorithm for solving optimization problems. Knowl.-Based Syst. 2016, 96, 120–133. [Google Scholar] [CrossRef]
- Li, L.; Sun, J.; Tseng, M.; Li, Z. Extreme learning machine optimized by whale optimization algorithm using insulated gate bipolar transistor module aging degree evaluation. Expert Syst. Appl. 2019, 127, 58–67. [Google Scholar] [CrossRef]
- Chen, W.; Hong, H.; Panahi, M.; Shahabi, H.; Wang, Y.; Shirzadi, A.; Pirasteh, S.; Alesheikh, A.A.; Khosravi, K.; Panahi, S. Spatial prediction of landslide susceptibility using gis-based data mining techniques of anfis with whale optimization algorithm (woa) and grey wolf optimizer (gwo). Appl. Sci. 2019, 9, 3755. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.A.; Sulaiman, M.; Aljohani, A.J.; Kumam, P.; Alrabaiah, H. Analysis of multi-phase flow through porous media for imbibition phenomena by using the LeNN-WOA-NM algorithm. IEEE Access 2020, 8, 196425–196458. [Google Scholar] [CrossRef]
- Cheng, L.; Tang, X.F. Improved sine cosine algorithm optimizing feature selection and data classification. J. Comput. Appl. 2021, 1–11. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=JSJY20210926007&DbName=CAPJ2021 (accessed on 7 September 2021).
- Farifteh, J.; Van der Meer, F.; Atzberger, C.; Carranza, E.J.M. Quantitative analysis of salt-affected soil reflectance spectra: A comparison of two adaptive methods (PLSR and ANN). Remote Sens. Environ. 2007, 110, 59–78. [Google Scholar] [CrossRef]
- Fourati, H.T.; Bouaziz, M.; Benzina, M.; Bouaziz, S. Modeling of soil salinity within a semi-arid region using spectral analysis. Arab. J. Geosci. 2015, 8, 11175–11182. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python; Manning Publications: New York, NY, USA, 2017. [Google Scholar]
- Zhang, J.; Zhang, Z.; Chen, J.; Chen, H.; Jin, J.; Han, J.; Wang, X.; Song, Z.; Wei, G. Estimating soil salinity with different fractional vegetation cover using remote sensing. Land Degrad. Dev. 2020, 32, 597–612. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Ding, J.; Kung, H.T.; Latif, A.; Johnson, V.C. Estimation of soil salt content (SSC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR), Northwest China, based on a Bootstrap-BP neural network model and optimal spectral indices. Sci. Total Environ. 2018, 615, 918–930. [Google Scholar] [CrossRef] [PubMed]
- McBratney, A.B.; Santos, M.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Arruda, G.P.D.; Demattê, J.A.M.; Chagas, C.D.S.; Fiorio, P.R.; Souza, A.B.E.; Fongaro, C.T. Digital soil mapping using reference area and artificial neural networks. Scientia Agric. 2016, 73, 266–273. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Peng, J.; Li, H.; Yin, C.; Liu, W.; Wang, T.; Zhang, H. Soil salinity mapping using machine learning algorithms with the Sentinel-2 MSI in arid areas, China. Remote Sens. 2021, 13, 305. [Google Scholar] [CrossRef]
- Naz, N.S.; Khan, M.A.; Abbas, S.; Ather, A.; Saqib, S. Intelligent routing between capsules empowered with deep extreme machine learning technique. SN Appl. Sci. 2020, 2, 108. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, T.; Wang, C.; Yu, Z.; Stach, R.; Mizaikoff, B.; Huang, G.; Wang, Q. NOx measurements in vehicle exhaust using advanced deep ELM networks. IEEE Trans. Instrum. Meas. 2021, 70, 1–10. [Google Scholar] [CrossRef]
- Bas, E.; Egrioglu, E.; Karahasan, O. A Pi-Sigma artificial neural network based on sine cosine optimization algorithm. Granular Comput. 2021. [Google Scholar] [CrossRef]
- Wang, F.; Yang, S.; Wei, Y.; Shi, Q.; Ding, J. Characterizing soil salinity at multiple depth using electromagnetic induction and remote sensing data with random forests: A case study in Tarim River Basin of southern Xinjiang, China. Sci. Total Environ. 2021, 754, 142030. [Google Scholar] [CrossRef]
