Desert Soil Salinity Inversion Models Based on Field In Situ Spectroscopy in Southern Xinjiang, China
Abstract
1. Introduction
2. Material and Methods
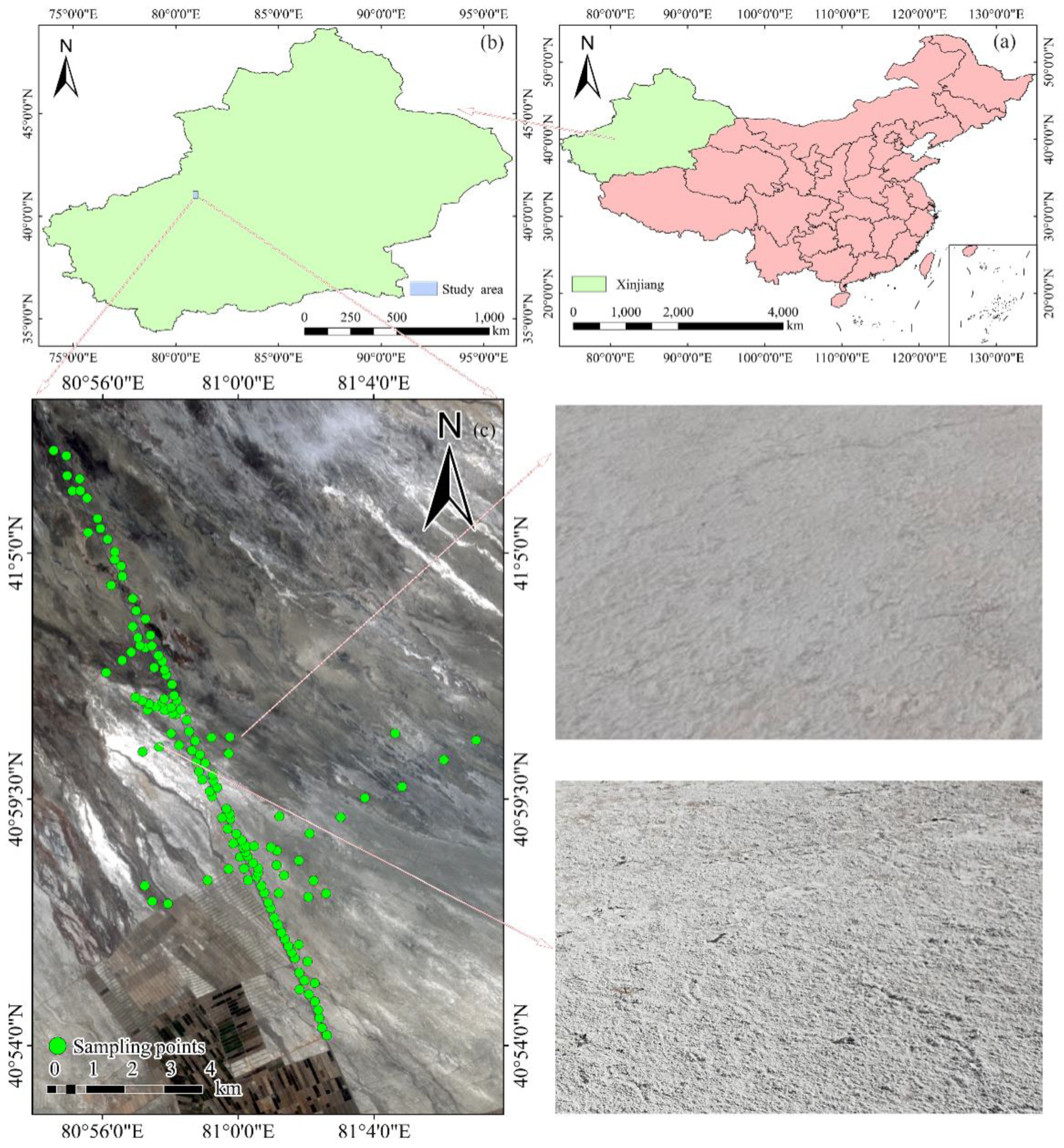
2.1. Study Area
2.2. Soil Sample Collection and Analysis
2.3. In Situ Spectral Data Acquisition and Pre-Processing
2.4. Feature Bands Selection
2.5. Model Establishment and Accuracy Evaluation
3. Results
3.1. Descriptive Statistics of SS
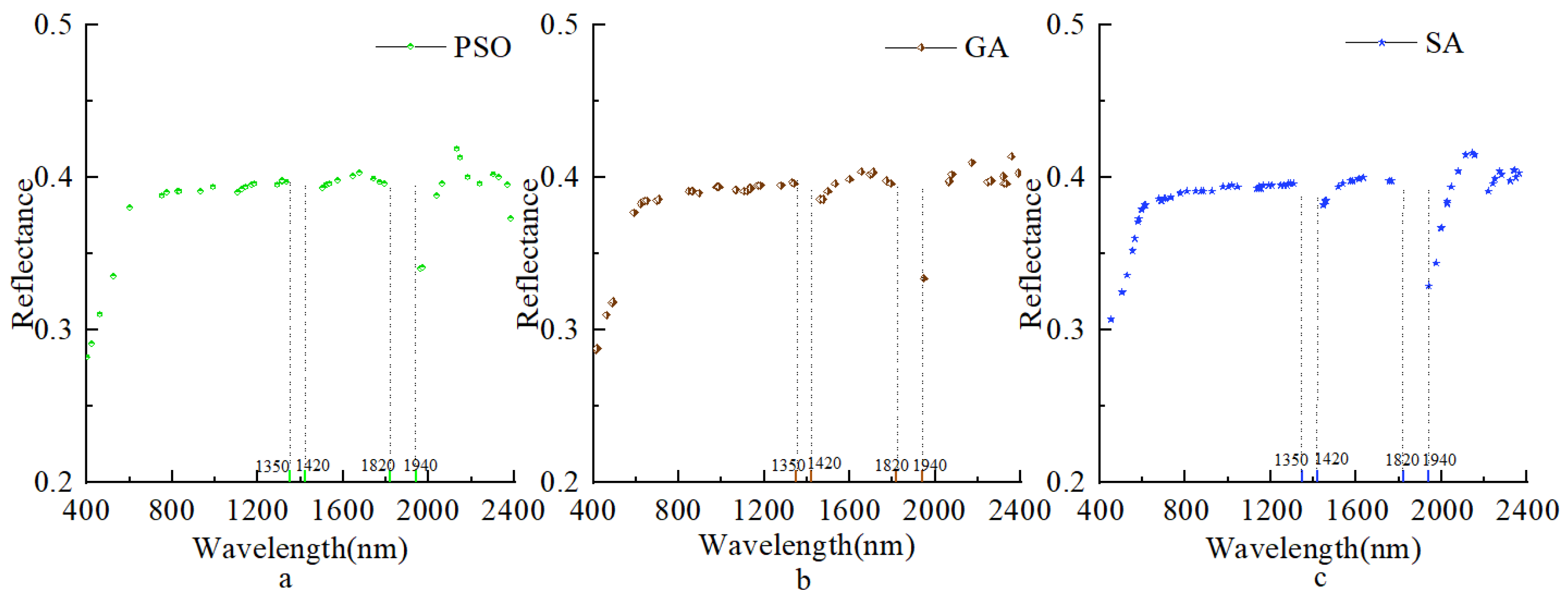
3.2. Feature Band Selection Based on the Different Methods
3.3. Predictive Regression Models
4. Discussion
4.1. Source of Uncertainty of Predicting SS Using Field In Situ Spectroscopy
4.2. Performance Comparison of Different Feature Band Selection Methods
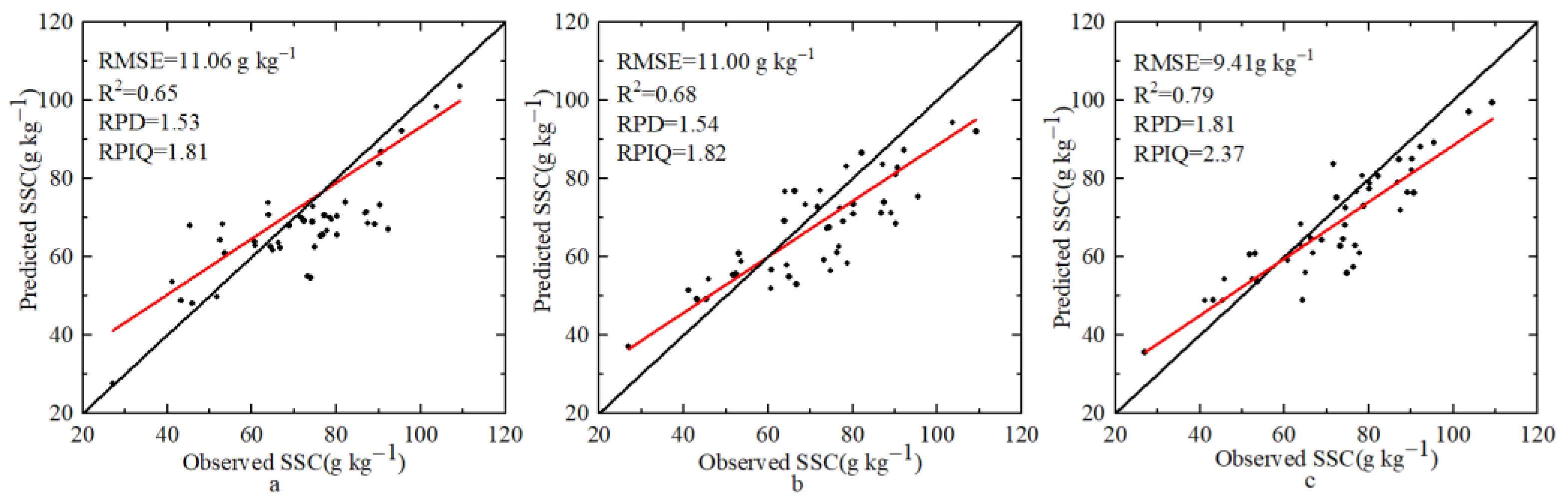
4.3. Comparison of ELM, BPNN, and CNN Estimation Models
4.4. Interpretability of the Selected Bands and Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Q.; Li, P.; Chen, X. Modeling salinity effects on soil reflectance under various moisture conditions and its inverse application: A laboratory experiment. Geoderma 2012, 170, 103–111. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.-G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef]
- Metternicht, G.I.; Zinck, J.A. Remote sensing of soil salinity: Potentials and constraints. Remote Sens. Environ. 2003, 85, 1–20. [Google Scholar] [CrossRef]
- Liu, L.-P.; Long, X.-H.; Shao, H.-B.; Liu, Z.-P.; Ya, T.; Zhou, Q.-Z.; Zong, J.-Q. Ameliorants improve saline-alkaline soils on a large scale in northern Jiangsu Province, China. Ecol. Eng. 2015, 81, 328–334. [Google Scholar]
- Peng, J.; Ji, W.; Ma, Z.; Li, S.; Chen, S.; Zhou, L.; Shi, Z. Predicting total dissolved salts and soluble ion concentrations in agricultural soils using portable visible near-infrared and mid-infrared spectrometers. Biosyst. Eng. 2016, 152, 94–103. [Google Scholar] [CrossRef]
- Hu, B.; Chen, S.; Hu, J.; Xia, F.; Xu, J.; Li, Y.; Shi, Z. Application of portable XRF and VNIR sensors for rapid assessment of soil heavy metal pollution. PLoS ONE 2017, 12, e0172438. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.R.; Das, B.; Gaikwad, B.; Murgaonkar, D.; Desai, A.; Morajkar, S.; Patel, K.P.; Kulkarni, R.M. Monitoring properties of the salt-affected soils by multivariate analysis of the visible and near-infrared hyperspectral data. Catena 2021, 198, 105041. [Google Scholar] [CrossRef]
- Bai, Z.; Xie, M.; Hu, B.; Luo, D.; Wan, C.; Peng, J.; Shi, Z. Estimation of Soil Organic Carbon Using Vis-NIR Spectral Data and Spectral Feature Bands Selection in Southern Xinjiang, China. Sensors 2022, 22, 6124. [Google Scholar] [CrossRef] [PubMed]
