Tree Detection and Species Classification in a Mixed Species Forest Using Unoccupied Aircraft System (UAS) RGB and Multispectral Imagery
Abstract
1. Introduction
- Establish whether individual tree detection based on deep learning can achieve better classification accuracies compared to canopy delineation through MRS.
- Identify whether oversegmentation through superpixels can be used to discriminate species to similar levels of accuracy as conventional image segmentation.
- Assess the importance of data layers and image features for species classification.
2. Methods
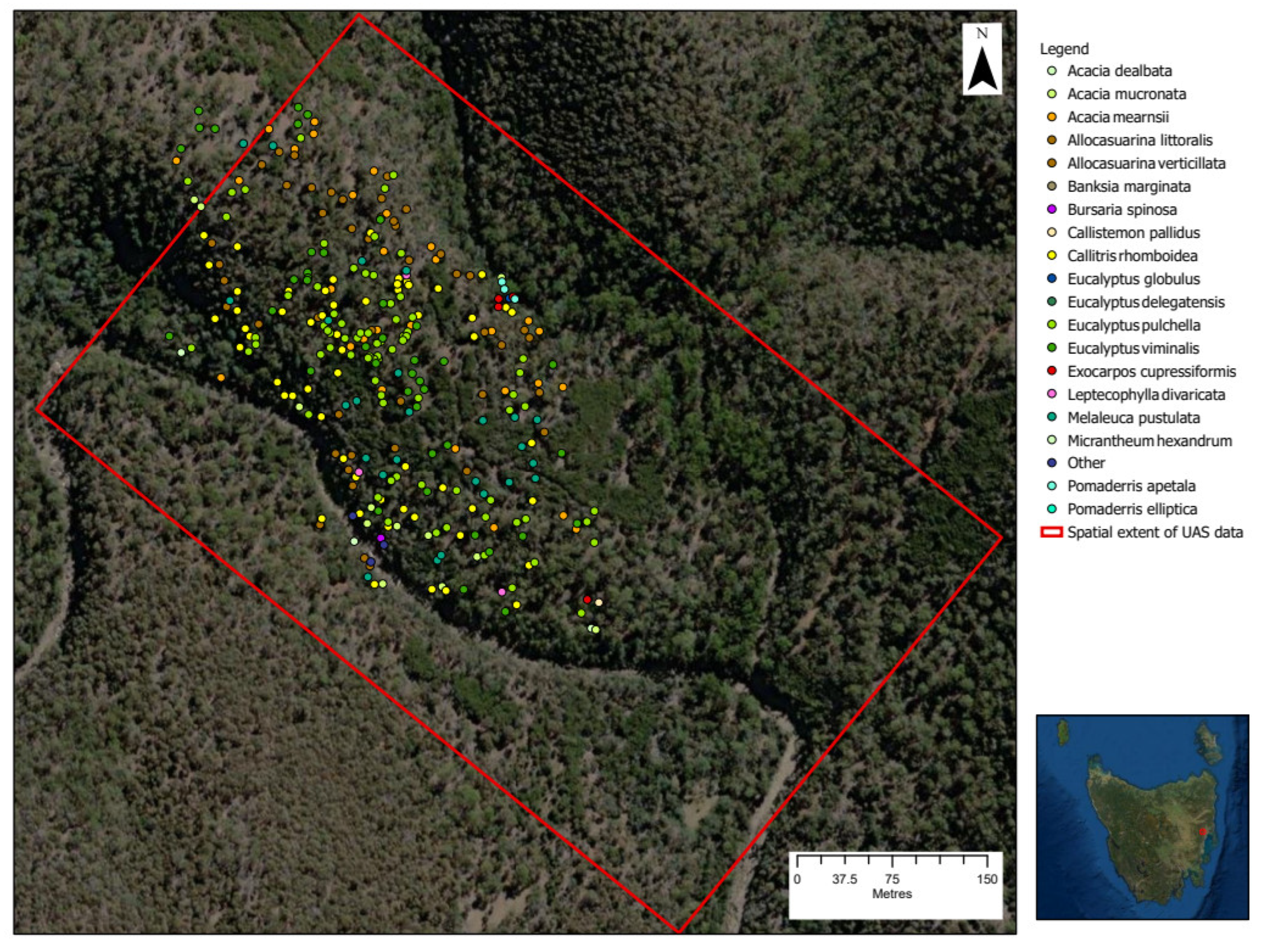
2.1. Study Site
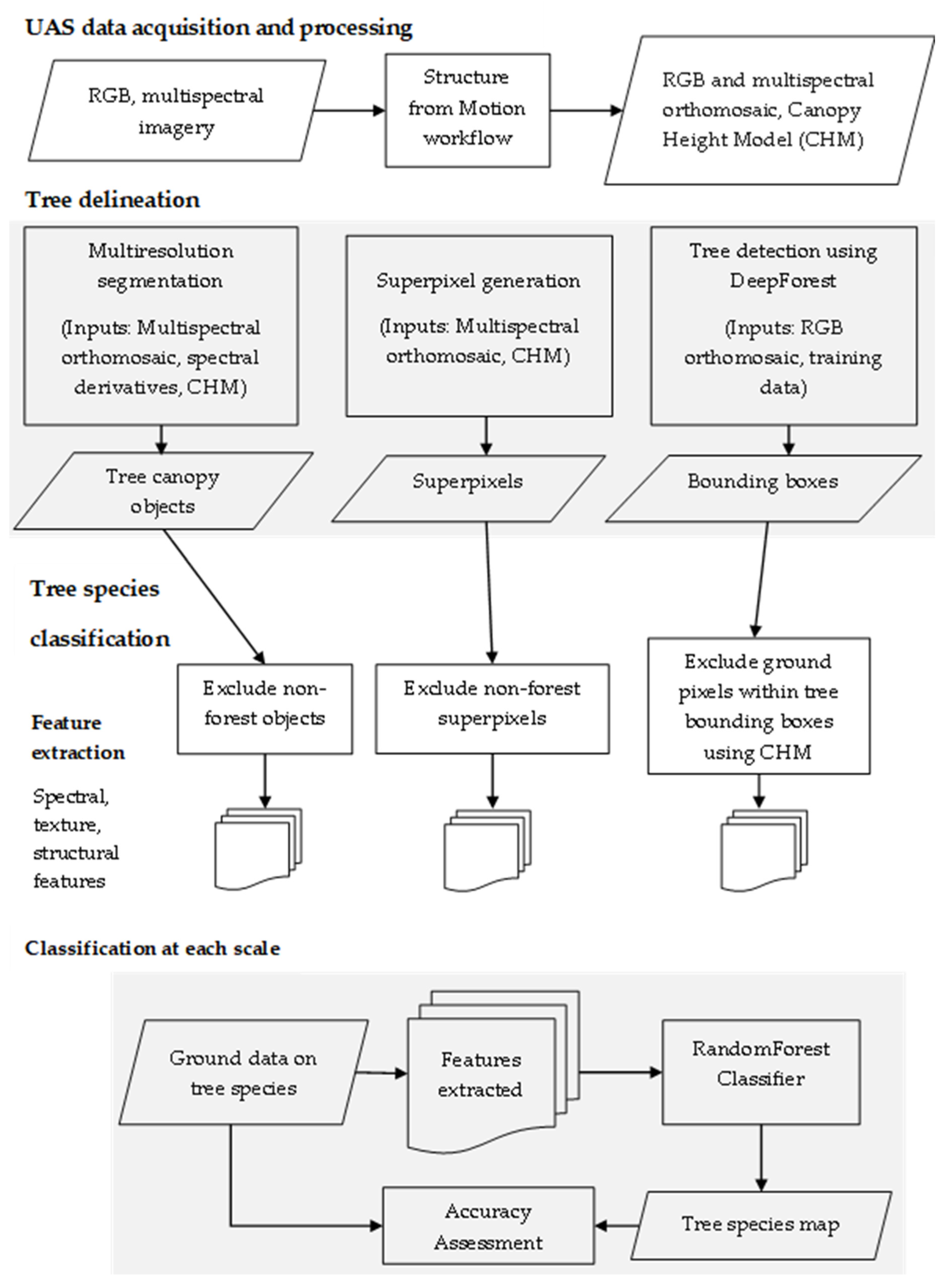