- Paz, M.C.; Farzamian, M.; Paz, A.M.; Castanheira, N.L.; Gonçalves, M.C.; Monteiro Santos, F. Assessing soil salinity dynamics using time-lapse electromagnetic conductivity imaging. Soil 2020, 6, 499–511. [Google Scholar] [CrossRef]
- Mougenot, B.; Pouget, M.; Epema, G.F. Remote sensing of salt affected soils. Remote Sens. Rev. 1993, 7, 241–259. [Google Scholar] [CrossRef]
- Everitt, J.H.; Gerbermann, A.H.; Cuellar, J.A. Distinguishing saline from nonsaline rangelands with Skylab imagery. Remote Sens. Earth Resour. 1977, 6, 51–65. [Google Scholar]
- Eastes, J.W. Spectral properties of halite-rich mineral mixtures: Implications for middle infrared remote sensing of highly saline environments. Remote Sens. Environ. 1989, 27, 289–303. [Google Scholar] [CrossRef]
- Khan, N.M.; Rastoskuev, V.V.; Sato, Y.; Shiozawa, S. Assessment of hydrosaline land degradation by using a simple approach of remote sensing indicators. Agric. Water Manag. 2005, 77, 96–109. [Google Scholar] [CrossRef]
- Douaoui, A.E.K.; Nicolas, H.; Walter, C. Detecting salinity hazards within a semiarid context by means of combining soil and remote-sensing data. Geoderma 2006, 134, 217–230. [Google Scholar] [CrossRef]
- Abbas, A.; Khan, S. Using remote sensing techniques for appraisal of irrigated soil salinity. In International Congress on Modelling and Simulation (MODSIM 2007)—Land, Water & Environmental Management: Integrated Systems for Sustainability, Christchurch, New Zealand, 10–13 December 2007; Modelling and Simulation Society of Australia and New Zealand: Canberra, Australia, 2007. [Google Scholar]
- Bannari, A.; Guedon, A.M.; El Harti, A.; Cherkaoui, F.Z.; El Ghmari, A. Characterization of slightly and moderately saline and sodic soils in irrigated agricultural land using simulated data of advanced land imaging (EO-1) sensor. Commun. Soil Sci. Plan. 2008, 39, 2795–2811. [Google Scholar] [CrossRef]
- Dehni, A.; Lounis, M. Remote Sensing Techniques for salt affected soil mapping: Application to the Oran Region of Algeria. Procedia Eng. 2012, 33, 188–198. [Google Scholar] [CrossRef] [Green Version]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; Texas A&M University: College Station, TX, USA, 1974. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Wu, W. The generalized difference vegetation index (GDVI) for dryland characterization. Remote Sens. 2014, 6, 1211–1233. [Google Scholar] [CrossRef] [Green Version]
- Goel, N.S.; Qin, W. Influences of canopy architecture on relationships between various vegetation indices and LAI and Fpar: A computer simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Zhang, L.; Qiao, N.; Baig, M.H.A.; Huang, C.; Lv, X.; Sun, X.; Zhang, Z. Monitoring vegetation dynamics using the universal normalized vegetation index (UNVI): An optimized vegetation index-VIUPD. Remote Sens. Lett. 2019, 10, 629–638. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Dalposso, G.H.; Uribe-Opazo, M.A.; Mercante, E.; Lamparelli, R.A. Spatial autocorrelation of NDVI and GVI indices derived from Landsat/TM images for soybean crops in the western of the state of Paraná in 2004/2005 crop season. Engenharia Agrícola 2013, 33, 525–537. [Google Scholar] [CrossRef] [Green Version]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Roujean, J.; Breon, F. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Bannari, A.; Asalhi, H.; Teillet, P.M. Transformed difference vegetation index (TDVI) for vegetation cover mapping. In IEEE International Geoscience and Remote Sensing Symposium; IEEE: Toronto, ON, Canada, 2005. [Google Scholar]
- Scudiero, E.; Skaggs, T.H.; Corwin, D.L. Regional scale soil salinity evaluation using Landsat 7, western San Joaquin Valley, California, USA. Geoderma Reg. 2014, 2, 82–90. [Google Scholar] [CrossRef]
- Xu, H.Q. A study on information extraction of water body with the modified normalized difference water index (MNDWI). J. Remote Sens. 2005, 5, 589–595. [Google Scholar]