- Biney, J.K.M.; Blöcher, J.R.; Bell, S.M.; Borůvka, M.; Vašát, R. Can in situ spectral measurements under disturbance-reduced environmental conditions help improve soil organic carbon estimation? Sci. Total Environ. 2022, 838, 156304. [Google Scholar] [CrossRef] [PubMed]
- Taghdis, S.; Farpoor, M.H.; Mahmoodabadi, M. Pedological assessments along an arid and semi-arid transect using soil spectral behavior analysis. Catena 2022, 214, 106288. [Google Scholar] [CrossRef]
- Farifteh, J.; Van der Meer, F.; Atzberger, C.; Carranza, E.J.M. Quantitative analysis of salt-affected soil reflectance spectra: A comparison of two adaptive methods (PLSR and ANN). Remote Sens. Environ. 2007, 110, 59–78. [Google Scholar] [CrossRef]
- Mashimbye, Z.E.; Cho, M.A.; Nell, J.P.; De Clercq, W.P.; Van Niekerk, A.; Turner, D.P. Model-Based Integrated Methods for Quantitative Estimation of Soil Salinity from Hyperspectral Remote Sensing Data: A Case Study of Selected South African Soils. Pedosphere 2012, 22, 640–649. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, B. Prediction of soil salinity with soil-reflected spectra: A comparison of two regression methods. Sci. Rep. 2019, 9, 5067. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, V.; Ardejani, F.D.; Yousefi, S.; Aryafar, A. Monitoring soil lead and zinc contents via combination of spectroscopy with extreme learning machine and other data mining methods. Geoderma 2018, 318, 29–41. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Chen, J.; Chen, H.; Jin, J.; Han, J.; Wang, X.; Song, Z.; Wei, G. Estimating soil salinity with different fractional vegetation cover using remote sensing. Land Degrad. Dev. 2021, 32, 597–612. [Google Scholar] [CrossRef]
- Ding, J.; Yu, D. Monitoring and evaluating spatial variability of soil salinity in dry and wet seasons in the Werigan-Kuqa Oasis, China, using remote sensing and electromagnetic induction instruments. Geoderma 2014, 235–236, 316–322. [Google Scholar] [CrossRef]
- Aldabaa, A.A.A.; Weindorf, D.C.; Chakraborty, S.; Sharma, A.; Li, B. Combination of proximal and remote sensing methods for rapid soil salinity quantification. Geoderma 2015, 239–240, 34–46. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Ji, R.; Wang, M.; Zheng, L. A deep learning-based method for screening soil total nitrogen characteristic wavelengths. Comput. Electron. Agric. 2021, 187, 106228. [Google Scholar] [CrossRef]