2.2. Data Acquisition and Processing
2.3. Tree Segmentation
2.3.1. Multiresolution Segmentation
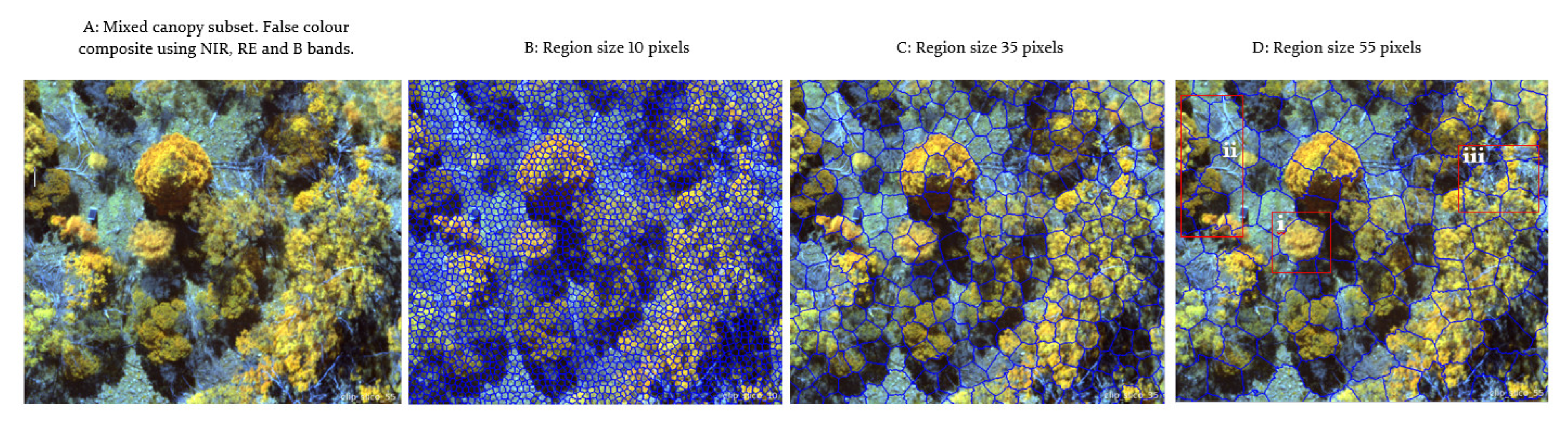
2.3.2. Superpixels
2.4. Tree detection Using DeepForest
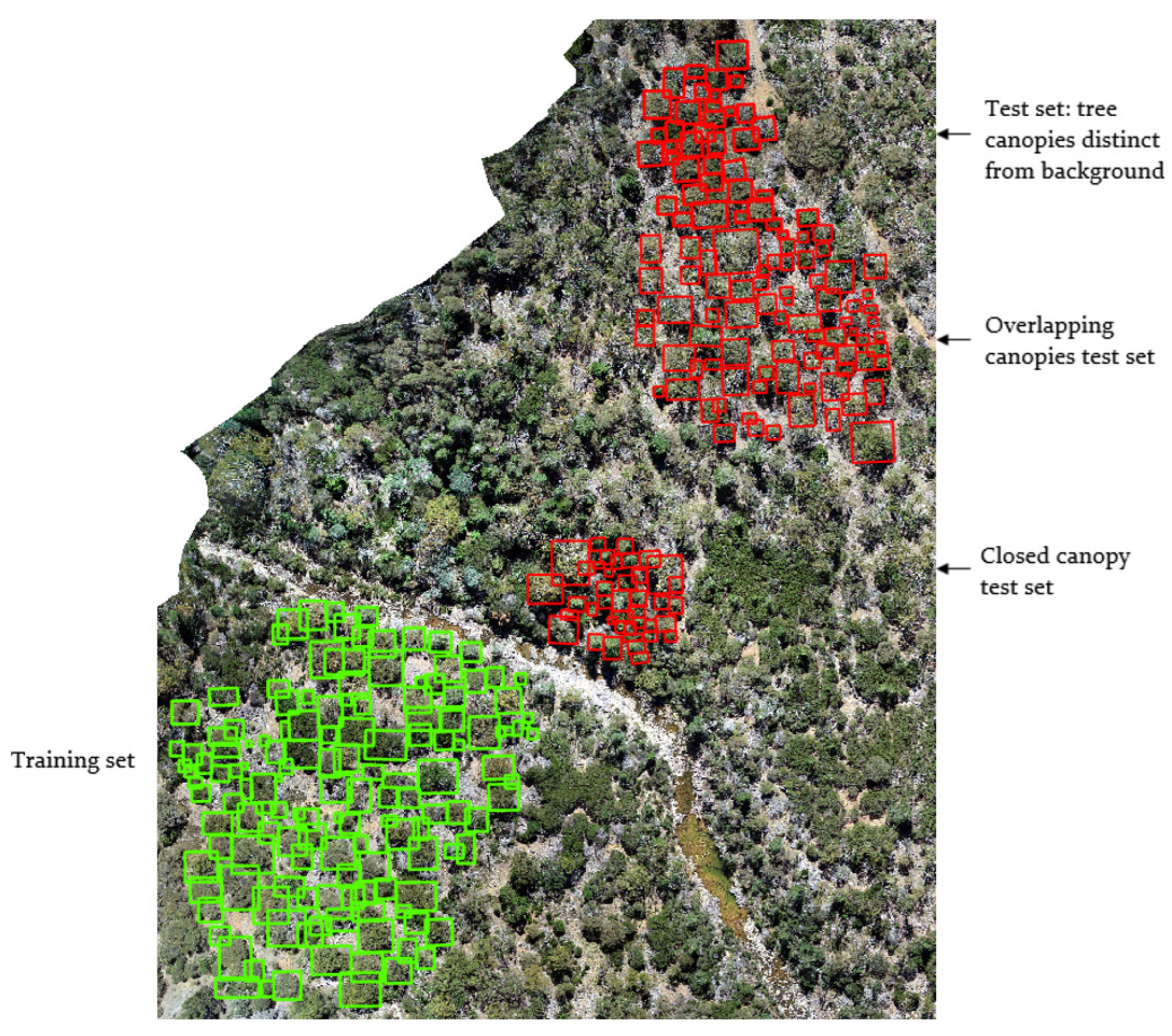
Validation
2.5. Tree Species Classification
2.5.1. Spectral Features
- 4.
