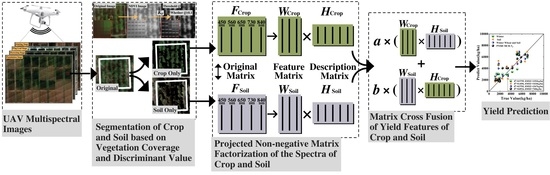
UAV Remote Sensing Prediction Method of Winter Wheat Yield Based on the Fused Features of Crop and Soil
Abstract
1. Introduction
2. Materials and Methods
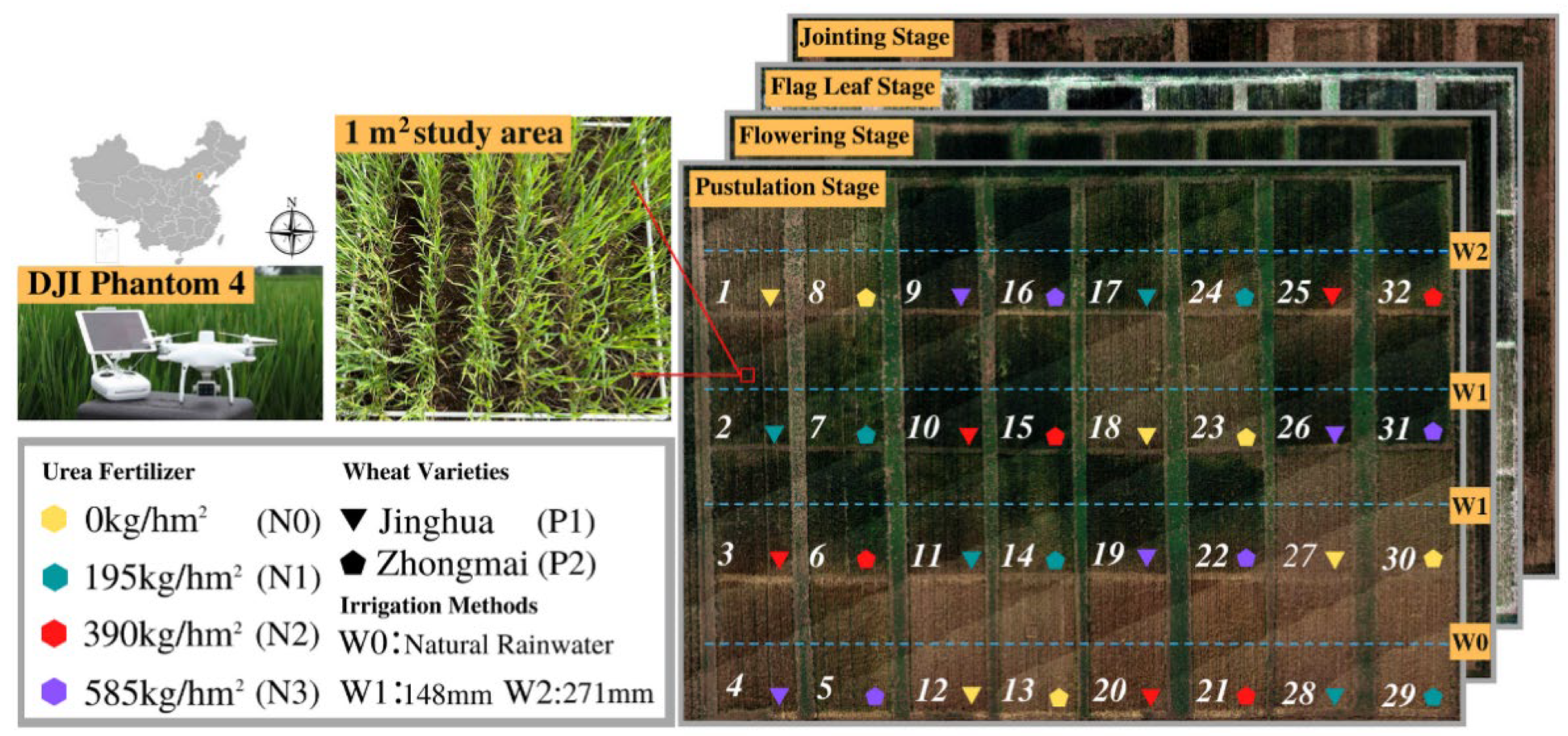
2.1. Experiment Design
Data Acquisition
2.2. Crop and Soil Piecewise Segmentation Method
2.2.1. Vegetation Coverage Determination Function
2.2.2. Discriminant Value Q for the Segmentation of Crop and Soil
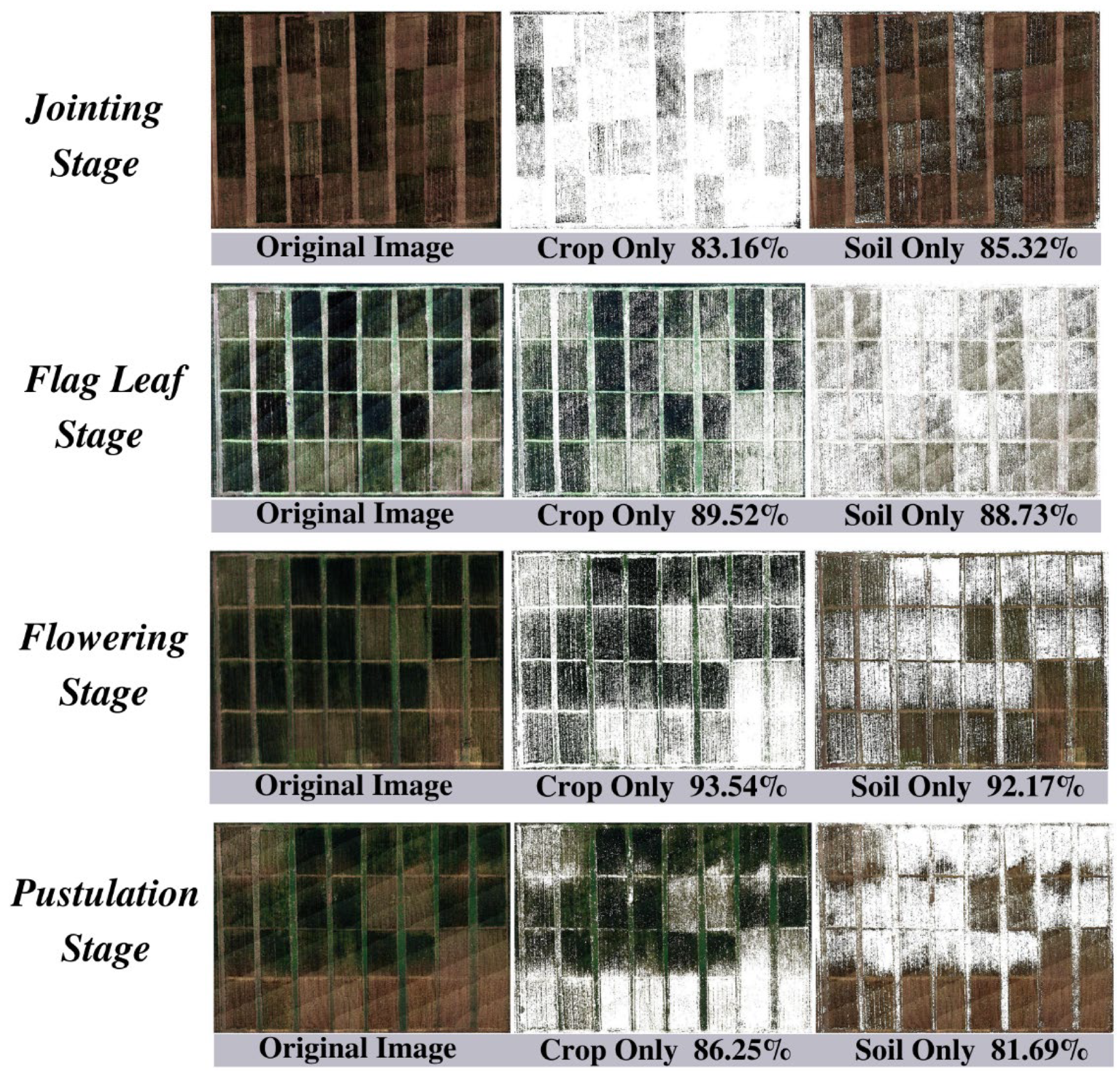