- Yang, J.; Wang, X.; Wang, R.; Wang, H. Combination of Convolutional Neural Networks and Recurrent Neural Networks for predicting soil properties using Vis-NIR spectroscopy. Geoderma 2020, 380, 114616. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Using deep learning to predict soil properties from regional spectral data. Geoderma Reg. 2019, 16, e00198. [Google Scholar] [CrossRef]
- Wang, J.; Peng, J.; Li, H.; Yin, C.; Liu, W.; Wang, T.; Zhang, H. Soil Salinity Mapping Using Machine Learning Algorithms with the Sentinel-2 MSI in Arid Areas, China. Remote Sens. 2021, 13, 305. [Google Scholar] [CrossRef]
- Peng, J.; Biswas, A.; Jiang, Q.; Zhao, R.; Hu, J.; Hu, B.; Shi, Z. Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 2019, 337, 1309–1319. [Google Scholar] [CrossRef]
- Wang, N.; Peng, J.; Xue, J.; Zhang, X.; Huang, J.; Biswas, A.; He, Y.; Shi, Z. A framework for determining the total salt content of soil profiles using time-series Sentinel-2 images and a random forest-temporal convolution network. Geoderma 2022, 409, 115656. [Google Scholar] [CrossRef]
- Stevens, A.; van Wesemael, B.; Bartholomeus, H.; Rosillon, D.; Tychon, B.; Ben-Dor, E. Laboratory, field and airborne spectroscopy for monitoring organic carbon content in agricultural soils. Geoderma 2008, 144, 395–404. [Google Scholar] [CrossRef]
- Jian, H.; Lin, Q.; Wu, J.; Fan, X.; Wang, X. Design of the color classification system for sunglass lenses using PCA-PSO-ELM. Measurement 2022, 189, 110498. [Google Scholar] [CrossRef]
- Ong, P.; Tung, I.-C.; Chiu, C.-F.; Tsai, I.-L.; Shih, H.-C.; Chen, S.; Chuang, Y.-K. Determination of aflatoxin B1 level in rice (Oryza sativa L.) through near-infrared spectroscopy and an improved simulated annealing variable selection method. Food Control 2022, 136, 108886. [Google Scholar] [CrossRef]
- Sajadi, S.; Fathi, A. Genetic algorithm based local and global spectral features extraction for ear recognition. Expert Syst. Appl. 2020, 159, 113639. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, S.; Zhang, Y.; Chen, Y.; Yu, L.; Liu, Y.; Liu, Y.; Cheng, H.; Liu, Y. Rapid identification of soil organic matter level via visible and near-infrared spectroscopy: Effects of two-dimensional correlation coefficient and extreme learning machine. Sci. Total Environ. 2018, 644, 1232–1243. [Google Scholar] [CrossRef]