- McFeeters, S.K. The use of the normalized difference water index (NDWI) in the delineation of open water features. Int. J. Remote Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Ghulam, A.; Qin, Q.; Zhan, Z. Designing of the perpendicular drought index. Environ. Geol. 2007, 52, 1045–1052. [Google Scholar] [CrossRef]
- Ghulam, A.; Qin, Q.; Teyip, T.; Li, Z. Modified perpendicular drought index (MPDI): A real-time drought monitoring method. ISPRS J. Photogramm. 2007, 62, 150–164. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Oldham, K.B.; Spanier, J. The Fractional Calculus; Academic Press: New York, NY, USA; London, UK, 1974. [Google Scholar]
- Podlubny, I. Fractional Differential Equations: An Introduction to Fractional Derivatives, Fractional Differential Equations, to Methods of Their Solution and Some of Their Applications; Academic Press: San Diego, CA, USA; Boston, MA, USA; New York, NY, USA; London, UK; Sydney, Austria; Tokyo, Japan; Toronto, ON, Canada, 1998. [Google Scholar]
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258. [Google Scholar] [CrossRef]
- Huang, G.; Zhu, Q.; Siew, C. Extreme learning machine: A new learning scheme of feedforward neural networks. In Proceedings of the 2004 IEEE International Joint Conference on Neural Networks (IEEE Cat. No.04CH37541), Budapest, Hungary, 25–29 July 2004; pp. 985–990. [Google Scholar] [CrossRef]
- Wang, X.C.; Shi, F.; Yu, L.; Li, Y. Analysis of 43 Cases of Neural Network in MATLAB; Beihang University Press: Beijing, China, 2013. [Google Scholar]
- Tissera, M.D.; McDonnell, M.D. Deep extreme learning machines: Supervised autoencoding architecture for classification. Neurocomputing 2016, 174, 42–49. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Liang, Z.; Zhu, S.; Correia, J.A.F.O.; De Jesus, A.M.P. PSO-BP neural network-based strain prediction of wind turbine blades. Materials 2019, 12, 1889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ding, J.; Yu, D.; Teng, D.; He, B.; Chen, X.; Ge, X.; Zhang, Z.; Wang, Y.; Yang, X.; et al. Machine learning-based detection of soil salinity in an arid desert region, Northwest China: A comparison between Landsat-8 OLI and Sentinel-2 MSI. Sci. Total Environ. 2020, 707, 136092. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.D.; Jiang, X.G.; Tang, L.L. Remote Sensing Image Application Processing and Analysis; Tsinghua University Press: Beijing, China, 2004. [Google Scholar]
- Wang, Q.; Li, P.; Chen, X. Modeling salinity effects on soil reflectance under various moisture conditions and its inverse application: A laboratory experiment. Geoderma 2012, 170, 103–111. [Google Scholar] [CrossRef]
- Bowers, S.A.; Smith, S.J. Spectrophotometric determination of soil-water content. Soil Sci. Soc. Am. J. 1972, 36, 978–980. [Google Scholar] [CrossRef]
- Baumgardner, M.F.; Silva, L.F.; Bieh, L.L.; Stoner, E.R. Reflectance properties of soils. Adv. Agron. 1985, 38, 1–44. [Google Scholar]
- Lindberg, J.D.; Snyder, D.G. Diffuse reflectance spectra of several clay-minerals. Am. Miner. 1972, 57, 485. [Google Scholar]
- Stoner, E.R.; Baumgardner, M.F. Characteristic variations in reflectance of surface soils. Soil Sci. Soc. Am. J. 1981, 45, 1161–1165. [Google Scholar] [CrossRef] [Green Version]
- Karmanov, I.I. Study of soils from spectral composition of reflected radiation. Soviet Soil Science-Ussr 1970, 2, 226. Available online: https://apps.webofknowledge.com/full_record.do?product=UA&search_mode=GeneralSearch&qid=2&SID=7CAMmr3NAVtWbMXpd7Y&page=1&doc=1 (accessed on 7 September 2021).
- Wen, Z.W. Discussion on the classification of saline soil in Xinjiang Province. Xinjiang Agric. Sci. 1963, 463–469. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CJFD&dbname=CJFD7984&filename=XJNX196312000&uniplatform=NZKPT&v=y4lGY2wIESlvvhpmFrpQ1a4rzvrukV5FeP-WT2plO6W2tANIbB838fafPUV7vzZD (accessed on 7 September 2021).