2.2.3. Accuracy Evaluation for Crop and Soil Segmentation
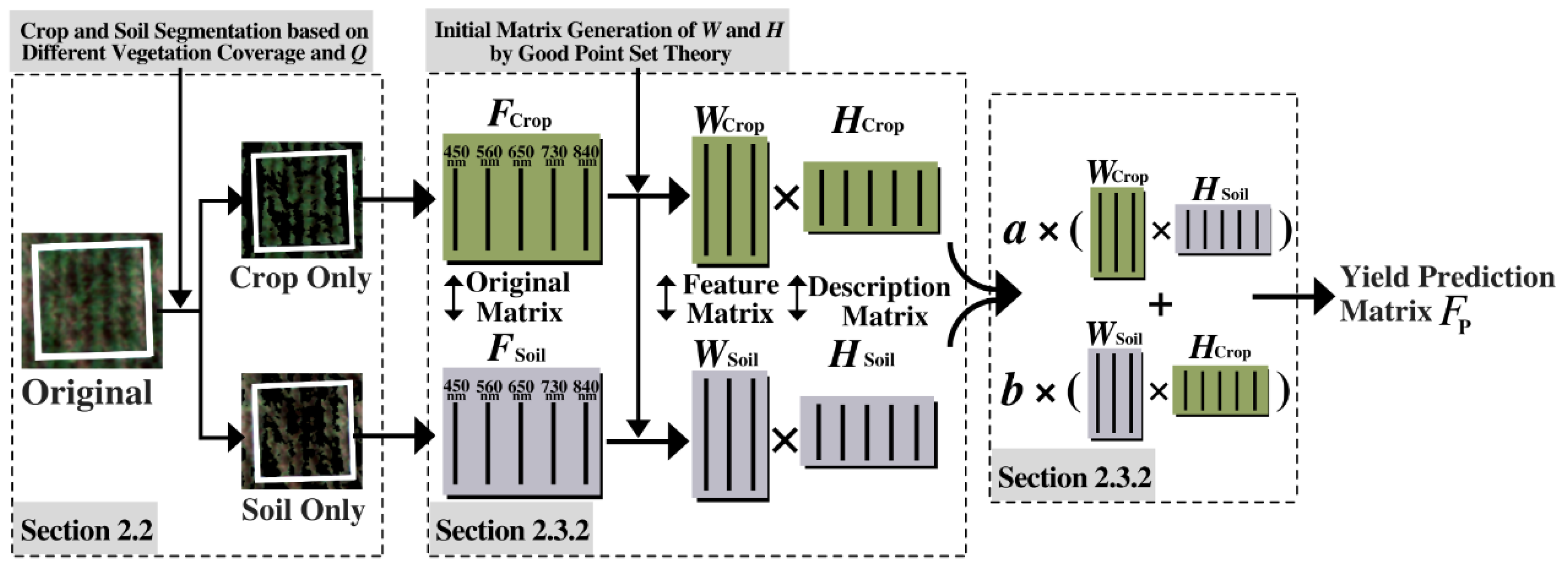
2.3. Projected Non-Negative Matrix Factorization and Matrix Cross Fusion
2.3.1. Matrix Initialization Based on Good Point Set
2.3.2. Projected Non-Negative Matrix Factorization Optimized by Good Point Set
2.3.3. Matrix Cross Fusin
2.4. Crop Yield Prediction Method
3. Results
3.1. Crop and Soil Multispectral Image Segmentation
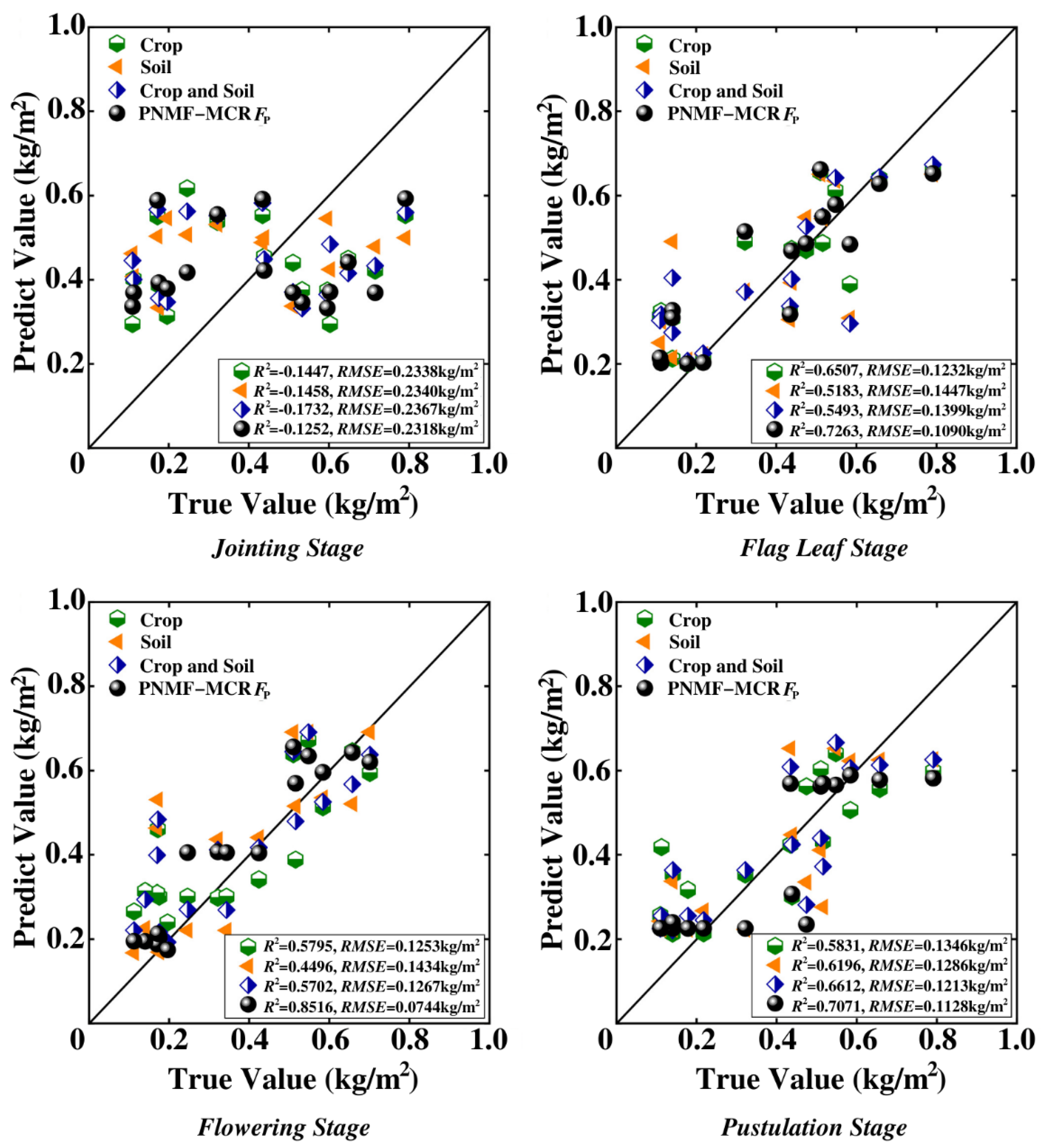
3.2. Crop Yield Prediction