- Ma, Q.; Teng, Y.; Li, C.; Jiang, L. Simultaneous quantitative determination of low-concentration ternary pesticide mixtures in wheat flour based on terahertz spectroscopy and BPNN. Food. Chem. 2022, 377, 132030. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Ding, J.; Kung, H.-T.; Latif, A.; Johnson, V.C. Estimation of soil salt content (SSC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR), Northwest China, based on a Bootstrap-BP neural network model and optimal spectral indices. Sci. Total Environ. 2018, 615, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wang, X.; Dou, H.; Li, H.; Lv, R.; Wu, Y.; Song, G.; Wang, W.; Zhai, R. Non-Uniform Synthetic Aperture Radiometer Image Reconstruction Based on Deep Convolutional Neural Network. Remote Sens. 2022, 14, 2359. [Google Scholar] [CrossRef]
- Chen, Y.; Li, L.; Whiting, M.; Chen, F.; Sun, Z.; Song, K.; Wang, Q. Convolutional neural network model for soil moisture prediction and its transferability analysis based on laboratory Vis-NIR spectral data. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102550. [Google Scholar] [CrossRef]
- Cui, C.; Fearn, T. Modern practical convolutional neural networks for multivariate regression: Applications to NIR calibration. Chemom. Intell. Lab. Syst. 2018, 182, 9–20. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, Y.; Chen, S.; Shen, R.; Hu, B.; Peng, J.; Wang, N.; Guo, L.; Zhuo, Z.; Yang, Y.; et al. Data mining of urban soil spectral library for estimating organic carbon. Geoderma 2022, 426, 116102. [Google Scholar] [CrossRef]
- Hu, B.; Xue, J.; Zhou, Y.; Shao, S.; Fu, Z.; Li, Y.; Chen, S.; Qi, L.; Shi, Z. Modelling bioaccumulation of heavy metals in soil-crop ecosystems and identifying its controlling factors using machine learning. Environ. Pollut. 2020, 262, 114308. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Žižala, D.; Saberioon, M.; Borůvka, L. Soil organic carbon and texture retrieving and mapping using proximal, airborne and Sentinel-2 spectral imaging. Remote Sens. Environ. 2018, 218, 89–103. [Google Scholar] [CrossRef]
- Lao, C.; Chen, J.; Zhang, Z.; Chen, Y.; Ma, Y.; Chen, H.; Gu, X.; Ning, J.; Jin, J.; Li, X. Predicting the contents of soil salt and major water-soluble ions with fractional-order derivative spectral indices and variable selection. Comput. Electron. Agric. 2021, 182, 106031. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Wang, S.; Liu, H.; Fu, G.; Xiong, Y. Spatial distribution of soil salinity and potential implications for soil management in the Manas River watershed. China. Soil Use Manag. 2020, 36, 93–103. [Google Scholar] [CrossRef]