- Spectra extracted from all pixels within the canopy.
- 5.
- Spectra extracted only from pixels with SR (which is NIR/R) above the mean value in the tree canopy. This was performed to address the mixing of bare ground and canopy gaps at all scales of analysis.
2.5.2. Texture Features
2.5.3. Structural Features
| Class | Feature | Formula | Study |
|---|---|---|---|
| Spectral (Mean and Standard Deviation) | All multispectral bands: B, G, R, RE, and NIR | ||
| Normalized ratios of scaled B, RE, and NIR | cx/(cNIR + cRE + cB) where cx is one of cNIR, cRE, cB relative reflectance stretched to RGB colour space (0–255) | [4] | |
| NDVI | (NIR − R)/(NIR + R) | ||
| Red Blue NDVI (RBNDVI) | (NIR − (R − B))/(NIR + (R − B)) | [43] | |
| Normalized Difference RedEdge Index (NDRE) | (NIR − RE)/(NIR + RE) | [29] | |
| Modified Canopy Chlorophyll Content Index (M3CI) | (NIR + R − RE)/ (NIR − R + RE) | ||
| Plant Senescence Reflectance Index (PSRI) | (R − G)/RE | ||
| Chlorophyll Index RedEdge (CIRE) | (NIR/RE) − 1 | ||
| Green NDVI | (G − NIR)/(G + NIR) | [20] | |
| Normalized Green Red Vegetation Index (NGRVI) | (G − R)/(G + R) | ||
| Normalized Green Blue Index (NGBI) | (G − B)/(G + B) | ||
| Normalized Red Blue Index (NRBI) | (R − B)/(R + B) | ||
| Simple Ratio | NIR/R | ||
| Green Vegetation Index | NIR/G | ||
| Object-level texture (Scales: superpixel, object) | GLCM co-occurrence measures on NIR band: mean, standard deviation, contrast, dissimilarity, homogeneity, angular