4. Discussion
4.1. The Comparison of the Proposed Segmentation Method with HSV−Based Method and Deep Learning−Based Method
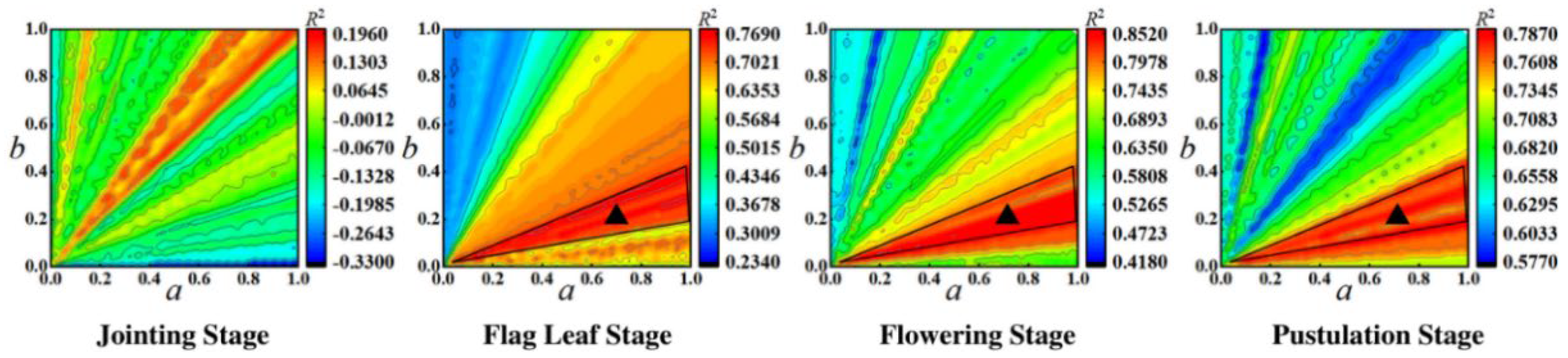
4.2. The Impact of Fusion Coefficients a and b on the Results of Yield Prediction
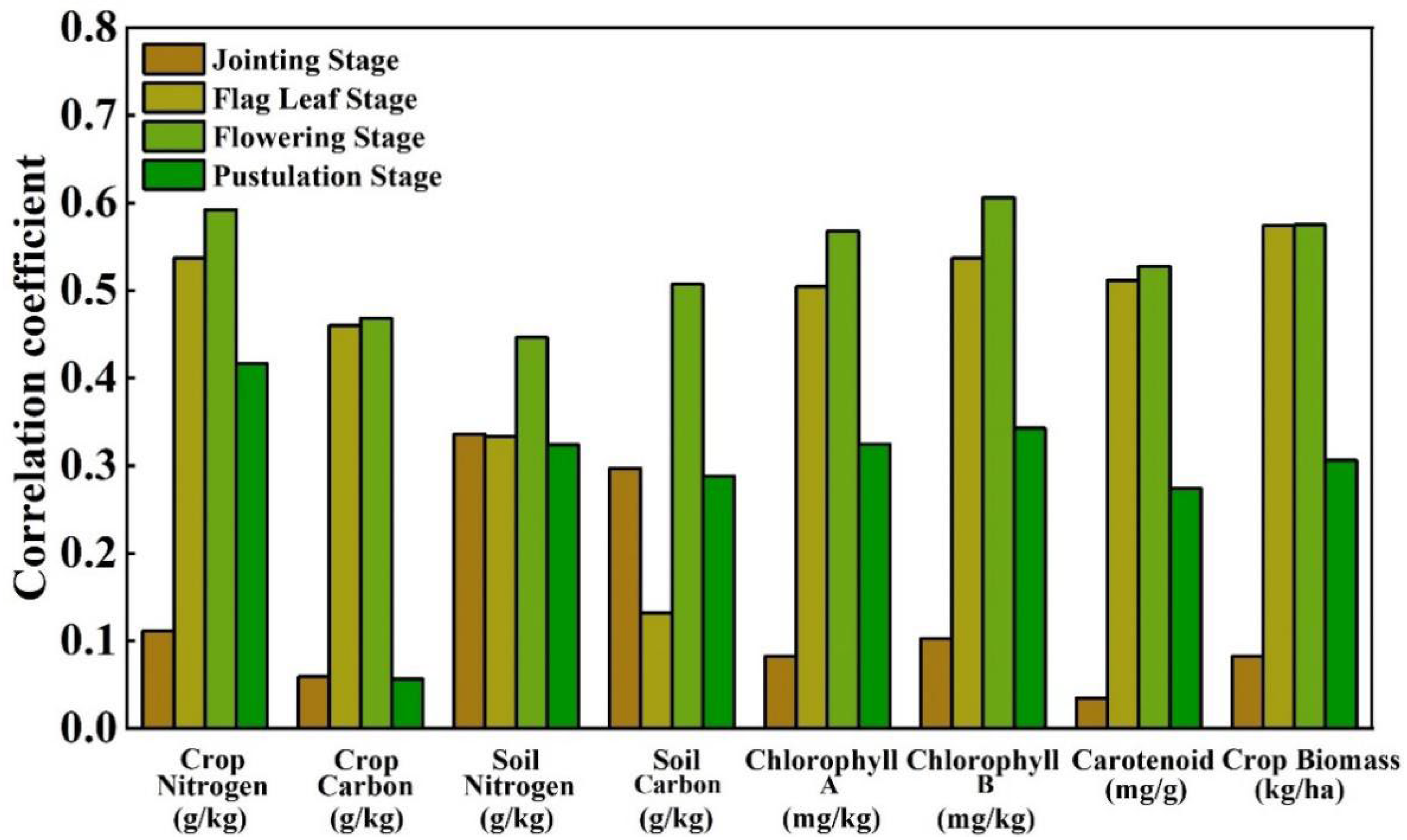
4.3. Correlation between FP and Other Biochemical Parameters
5. Conclusions
- The complex and changeable farmland environment poses a challenge to accurately identify crop and soil using remote sensing data. On the basis of different vegetation coverage index Kv, a segmentation discriminant Q was proposed to achieve the accurate segmentation of crop and soil. The experimental results have showed that it is completely feasible to determine whether a pixel is a crop by determining whether the Q of the pixel is greater than or equal to 0.1. This research will facilitate the accurate pixel-level identification of crop and soil in practice in remote sensing platform.