- Li, L.Q.; Wang, Z.Q. Salt-affected soils type and saline-geochemical features in Qaidam Basin. Acta Pedol Sin. 1990, 27, 43–53. [Google Scholar]
- Yu, R.P.; Wang, Z.Q.; Zhu, S.Q. Chinese Saline Soil; Science Press: Beijing, China, 1993. [Google Scholar]
- Farifteh, J.; van der Meer, F.; van der Meijde, M.; Atzberger, C. Spectral characteristics of salt-affected soils: A laboratory experiment. Geoderma 2008, 145, 196–206. [Google Scholar] [CrossRef]
- Hunt, G.R.; Salisbury, J.W.; Lenhoff, C.J. Visible and near-infrared spectra of minerals and rocks: V. Halides, phosphates, arsenates, vanadates and borates. Mod. Geol. 1972, 3, 121–132. [Google Scholar]
- Metternicht, G.I.; Zinck, J.A. Remote sensing of soil salinity: Potentials and constraints. Remote Sens. Environ. 2003, 85, 1–20. [Google Scholar] [CrossRef]
- Cheng, Q.; Zhong, F.; Wang, P. Potential linkages of extreme climate events with vegetation and large-scale circulation indices in an endorheic river basin in northwest China. Atmos. Res. 2021, 247, 105256. [Google Scholar] [CrossRef]
- An, Q.; He, H.; Gao, J.; Nie, Q.; Cui, Y.; Wei, C.; Xie, X. Analysis of temporal-spatial variation characteristics of drought: A case study from Xinjiang, China. Water 2020, 12, 741. [Google Scholar] [CrossRef] [Green Version]
- Tilley, D.R.; Ahmed, M.; Son, J.H.; Badrinarayanan, H. Hyperspectral reflectance response of freshwater macrophytes to salinity in a brackish subtropical marsh. J. Environ. Qual. 2007, 36, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zeng, S.; Gao, Y.; Ouyang, Z.; Li, B.; Fang, C.; Zhao, B. Using hyperspectral vegetation indices as a proxy to monitor soil salinity. Ecol. Indic. 2011, 11, 1552–1562. [Google Scholar] [CrossRef]
- Ding, J.; Yu, D. Monitoring and evaluating spatial variability of soil salinity in dry and wet seasons in the Werigan–Kuqa Oasis, China, using remote sensing and electromagnetic induction instruments. Geoderma 2014, 235–236, 316–322. [Google Scholar] [CrossRef]
- Akramkhanov, A.; Martius, C.; Park, S.J.; Hendrickx, J.M.H. Environmental factors of spatial distribution of soil salinity on flat irrigated terrain. Geoderma 2011, 163, 55–62. [Google Scholar] [CrossRef]
- Wang, F.; Shi, Z.; Biswas, A.; Yang, S.; Ding, J. Multi-algorithm comparison for predicting soil salinity. Geoderma 2020, 365, 114211. [Google Scholar] [CrossRef]
- Rao, B.; Sankar, T.R.; Dwivedi, R.S.; Thammappa, S.S.; Venkataratnam, L.; Sharma, R.C.; Das, S.N. Spectral behavior of salt-affected soils. Int. J. Remote Sens. 1995, 16, 2125–2136. [Google Scholar] [CrossRef]
- Singh, R.P.; Sirohi, A. Spectral reflectance properties of different types of soil surfaces. ISPRS J. Photogramm. 1994, 49, 34–40. [Google Scholar] [CrossRef]
- Allbed, A.; Kumar, L.; Aldakheel, Y.Y. Assessing soil salinity using soil salinity and vegetation indices derived from IKONOS high-spatial resolution imageries: Applications in a date palm dominated region. Geoderma 2014, 230, 1–8. [Google Scholar] [CrossRef]