- Li, M.; Feng, Y.; Yu, Y.; Zhang, T.; Yan, C.; Tang, H.; Sheng, Q.; Li, H. Quantitative analysis of polycyclic aromatic hydrocarbons in soil by infrared spectroscopy combined with hybrid variable selection strategy and partial least squares. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 257, 119771. [Google Scholar] [CrossRef]
- Zhu, C.M.; Ding, J.L.; Zhang, Z.P.; Wang, Z. Exploring the potential of UAV hyperspectral image for estimating SS: Effects of optimal band combination algorithm and random forest. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 279, 121416. [Google Scholar] [CrossRef] [PubMed]
- Sidike, A.; Zhao, S.; Wen, Y. Estimating soil salinity in Pingluo County of China using QuickBird data and soil reflectance spectra. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 156–175. [Google Scholar] [CrossRef]
- Wei, Q.; Nurmemet, I.; Gao, M.; Xie, B. Inversion of Soil Salinity Using Multisource Remote Sensing Data and Particle Swarm Machine Learning Models in Keriya Oasis, Northwestern China. Remote Sens. 2022, 14, 512. [Google Scholar] [CrossRef]
- Yu, W.; Hong, Y.; Chen, S.; Chen, Y.; Zhou, L. Comparing Two Different Development Methods of External Parameter Orthogonalization for Estimating Organic Carbon from Field-Moist Intact Soils by Reflectance Spectroscopy. Remote Sens. 2022, 14, 1303. [Google Scholar] [CrossRef]
- Li, S.; Shi, Z.; Chen, S.; Ji, W.; Zhou, L.; Yu, W.; Webster, R. In situ measurements of organic carbon in soil profiles using vis-NIR spectroscopy on the Qinghai-Tibet plateau. Environ. Sci. Technol. 2015, 49, 4980–4987. [Google Scholar] [CrossRef]
- Farifteh, J.; Van der Meer, F.; Van der Meijde, M.; Atzberger, C. Spectral characteristics of salt-affected soils: A laboratory experiment. Geoderma 2008, 145, 196–206. [Google Scholar] [CrossRef]
- Piekarczyk, J.; Kaźmierowski, C.; Królewicz, S.; Cierniewski, J. Effects of soil surface roughness on soil reflectance measured in laboratory and outdoor conditions. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 9, 827–834. [Google Scholar] [CrossRef]
- Nocita, M.; Kooistra, L.; Bachmann, M.; Müller, A.; Powell, M.; Weel, S. Predictions of soil surface and topsoil organic carbon content through the use of laboratory and field spectroscopy in the Albany Thicket Biome of Eastern Cape Province of South Africa. Geoderma 2011, 167–168, 295–302. [Google Scholar] [CrossRef]
- Rossel, R.A.V.; Cattle, S.R.; Ortega, A.; Fouad, Y. In situ measurements of soil colour, mineral composition and clay content by vis-NIR spectroscopy. Geoderma 2009, 150, 253–266. [Google Scholar] [CrossRef]