- The significance of synthetically considering crop and soil for yield research is to reduce bias compared to crop−only yield prediction. The PNMF−MCF can effectively fuse the yield features of crop and soil, and then achieve high precision yield prediction. Compared to the existing UAV based wheat yield studies [55,56,57,58,59], the method proposed in this manuscript obtained a better yield prediction performance. The experimental results show that the flowering stage is the best time to perform PNMF−MCF, because not only the flowering period is the most metabolically active stage, with intense photosynthesis shaping the basis of yield, but also the sealing ridge is not completed in this period and thus crop and soil spectral information from UAV images could be better captured to achieve adequate utilization.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Zhang, G.; Liu, S.; Dong, Y.; Liao, Y.; Han, J. A Nitrogen Fertilizer Strategy for Simultaneously Increasing Wheat Grain Yield and Protein Content: Mixed Application of Controlled-Release Urea and Normal Urea. Field Crop. Res. 2022, 277, 108405. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, Q. Winter Wheat Yield Estimation with Ground Based Spectral Information. In Proceedings of the IGARSS 2018—2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 6863–6866. [Google Scholar]
- Islam, Z.; Kokash, D.M.; Babar, M.S.; Uday, U.; Hasan, M.M.; Rackimuthu, S.; Essar, M.Y.; Nemat, A. Food Security, Conflict, and COVID-19: Perspective from Afghanistan. Am. J. Trop. Med. Hyg. 2022, 106, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qin, Q.; Ren, H.; Sun, Y.; Li, M.; Zhang, T.; Ren, S. Optimal Hyperspectral Characteristics Determination for Winter Wheat Yield Prediction. Remote Sens. 2018, 10, 2015. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Lipping, T. Crop Yield Prediction with Deep Convolutional Neural Networks. Comput. Electron. Agric. 2019, 163, 104859. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Zha, Y.; Zhu, P. Deep Convolutional Neural Networks for Rice Grain Yield Estimation at the Ripening Stage Using UAV-Based Remotely Sensed Images. Field Crop. Res. 2019, 235, 142–153. [Google Scholar] [CrossRef]
- Zhang, Y.; Hui, J.; Qin, Q.; Sun, Y.; Zhang, T.; Sun, H.; Li, M. Transfer-Learning-Based Approach for Leaf Chlorophyll Content Estimation of Winter Wheat from Hyperspectral Data. Remote Sens. Environ. 2021, 267, 112724. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine Learning Approaches for Crop Yield Prediction and Nitrogen Status Estimation in Precision Agriculture: A Review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- West, H.; Quinn, N.; Horswell, M. Remote Sensing for Drought Monitoring & Impact Assessment: Progress, Past Challenges and Future Opportunities. Remote Sens. Environ. 2019, 232, 111291. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, Q.; Chen, X.; Bao, A.; Zhang, J.; Wang, Y. Large Scale Agricultural Plastic Mulch Detecting and Monitoring with Multi-Source Remote Sensing Data: A Case Study in Xinjiang, China. Remote Sens. 2019, 11, 2088. [Google Scholar] [CrossRef]
- Sun, C.; Feng, L.; Zhang, Z.; Ma, Y.; Crosby, T.; Naber, M.; Wang, Y. Prediction of End-Of-Season Tuber Yield and Tuber Set in Potatoes Using In-Season UAV-Based Hyperspectral Imagery and Machine Learning. Sensors 2020, 20, 5293. [Google Scholar] [CrossRef]
- Iizuka, K.; Itoh, M.; Shiodera, S.; Matsubara, T.; Dohar, M.; Watanabe, K. Advantages of Unmanned Aerial Vehicle (UAV) Photogrammetry for Landscape Analysis Compared with Satellite Data: A Case Study of Postmining Sites in Indonesia. Cogent Geosci. 2018, 4, 1498180. [Google Scholar] [CrossRef]
- Bian, C.; Shi, H.; Wu, S.; Zhang, K.; Wei, M.; Zhao, Y.; Sun, Y.; Zhuang, H.; Zhang, X.; Chen, S. Prediction of Field-Scale Wheat Yield Using Machine Learning Method and Multi-Spectral UAV Data. Remote Sens. 2022, 14, 1474. [Google Scholar] [CrossRef]