second moment, entropy, and correlation. | Direction 135° to match the pixel-level texture calculated with direction shift X, Y: 1, 1. In the eCognition implementation of Haralick texture, 0° represents the vertical direction and 90° represents the horizontal direction. | [47,48] |
| Pixel-level texture (Scale: bounding box) | As above. ‘Mean’ of each measure extracted from bounding box. | Window size 5 pixels, direction shift: X,Y: 1,1. | [47] |
| CHM | Mean and standard deviation | CHM | |
| Structural metrics (Scales: object, bounding box) | Metrics derived from the photogrammetric point cloud: canopy cover, skewness, minimum, square of average height, average, maximum, standard deviation, kurtosis, and vertical complexity. | Default height cut-off of 1.3 m | |
| Shape (Scale: object) | Border index, roundness, compactness, shape index, rectangular fit, and asymmetry | Object-level features |
2.5.4. RandomForest Classification
| Scale of Analysis | Feature Combination | Classes |
|---|---|---|
| Superpixels | Spectral, CHM | Acacia mearnsii, Allocasuarina spp., Callitris rhomboidea, Eucalyptus pulchella, Eucalyptus viminalis, Melaleuca pustulata, ‘Other Vegetation’ |
| Spectral, CHM, object-level Texture | ||
| MRS | Spectral, CHM metrics | |
| Spectral, CHM, object-level Texture | ||
| Spectral, CHM, shape | ||
| Spectral, CHM, PPC metrics | ||
| Spectral, CHM, object-level Texture, shape, PPC metrics | ||