- Gutierrez, M.; Johnson, E. Temporal variations of natural soil salinity in an arid environment using satellite images. J. S. Am. Earth Sci. 2010, 30, 46–57. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Minasny, B.; Sarmadian, F.; Malone, B.P. Digital mapping of soil salinity in Ardakan region, central Iran. Geoderma 2014, 213, 15–28. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Zhang, Z.; Chen, H.; Li, X.; Wang, M.; Chai, H. Quantitatively estimating main soil water-soluble salt ions content based on Visible-near infrared wavelength selected using GC, SR and VIP. PeerJ 2019, 7, e6310. [Google Scholar] [CrossRef]
- Yang, N.; Yang, S.; Cui, W.; Zhang, Z.; Zhang, J.; Chen, J.; Ma, Y.; Lao, C.; Song, Z.; Chen, Y. Effect of spring irrigation on soil salinity monitoring with UAV-borne multispectral sensor. Int. J. Remote Sens. 2021, 42, 8952–8978. [Google Scholar] [CrossRef]
- Hagiwara, K.; Fukumizu, K. Relation between weight size and degree of over-fitting in neural network regression. Neural Netw. 2008, 21, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Biswas, A.; Jiang, Q.S.; Zhao, R.Y.; Hu, J.; Hu, B.F.; Shi, Z. Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 2019, 337, 1309–1319. [Google Scholar] [CrossRef]
- Chi, Y.; Sun, J.; Liu, W.; Wang, J.; Zhao, M. Mapping coastal wetland soil salinity in different seasons using an improved comprehensive land surface factor system. Ecol. Indic. 2019, 107, 105517. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Hu, Y.; Tian, S.; Liu, D. Soil moisture and salinity inversion based on new remote sensing index and neural network at a salina-alkaline wetland. Water 2021, 13, 2762. [Google Scholar] [CrossRef]
- Ghamisi, P.; Plaza, J.; Chen, Y.; Li, J.; Plaza, A.J. Advanced spectral classifiers for hyperspectral images: A review. IEEE Geosci. Remote Sens.Mag. 2017, 5, 8–32. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, F.; Kung, H.; Johnson, V.C.; Latif, A. Extracting soil salinization information with a fractional-order filtering algorithm and grid-search support vector machine (GS-SVM) model. Int. J. Remote Sens. 2020, 41, 953–973. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, Y.; Chen, Y.; Liu, Y.; Yu, L.; Liu, Y.; Cheng, H. Application of fractional-order derivative in the quantitative estimation of soil organic matter content through visible and near-infrared spectroscopy. Geoderma 2019, 337, 758–769. [Google Scholar] [CrossRef]
- Wang, M.; Chen, H.; Li, H.; Cai, Z.; Zhao, X.; Tong, C.; Li, J.; Xu, X. Grey wolf optimization evolving kernel extreme learning machine: Application to bankruptcy prediction. Eng. Appl. Artif. Intel. 2017, 63, 54–68. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, D.; Gravina, R.; Fortino, G.; Jiang, Y.; Tang, K. Kernel fusion based extreme learning machine for cross-location activity recognition. Inform. Fusion. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Xu, Z.; Yao, M.; Wu, Z.; Dai, W. Incremental regularized extreme learning machine and it׳ s enhancement. Neurocomputing 2016, 174, 134–142. [Google Scholar] [CrossRef]