- Ramos, A.P.M.; Osco, L.P.; Furuya, D.E.G.; Gonçalves, W.N.; Santana, D.C.; Teodoro, L.P.R.; da Silva, C.A., Jr.; Capristo-Silva, G.F.; Li, J.; Baio, F.H.R.; et al. A Random Forest Ranking Approach to Predict Yield in Maize with Uav-Based Vegetation Spectral Indices. Comput. Electron. Agric. 2020, 178, 105791. [Google Scholar] [CrossRef]
- Shvorov, S.; Lysenko, V.; Pasichnyk, N.; Opryshko, O.; Komarchuk, D.; Rosamakha, Y.; Rudenskyi, A.; Lukin, V.; Martsyfei, A. The Method of Determining the Amount of Yield Based on the Results of Remote Sensing Obtained Using UAV on the Example of Wheat. In Proceedings of the 2020 IEEE 15th International Conference on Advanced Trends in Radioelectronics, Telecommunications and Computer Engineering (TCSET), Lviv-Slavske, Ukraine, 25–29 February 2020; pp. 245–248. [Google Scholar]
- Dyson, J.; Mancini, A.; Frontoni, E.; Zingaretti, P. Deep Learning for Soil and Crop Segmentation from Remotely Sensed Data. Remote Sens. 2019, 11, 1859. [Google Scholar] [CrossRef]
- Bodner, G.; Nakhforoosh, A.; Arnold, T.; Leitner, D. Hyperspectral Imaging: A Novel Approach for Plant Root Phenotyping. Plant Method. 2018, 14, 84. [Google Scholar] [CrossRef]
- Su, W.-H. Advanced Machine Learning in Point Spectroscopy, RGB and Hyperspectral-Imaging for Automatic Discriminations of Crops and Weeds: A Review. Smart Cities 2020, 3, 767–792. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.J.; Burgos-Artizzu, X.P.; Ribeiro, A. Automatic Segmentation of Relevant Textures in Agricultural Images. Comput. Electron. Agric. 2011, 75, 75–83. [Google Scholar] [CrossRef]
- Castillo-Martínez, M.Á.; Gallegos-Funes, F.J.; Carvajal-Gámez, B.E.; Urriolagoitia-Sosa, G.; Rosales-Silva, A.J. Color Index Based Thresholding Method for Background and Foreground Segmentation of Plant Images. Comput. Electron. Agric. 2020, 178, 105783. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef]
- Syers, J.K. Managing Soils for Long-Term Productivity. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1997, 352, 1011–1021. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, X.; Tu, S.; Lindström, K. Soil Microbial Biomass, Crop Yields, and Bacterial Community Structure as Affected by Long-Term Fertilizer Treatments under Wheat-Rice Cropping. Eur. J. Soil Biol. 2009, 45, 239–246. [Google Scholar] [CrossRef]
- Yu, D.; Zha, Y.; Shi, L.; Jin, X.; Hu, S.; Yang, Q.; Huang, K.; Zeng, W. Improvement of Sugarcane Yield Estimation by Assimilating UAV-Derived Plant Height Observations. Eur. J. Agron. 2020, 121, 126159. [Google Scholar] [CrossRef]
- Yuan, N.; Gong, Y.; Fang, S.; Liu, Y.; Duan, B.; Yang, K.; Wu, X.; Zhu, R. UAV Remote Sensing Estimation of Rice Yield Based on Adaptive Spectral Endmembers and Bilinear Mixing Model. Remote Sens. 2021, 13, 2190. [Google Scholar] [CrossRef]
- Ashapure, A.; Jung, J.; Chang, A.; Oh, S.; Yeom, J.; Maeda, M.; Maeda, A.; Dube, N.; Landivar, J.; Hague, S.; et al. Developing a Machine Learning Based Cotton Yield Estimation Framework Using Multi-Temporal UAS Data. ISPRS J. Photogramm. Remote Sens. 2020, 169, 180–194. [Google Scholar] [CrossRef]
- Bhanumathi, S.; Vineeth, M.; Rohit, N. Crop Yield Prediction and Efficient Use of Fertilizers. In Proceedings of the 2019 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 4–6 April 2019; pp. 769–773. [Google Scholar]
- Ines, A.V.M.; Das, N.N.; Hansen, J.W.; Njoku, E.G. Assimilation of Remotely Sensed Soil Moisture and Vegetation with a Crop Simulation Model for Maize Yield Prediction. Remote Sens. Environ. 2013, 138, 149–164. [Google Scholar] [CrossRef]
- Lin, C.-J. Projected Gradient Methods for Nonnegative Matrix Factorization. Neural Comput. 2007, 19, 2756–2779. [Google Scholar] [CrossRef]
- Xiao, C.; Cai, Z.; Wang, Y. Incorporating Good Nodes Set Principle into Evolution Strategy for Constrained Optimization. In Proceedings of the Third International Conference on Natural Computation (ICNC 2007), Haikou, China, 24–27 August 2007; pp. 243–247. [Google Scholar]
- Zhang, X.; Pei, D.; Chen, S. Root Growth and Soil Water Utilization of Winter Wheat in the North China Plain. Hydrol. Process. 2004, 18, 2275–2287. [Google Scholar] [CrossRef]