- Vohland, M.; Ludwig, B.; Seidel, M.; Hutengs, C. Quantification of soil organic carbon at regional scale: Benefits of fusing vis-NIR and MIR diffuse reflectance data are greater for in situ than for laboratory-based modelling approaches. Geoderma 2022, 405, 115426. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, D.; Yang, L.; Ma, Y.; Cui, T.; He, X.; Du, Z. Developing a generalized vis-NIR prediction model of soil moisture content using external parameter orthogonalization to reduce the effect of soil type. Geoderma 2022, 419, 115877. [Google Scholar] [CrossRef]
- Jin, P.; Li, P.; Wang, Q.; Pu, Z. Developing and applying novel spectral feature parameters for classifying soil salt types in arid land. Ecol. Indic. 2015, 54, 116–123. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.; Yu, D.; Ma, X.; Zhang, Z.; Ge, X.; Teng, D.; Li, X.; Liang, J.; Lizaga, I.; et al. Capability of Sentinel-2 MSI data for monitoring and mapping of soil salinity in dry and wet seasons in the Ebinur Lake region, Xinjiang, China. Geoderma 2019, 353, 172–187. [Google Scholar] [CrossRef]
- Eappen, G.; Shankar, T. Hybrid PSO-GSA for energy efficient spectrum sensing in cognitive radio network. Phys. Commun. 2020, 40, 101091. [Google Scholar] [CrossRef]
- Wan, T.; Bai, Y.; Wang, T.; Wei, Z. BPNN-based optimal strategy for dynamic energy optimization with providing proper thermal comfort under the different outdoor air temperatures. Appl. Energy 2022, 313, 118899. [Google Scholar] [CrossRef]
- Tsakiridis, N.L.; Keramaris, K.D.; Theocharis, J.B.; Zalidis, G.C. Simultaneous prediction of soil properties from VNIR-SWIR spectra using a localized multi-channel 1-D convolutional neural network. Geoderma 2020, 367, 114208. [Google Scholar] [CrossRef]
- Xiao, D.; Vu, Q.H.; Le, B.T. Salt content in saline-alkali soil detection using visible-near infrared spectroscopy and a 2D deep learning. Microchem. J. 2021, 165, 106182. [Google Scholar] [CrossRef]
- Romero, D.J.; Ben-Dor, E.; Demattê, J.A.; e Souza, A.B.; Vicente, L.E.; Tavares, T.R.; Martello, M.; Strabeli, T.F.; da Silva Barros, P.P.; Fiorio, P.R.; et al. Internal soil standard method for the Brazilian soil spectral library: Performance and proximate analysis. Geoderma 2018, 312, 95–103. [Google Scholar] [CrossRef]
- Liu, Y.; Deng, C.; Lu, Y.; Shen, Q.; Zhao, H.; Tao, Y.; Pan, X. Evaluating the characteristics of soil vis-NIR spectra after the removal of moisture effect using external parameter orthogonalization. Geoderma 2020, 376, 114568. [Google Scholar] [CrossRef]