- Nabiollahi, K.; Taghizadeh-Mehrjardi, R.; Shahabi, A.; Heung, B.; Amirian-Chakan, A.; Davari, M.; Scholten, T. Assessing agricultural salt-affected land using digital soil mapping and hybridized random forests. Geoderma 2021, 385, 114858. [Google Scholar] [CrossRef]
- Taghadosi, M.M.; Hasanlou, M.; Eftekhari, K. Soil salinity mapping using dual-polarized SAR Sentinel-1 imagery. Int. J. Remote Sens. 2019, 40, 237–252. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, L.; Sun, R.; Zhu, D.; Zhang, C.; Chen, Q. Retrieval of the soil salinity from Sentinel-1 Dual-Polarized SAR data based on deep neural network regression. IEEE Geosci. Remote Sens. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Hoa, P.; Giang, N.; Binh, N.; Hai, L.; Pham, T.; Hasanlou, M.; Tien Bui, D. Soil salinity mapping using SAR Sentinel-1 data and advanced machine learning algorithms: A case study at Ben Tre Province of the Mekong River Delta (Vietnam). Remote Sens. 2019, 11, 128. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Hellwich, O.; Luo, G.; Chen, C.; He, H.; Ochege, F.U.; Van de Voorde, T.; Kurban, A.; de Maeyer, P. A global meta-analysis of soil salinity prediction integrating satellite remote sensing, soil sampling, and machine learning. IEEE Trans. Geosci. Remote Sens. 2021, 1–15. Available online: https://ieeexplore.ieee.org/document/9538387/ (accessed on 7 September 2021). [CrossRef]
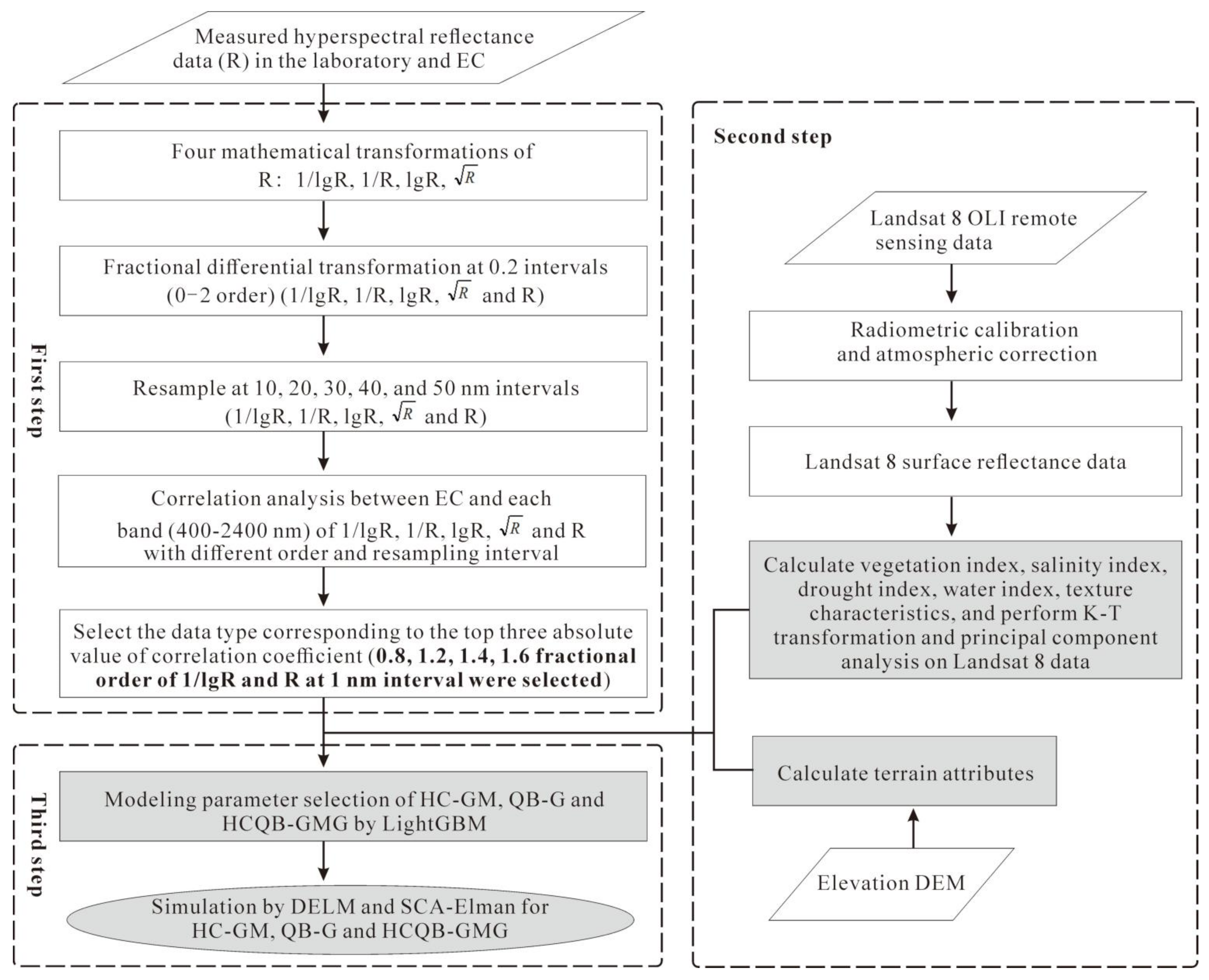
| Land Surface Parameters | Abbreviation | Formula | References |
|---|---|---|---|
| Salinity index | SI | (B4 × B2)0.5 | [30] |
| Normalized differential salinity index | NDSI | (B4 − B5)/(B4 + B5) | [30] |
| Salinity index 1 | SI1 | (B4 × B3)0.5 | [31] |
| Salinity index 2 | SI2 | [(B5)2 + (B4)2 + (B3)2]0.5 | [31] |
| Salinity index 3 | SI3 | [(B4)2 + (B3)2]0.5 | [31] |
| Salinity index I | S1 | B2/B4 | [32] |
| Salinity index II | S2 | (B2 − B4)/(B2 + B4) | [32] |
| Salinity index III | S3 | B3 × B4/B2 | [32] |
| Salinity index IV | S4 | B2 × B4/B3 | [32] |
| Salinity index V | S5 | B4 × B5/B3 | [32] |
| Salinity index VI | S6 | B6/B7 | [33] |
| Salinity index VII | S7 | (B6 − B7)/(B6 + B7) | [33] |
| Salinity index VIII | S8 | B6 − B7 | [33] |
| Salinity index IX | S9 | (B6 × B7 − B7 × B7)/B6 | [33] |
| Intensity index 1 | Int1 | (B3 + B4)/2 | [31] |
| Intensity index 2 | Int2 | (B3 + B4 + B5)/2 | [31] |
| Vegetation soil salinity index | VSSI | 2 × B3 − 5 × (B4 + B5) | [34] |
| Land Surface Parameters | Abbreviation | Formula | References |
|---|---|---|---|
| Normalized difference vegetation index | NDVI * | (B5 − B4)/(B5 + B4) | [35] |
| Enhanced vegetation index | EVI * | (1 + L) ×(B5 − B4)/(B5 + C1 × B4− C2 × B2 + L), L is the background adjustment parameter and C1 and C2 are the atmospheric correction parameters | [36] |
| Generalized difference vegetation index | GDVI | (B52 − B42)/(B52 + B42) | [37] |
| Non-linear vegetation index | NLI | (B52 − B4)/(B52 + B4) | [38] |
| Modified soil adjusted vegetation index | MSAVI | {(2 × B5-1) − [(2 × B5 + 1) ×(2 × B5 + 1) − 8 × (B5 − b4)]0.5}/2 | [39] |