- Stewart, B.A.; Porter, L.K.; Beard, W.E. Determination of Total Nitrogen and Carbon in Soils by a Commercial Dumas Apparatus. Soil Sci. Soc. Am. J. 1964, 28, 366–368. [Google Scholar] [CrossRef]
- Guo, Y.; Yin, G.; Sun, H.; Wang, H.; Chen, S.; Senthilnath, J.; Wang, J.; Fu, Y. Scaling Effects on Chlorophyll Content Estimations with RGB Camera Mounted on a UAV Platform Using Machine-Learning Methods. Sensors 2020, 20, 5130. [Google Scholar] [CrossRef]
- Wellburn, A.R. The Spectral Determination of Chlorophylls a and b, as Well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of Biomass in Wheat Using Random Forest Regression Algorithm and Remote Sensing Data. Crop J. 2016, 4, 212–219. [Google Scholar] [CrossRef]
- Carfagna, E.; Gallego, F.J. Using Remote Sensing for Agricultural Statistics. Int. Stat. Rev. 2006, 73, 389–404. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The Red Edge Position and Shape as Indicators of Plant Chlorophyll Content, Biomass and Hydric Status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Jakubauskas, M.E.; Legates, D.R.; Kastens, J.H. Crop Identification Using Harmonic Analysis of Time-Series AVHRR NDVI Data. Comput. Electron. Agric. 2002, 37, 127–139. [Google Scholar] [CrossRef]
- Huang, J.; Wang, H.; Dai, Q.; Han, D. Analysis of NDVI Data for Crop Identification and Yield Estimation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 4374–4384. [Google Scholar] [CrossRef]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A Commentary Review on the Use of Normalized Difference Vegetation Index (NDVI) in the Era of Popular Remote Sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Yan, J.; Wang, H.; Yan, M.; Diao, W.; Sun, X.; Li, H. IoU-Adaptive Deformable R-CNN: Make Full Use of IoU for Multi-Class Object Detection in Remote Sensing Imagery. Remote Sens. 2019, 11, 286. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the Parts of Objects by Non-Negative Matrix Factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Bertsekas, D.P. Nonlinear Programming. J. Oper. Res. Soc. 1997, 48, 334. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Y.; Zhang, H.; Li, Z.; Li, M.; Wu, J.; Liu, K. Winter Wheat and Soil Total Nitrogen Integrated Monitoring Based on Canopy Hyperspectral Feature Selection and Fusion. Comput. Electron. Agric. 2022, 201, 107285. [Google Scholar] [CrossRef]
- Liu, Y.; Qian, J.; Yue, H. Comprehensive Evaluation of Sentinel-2 Red Edge and Shortwave-Infrared Bands to Estimate Soil Moisture. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 7448–7465. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, Y.; Xu, K.; Pan, Y. Effects of Drought on Vegetation Productivity of Farmland Ecosystems in the Drylands of Northern China. Remote Sens. 2021, 13, 1179. [Google Scholar] [CrossRef]
- Saptoro, A.; Tadé, M.O.; Vuthaluru, H. A Modified Kennard-Stone Algorithm for Optimal Division of Data for Developing Artificial Neural Network Models. Chem. Prod. Process Model. 2012, 7. [Google Scholar] [CrossRef]
- Pal, M. Random Forest Classifier for Remote Sensing Classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Worland, A.J. The Influence of Flowering Time Genes on Environmental Adaptability in European Wheats. Euphytica 1996, 89, 49–57. [Google Scholar] [CrossRef]
- Shaik, K.B.; Ganesan, P.; Kalist, V.; Sathish, B.S.; Jenitha, J.M.M. Comparative Study of Skin Color Detection and Segmentation in HSV and YCbCr Color Space. Procedia Comput. Sci. 2015, 57, 41–48. [Google Scholar] [CrossRef]
- Guo, Y.; Liao, J.; Shen, G. A Deep Learning Model with Capsules Embedded for High-Resolution Image Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 214–223. [Google Scholar] [CrossRef]
- Bakht, J.; Shafi, M.; Jan, M.T.; Shah, Z. Influence of Crop Residue Management, Cropping System and N Fertilizer on Soil N and C Dynamics and Sustainable Wheat (Triticum Aestivum L.) Production. Soil Tillage Res. 2009, 104, 233–240. [Google Scholar] [CrossRef]
- Rüdiger, W.; Thümmler, F. Phytochrome, the Visual Pigment of Plants. Angew. Chem. Int. Ed. Engl. 1991, 30, 1216–1228. [Google Scholar] [CrossRef]