| Layer | Type | Kernel Size | Filters | Activation |
|---|---|---|---|---|
| 1 | Input | —— | —— | —— |
| 2 | Conv1 | 10 × 1 | 10 | ReLU |
| 3 | Maxpooling1 | 1 × 1 | —— | —— |
| 4 | Conv2 | 5 × 1 | 21 | ReLU |
| 5 | Maxpooling2 | 1 × 1 | —— | —— |
| 6 | Conv3 | 2 × 1 | 42 | ReLU |
| 7 | Maxpooling3 | 1 × 1 | —— | —— |
| 8 | Fully-connected1 | —— | —— | ReLU |
| 9 | Fully-connected2 | —— | —— | ReLU |
| 10 | Output | —— | —— | ReLU |
| Dataset | Number | Mean | Max | Min | SD | CV (%) |
|---|---|---|---|---|---|---|
| Calibration | 90 | 68.72 | 109.24 | 27.01 | 18.40 | 26.77 |
| Validation | 45 | 68.78 | 109.18 | 28.07 | 18.50 | 26.90 |
| Entire | 135 | 68.74 | 109.24 | 27.01 | 18.36 | 26.71 |
| FEA | Model | Calibration | Validation | ||||
|---|---|---|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | RPD | RPIQ | ||
| ELM | 0.50 | 13.21 | 0.46 | 13.57 | 1.25 | 1.40 | |
| R | BPNN | 0.59 | 11.96 | 0.51 | 12.86 | 1.32 | 1.46 |
| CNN | 0.63 | 11.40 | 0.57 | 11.62 | 1.46 | 1.49 | |
| ELM | 0.64 | 11.24 | 0.55 | 11.90 | 1.42 | 1.63 | |
| GA | BPNN | 0.70 | 10.44 | 0.60 | 11.26 | 1.51 | 1.79 |
| CNN | 0.76 | 9.85 | 0.68 | 11.00 | 1.54 | 1.82 | |
| ELM | 0.59 | 12.25 | 0.52 | 12.69 | 1.34 | 1.48 | |
| PSO | BPNN | 0.68 | 11.05 | 0.61 | 11.18 | 1.52 | 1.75 |
| CNN | 0.73 | 10.25 | 0.65 | 11.06 | 1.53 | 1.81 | |
| ELM | 0.65 | 11.13 | 0.57 | 11.61 | 1.46 | 1.66 | |
| SA | BPNN | 0.71 | 10.97 | 0.63 | 11.16 | 1.52 | 1.85 |
| CNN | 0.84 | 8.64 | 0.79 | 9.41 | 1.81 | 2.37 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Xie, M.; Hu, B.; Jiang, Q.; Shi, Z.; He, Y.; Peng, J. Desert Soil Salinity Inversion Models Based on Field In Situ Spectroscopy in Southern Xinjiang, China. Remote Sens. 2022, 14, 4962. https://doi.org/10.3390/rs14194962
Wang Y, Xie M, Hu B, Jiang Q, Shi Z, He Y, Peng J. Desert Soil Salinity Inversion Models Based on Field In Situ Spectroscopy in Southern Xinjiang, China. Remote Sensing. 2022; 14(19):4962. https://doi.org/10.3390/rs14194962
Chicago/Turabian StyleWang, Yu, Modong Xie, Bifeng Hu, Qingsong Jiang, Zhou Shi, Yinfeng He, and Jie Peng. 2022. "Desert Soil Salinity Inversion Models Based on Field In Situ Spectroscopy in Southern Xinjiang, China" Remote Sensing 14, no. 19: 4962. https://doi.org/10.3390/rs14194962
APA StyleWang, Y., Xie, M., Hu, B., Jiang, Q., Shi, Z., He, Y., & Peng, J. (2022). Desert Soil Salinity Inversion Models Based on Field In Situ Spectroscopy in Southern Xinjiang, China. Remote Sensing, 14(19), 4962. https://doi.org/10.3390/rs14194962