| Universal normalized vegetation index | UNVI | R(i) → [Cw × Pw (i) + Cv × Pv (i) + Cs × Ps (i) + C4 × P4 (i)], where i is the band number, R(i) is the spectrum under the i band of the ground object, Pw, Pv, Ps and P4 respectively represent the normalized reflectance value of the four reference samples; Cw, Cv, Cs, C4 represent the UPDM coefficient corresponding to each sample. | [40] |
| Atmospherically resistant vegetation index | ARVI * | {B5 − [B4 − γ × (B2 − B4)]}/{B5 + [B4 − γ × (B2 − B4)]}, γ is the correction coefficient of atmospheric radiation | [41] |
| Difference vegetation index | DVI | B5 − B4 | [42] |
| Green vegetation index | GVI * | −0.2848 × B2 − 0.2435 × B3 − 0.5436 × B4 + 0.7243 × B5 + 0.0840 × B6 − 0.1800 × B7 | [43] |
| Optimized soil adjusted vegetation index | OSAVI * | (B5 − B4)/(B5 + B4 + θ), θ is the soil regulation parameter that has nothing to do with vegetation coverage conditions | [44] |
| Renormalized difference vegetation index | RDVI * | (NDVI × DVI)0.5 | [45] |
| Soil adjusted vegetation index | SAVI * | (1 + L)(B5 − B4)/(B5 + B4 + L), L is the soil brightness index | [46] |
| Transformed difference vegetation index | TDVI * | 1.5 × [(B5 − B4)/(B5^2 + B4 + 0.5)0.5] | [47] |
| Canopy response salinity index | CRSI | [(B5 × B4 − B3 × B2)/(B5 × B4 + B3 × B2)]0.5 | [48] |
| Auxiliary Data | Land Surface Parameters | Abbreviation | Formula | References |
|---|---|---|---|---|
| Water index | Modified normalized difference water index | MNDWI * | (B3 − B6)/(B3 + B6) | [49] |
| Normalized difference water index | NDWI | (B3 − B5)/(B3 + B5) | [50] | |
| Remote Sensing data | Kauth–Thomas transformation (Brightness, Greenness, Wetness) | K–T transformation | ||
| Principal component analysis (PC1–PC7) | PCA | |||
| Texture (Mean, Variance, Homogeneity, Contrast, Dissimilarity, Entropy, Second, Correlation) | T | |||
| Brightness Index | BI | (B42 + B52)0.5 | [30] | |
| Drought index | Perpendicular drought index | PDI | (B4 + M × B5)/(1 + B42)0.5, M is the slope of the soil baseline, M of HC–GM is 0.189, M of QB–G is 0.32 | [51] |
| Modified perpendicular drought index | MPDI | (B4 + M4 × B5 − fv × 0.55)/[(1 − fv) × (1 + M 2)0.5], M is the slope of the soil baseline, M of HC–GM is 0.189, M of QB–G is 0.32, fv = 1 − ((NDVImax − NDVI)/(NDVImax − NDVI min))0.6175 | [52] |
| Land Surface Parameters | References | Land Surface Parameters | References |
|---|---|---|---|
| Elevation DEM | SAGA GIS | General curvature | SAGA GIS |
| Vertical distance to channel network | SAGA GIS | Flow-line curvature | SAGA GIS |
| Valley depth | SAGA GIS | Flow width | SAGA GIS |
| Total curvature | SAGA GIS | Cross-sectional curvature | SAGA GIS |
| Topographic wetness index | SAGA GIS | Convergence index | SAGA GIS |
| Standardized height | SAGA GIS | Closed depressions | SAGA GIS |
| Slope height | SAGA GIS | Channel network base level | SAGA GIS |
| Relative slope position | SAGA GIS | Channel network distance | SAGA GIS |
| Profile curvature | SAGA GIS | Catchment area | SAGA GIS |
| Normalized height | SAGA GIS | LS factor | SAGA GIS |
| Plan curvature | SAGA GIS | Aspect | SAGA GIS |
| Mid-slope position | SAGA GIS | Slope | SAGA GIS |