- Thelemann, R.; Johnson, G.; Sheaffer, C.; Banerjee, S.; Cai, H.; Wyse, D. The Effect of Landscape Position on Biomass Crop Yield. Agron. J. 2010, 102, 513–522. [Google Scholar] [CrossRef]
- Fei, S.; Hassan, M.A.; Xiao, Y.; Su, X.; Chen, Z.; Cheng, Q.; Duan, F.; Chen, R.; Ma, Y. UAV-Based Multi-Sensor Data Fusion and Machine Learning Algorithm for Yield Prediction in Wheat. Precis. Agric. 2022, 1–26. [Google Scholar] [CrossRef]
- Feng, H.; Tao, H.; Fan, Y.; Liu, Y.; Li, Z.; Yang, G.; Zhao, C. Comparison of Winter Wheat Yield Estimation Based on Near-Surface Hyperspectral and UAV Hyperspectral Remote Sensing Data. Remote Sens. 2022, 14, 4158. [Google Scholar] [CrossRef]
- Fu, Z.; Jiang, J.; Gao, Y.; Krienke, B.; Wang, M.; Zhong, K.; Cao, Q.; Tian, Y.; Zhu, Y.; Cao, W.; et al. Wheat Growth Monitoring and Yield Estimation Based on Multi-Rotor Unmanned Aerial Vehicle. Remote Sens. 2020, 12, 508. [Google Scholar] [CrossRef]
- Du, M.; Noguchi, N. Monitoring of Wheat Growth Status and Mapping of Wheat Yield’s within-Field Spatial Variations Using Color Images Acquired from UAV-Camera System. Remote Sens. 2017, 9, 289. [Google Scholar] [CrossRef]
- Yang, S.; Hu, L.; Wu, H.; Ren, H.; Qiao, H.; Li, P.; Fan, W. Integration of Crop Growth Model and Random Forest for Winter Wheat Yield Estimation from UAV Hyperspectral Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 6253–6269. [Google Scholar] [CrossRef]

| Nutrients | Materials | Instruments | Methods |
|---|---|---|---|
| Crop Nitrogen Crop Carbon | 20 healthy crops in each 1 m2 study area. The leaves, stalks and leaf sheaths were dried, chopped and mixed. | Dumas Automatic Tester (Primacs SN-100) | Dumas high-temperature combustion method. |
| Soil Nitrogen Soil Carbon | At 30 cm below the each collected crop, 1 kg soil was collected, dried and sieved. | Dumas Automatic Tester (Primacs SN-100) | Dumas high-temperature combustion method. |
| Chlorophyll A Chlorophyll B Carotenoid | Fresh leaves of the collected crop were cut and then ground into a homogenate by adding acetone. | Spectrophotometer (UV-1700) | Measure the absorbance of the homogenate at 470 nm, 633 nm and 645 nm; calculate the content according to the Lambert–Beer law. |
| Crop Biomass | 20 healthy crops in each 1 m2 study area. The leaves, stalks and leaf sheaths were dried, chopped and mixed. | Scale | Total weight of the dried crop divided by the sampling area. |
| Method | Growth Stages | Segmentation Accuracy of Crop | Segmentation Accuracy of Soil |
|---|---|---|---|
| Crop and Soil Piecewise Segmentation Method | Jointing Stage | 82.57% | 84.65% |
| Flag Leaf Stage | 88.32% | 89.03% | |
| Flowering Stage | 94.22% | 92.54% | |
| Pustulation Stage | 88.28% | 83.27% | |
| HSV | Jointing Stage | 79.23% | 76.47% |
| Flag Leaf Stage | 74.43% | 79.28% | |
| Flowering Stage | 72.25% | 65.99% | |
| Pustulation Stage | 70.07% | 68.55% | |
| Deep Learning | Jointing Stage | 81.45% | 83.72% |
| Flag Leaf Stage | 77.62% | 80.15% | |
| Flowering Stage | 83.33% | 84.56% | |
| Pustulation Stage | 79.59% | 82.26% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Z.; Zhang, Y.; Liu, K.; Li, Z.; Li, M.; Zhang, H.; Wu, J. UAV Remote Sensing Prediction Method of Winter Wheat Yield Based on the Fused Features of Crop and Soil. Remote Sens. 2022, 14, 5054. https://doi.org/10.3390/rs14195054
Tian Z, Zhang Y, Liu K, Li Z, Li M, Zhang H, Wu J. UAV Remote Sensing Prediction Method of Winter Wheat Yield Based on the Fused Features of Crop and Soil. Remote Sensing. 2022; 14(19):5054. https://doi.org/10.3390/rs14195054
Chicago/Turabian StyleTian, Zezhong, Yao Zhang, Kaidi Liu, Zhenhai Li, Minzan Li, Haiyang Zhang, and Jiangmei Wu. 2022. "UAV Remote Sensing Prediction Method of Winter Wheat Yield Based on the Fused Features of Crop and Soil" Remote Sensing 14, no. 19: 5054. https://doi.org/10.3390/rs14195054
APA StyleTian, Z., Zhang, Y., Liu, K., Li, Z., Li, M., Zhang, H., & Wu, J. (2022). UAV Remote Sensing Prediction Method of Winter Wheat Yield Based on the Fused Features of Crop and Soil. Remote Sensing, 14(19), 5054. https://doi.org/10.3390/rs14195054