| Longitudinal curvature | SAGA GIS | Analytical hillshading | SAGA GIS |
| Area | Datasets | Sample Numbers | Maximum (mS·cm−1) | Minimum (mS·cm−1) | Mean (mS·cm−1) | Median (mS·cm−1) | SD—Standard Deviation (mS·cm−1) | CV—Coefficient of Variation |
|---|---|---|---|---|---|---|---|---|
| HC–GM | Calibration | 57 | 47.67 | 0.05 | 6.93 | 1.95 | 11.35 | 1.64 |
| Validation | 29 | 57.40 | 0.07 | 10.29 | 4.76 | 15.15 | 1.47 | |
| All | 86 | 57.40 | 0.05 | 8.06 | 2.93 | 12.77 | 1.58 | |
| QB–G | Calibration | 53 | 131.77 | 0.06 | 32.11 | 19.12 | 35.29 | 1.10 |
| Validation | 26 | 94.05 | 0.09 | 28.50 | 10.74 | 30.83 | 1.08 | |
| All | 79 | 131.77 | 0.06 | 30.92 | 14.34 | 33.73 | 1.09 | |
| HCQB–GMG | Calibration | 110 | 131.77 | 0.05 | 17.67 | 4.47 | 27.59 | 1.56 |
| Validation | 55 | 94.05 | 0.06 | 21.68 | 7.12 | 27.41 | 1.26 | |
| All | 165 | 131.77 | 0.05 | 19.01 | 5.11 | 27.51 | 1.45 |
| Area | Model | Calibration | Validation | ||||
|---|---|---|---|---|---|---|---|
| MAEc | RMSEc | MAEv | RMSEv | ||||
| HC–GM | SCA–Elman | 0.06 | 0.11 | 0.67 | 0.13 | 0.18 | 0.62 |
| DELM | 0.07 | 0.09 | 0.80 | 0.17 | 0.21 | 0.51 | |
| QB–G | SCA–Elman | 0.09 | 0.13 | 0.79 | 0.11 | 0.15 | 0.60 |
| DELM | 0.07 | 0.09 | 0.88 | 0.14 | 0.18 | 0.49 | |
| HCQB–GMG | SCA–Elman | 0.08 | 0.11 | 0.71 | 0.11 | 0.15 | 0.59 |
| DELM | 0.06 | 0.08 | 0.85 | 0.10 | 0.13 | 0.66 | |
| Region | Surrogate Parameter of Salinity | Vegetation Indexes | Salinity Indexes | Terrain Attributes | PCA | References |
|---|---|---|---|---|---|---|
| Wensu county of southern Xinjiang Province, China | EC | 0.25–0.42 (mean: 0.31) | 0.01–0.29 (mean: 0.11) | 0.01–0.43 (mean: 0.09) | 0.01–0.33 (mean: 0.13) | [91] |
| The Yellow River delta of China | EC | 0.51–0.70 (mean: 0.61) | 0.08–0.52 (mean: 0.31) | - | - | [92] |
| The Kuqa oasis in the northwestern part of Tarim Basin, China | EC | 0.44–0.80 (mean: 0.55) | - | 0.04–0.41 (mean: 0.20) | - | [82] |
| The Ebinur Lake in Xinjiang, China | EC | 0.27–0.36 (mean: 0.34) | 0.13–0.65 (mean: 0.43) | - | - | [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Duan, H.; Liao, J.; Guo, P.; Huang, C.; Xue, X. Estimation of Soil Salinization by Machine Learning Algorithms in Different Arid Regions of Northwest China. Remote Sens. 2022, 14, 347. https://doi.org/10.3390/rs14020347
Jiang X, Duan H, Liao J, Guo P, Huang C, Xue X. Estimation of Soil Salinization by Machine Learning Algorithms in Different Arid Regions of Northwest China. Remote Sensing. 2022; 14(2):347. https://doi.org/10.3390/rs14020347
Chicago/Turabian StyleJiang, Xiaofang, Hanchen Duan, Jie Liao, Pinglin Guo, Cuihua Huang, and Xian Xue. 2022. "Estimation of Soil Salinization by Machine Learning Algorithms in Different Arid Regions of Northwest China" Remote Sensing 14, no. 2: 347. https://doi.org/10.3390/rs14020347
APA StyleJiang, X., Duan, H., Liao, J., Guo, P., Huang, C., & Xue, X. (2022). Estimation of Soil Salinization by Machine Learning Algorithms in Different Arid Regions of Northwest China. Remote Sensing, 14(2), 347. https://doi.org/10.3390/rs14020347