| DeepForest | Spectral, CHM metrics | |
| Spectral, CHM, pixel-level Texture | ||
| Spectral, CHM, PPC metrics | ||
| Spectral, CHM, pixel-level Texture, PPC metrics |
2.5.5. Feature Importance
2.5.6. Classification Accuracy Assessment
3. Results
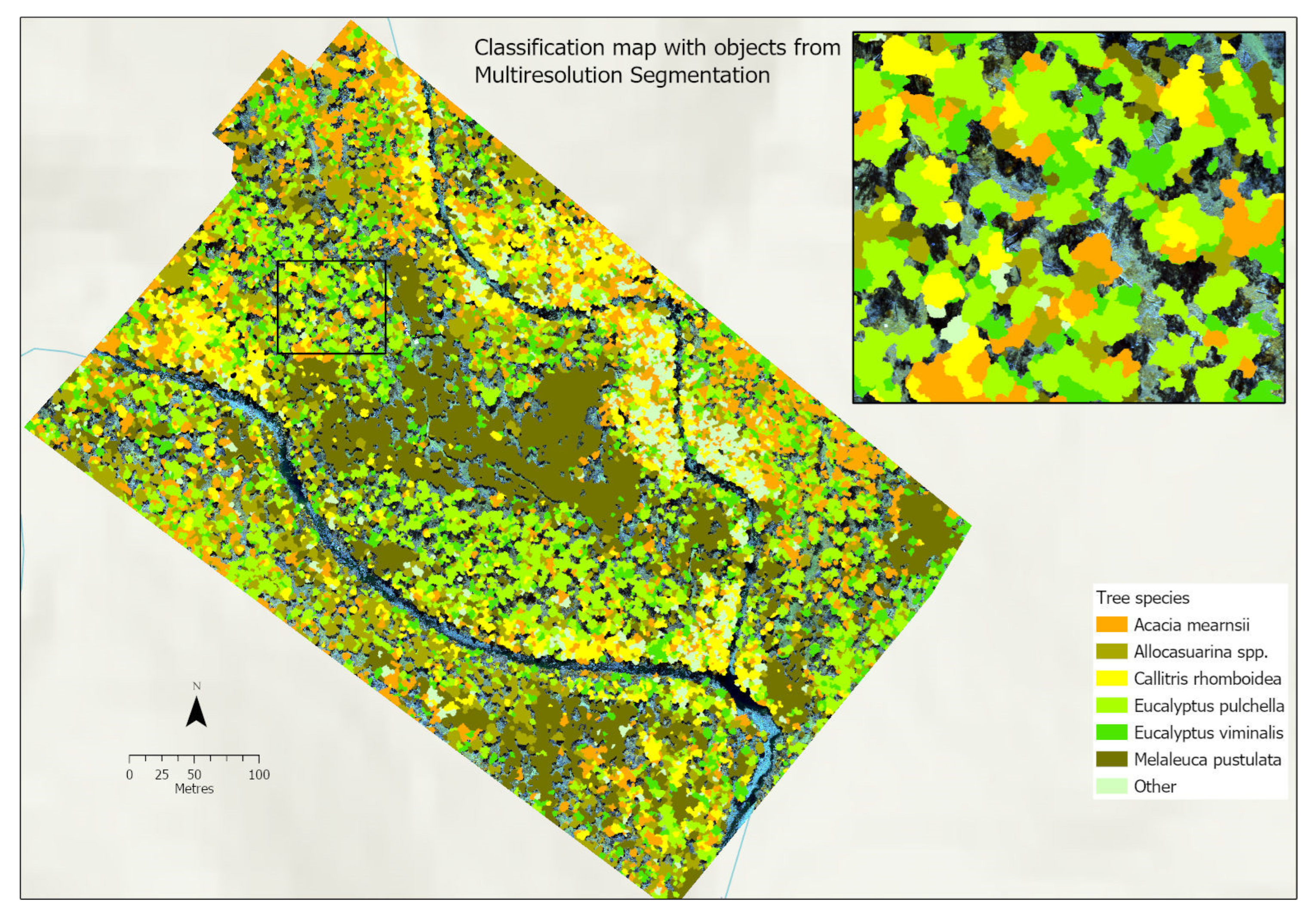
3.1. Multiresolution Segmentation
3.2. Superpixel Generation
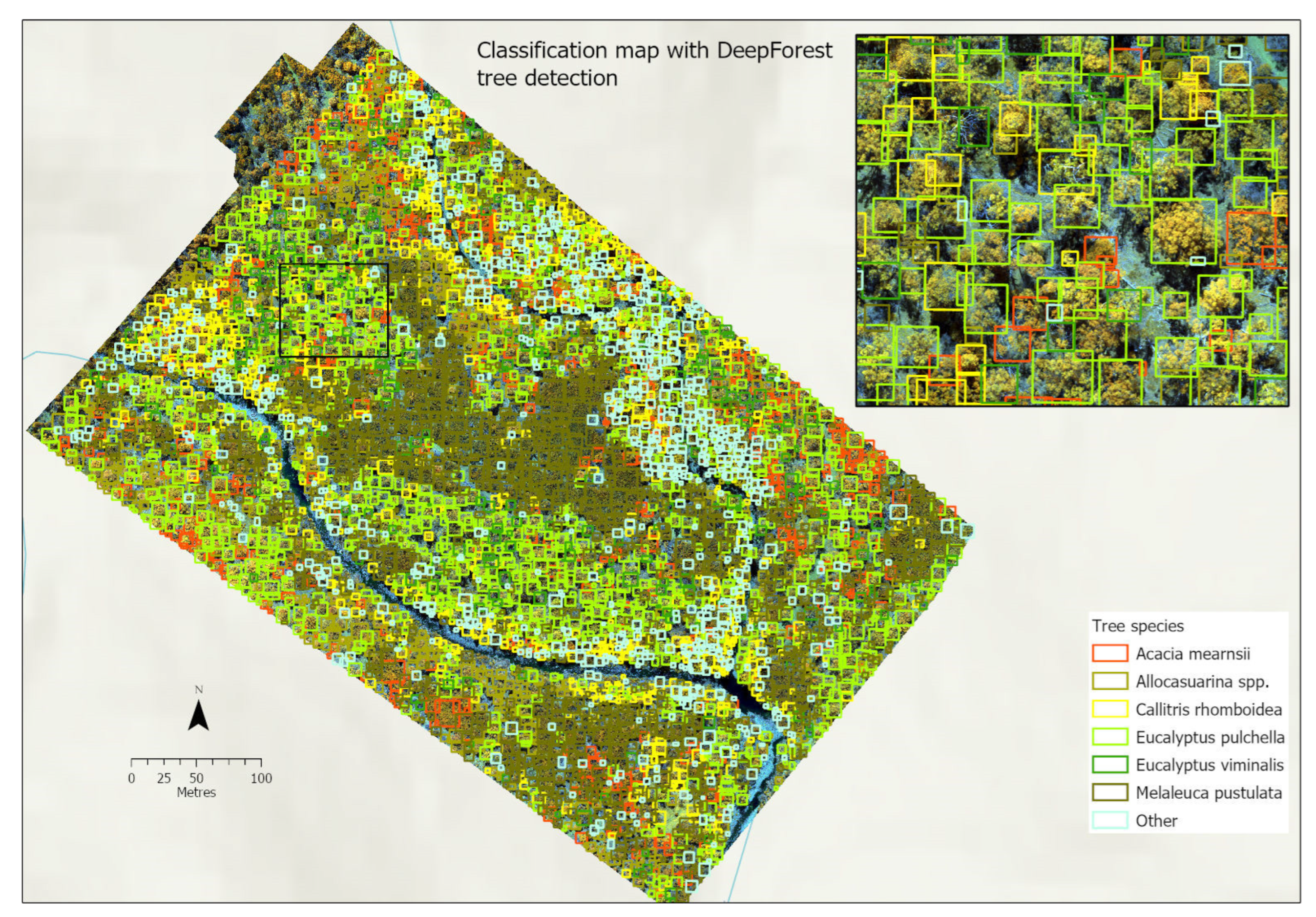
3.3. DeepForest
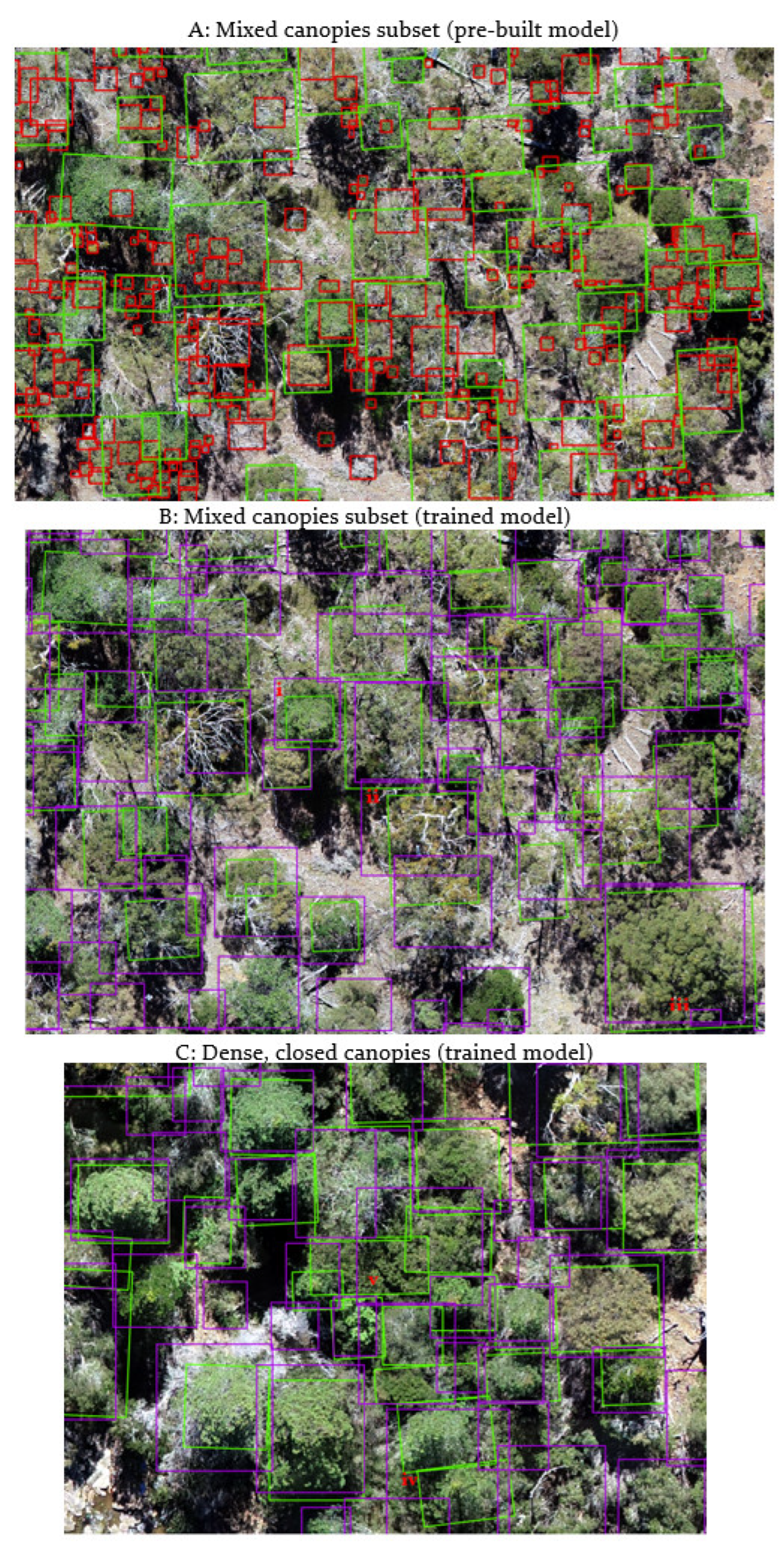
3.3.1. Pre-Built Model
3.3.2. Trained Model
3.3.3. Tree Detection in Mixed and Closed Canopies
3.4. RandomForest Classification
3.4.1. Classification Accuracy
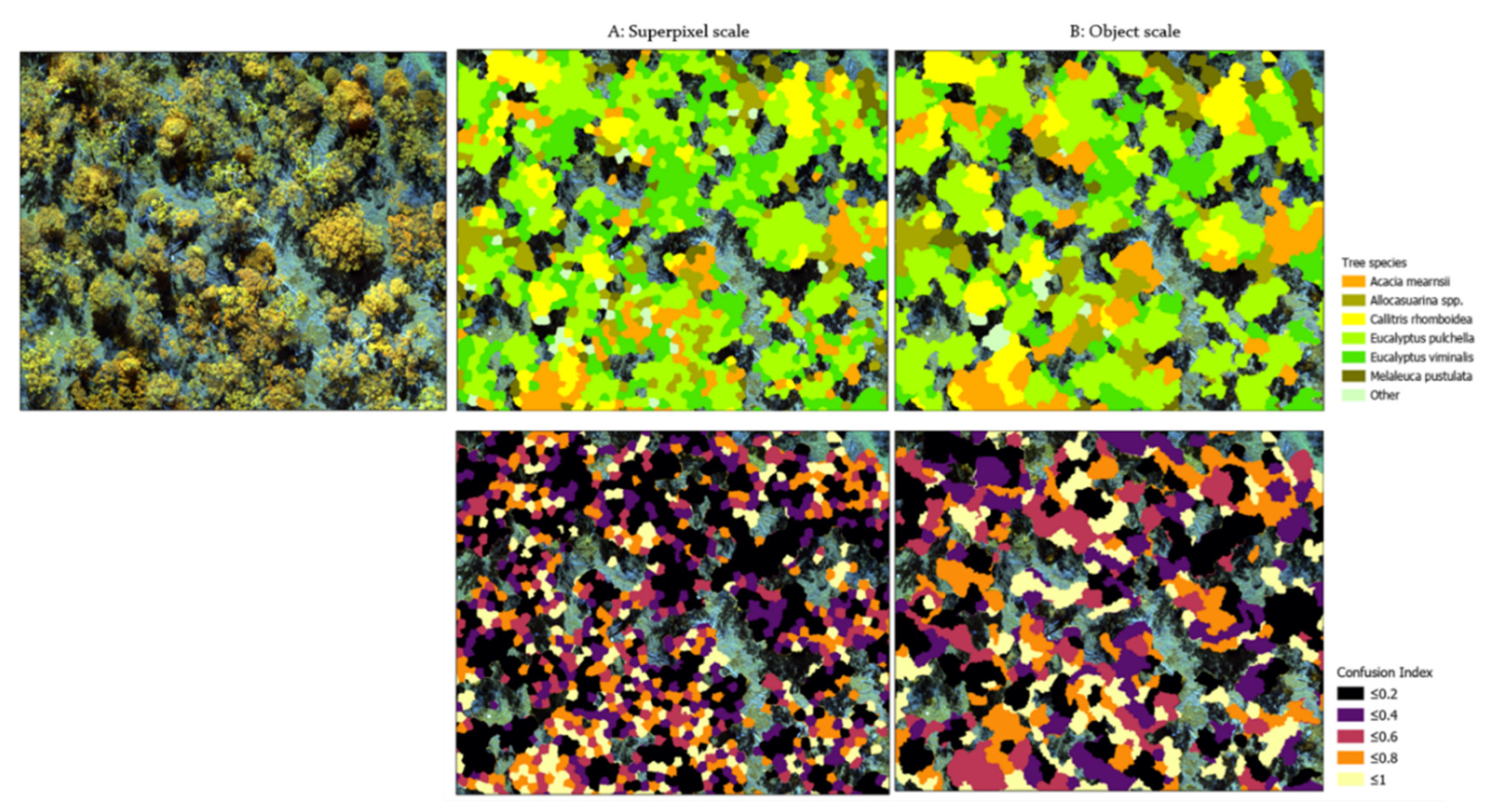
3.4.2. Class-Specific Accuracies
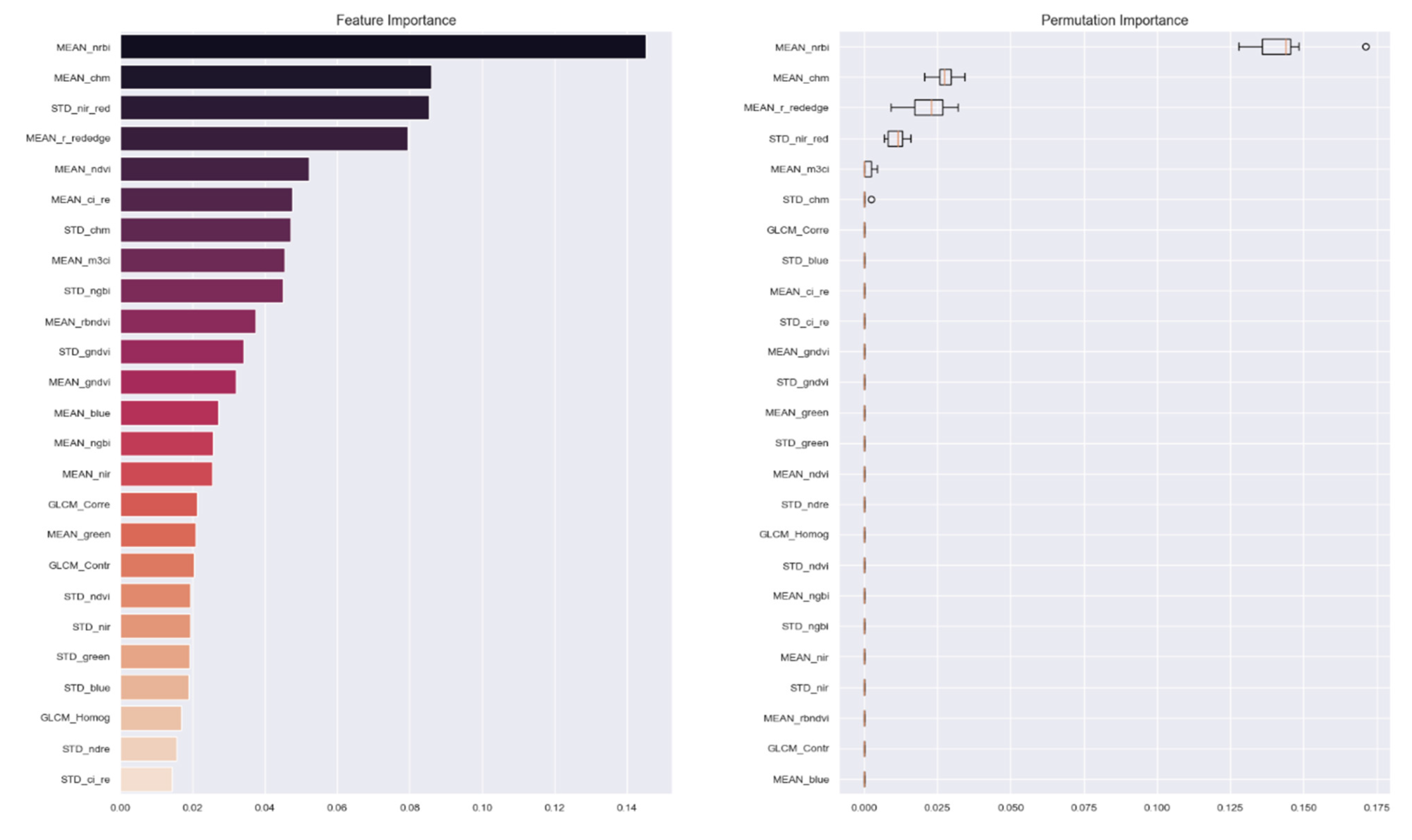
3.4.3. Important Features
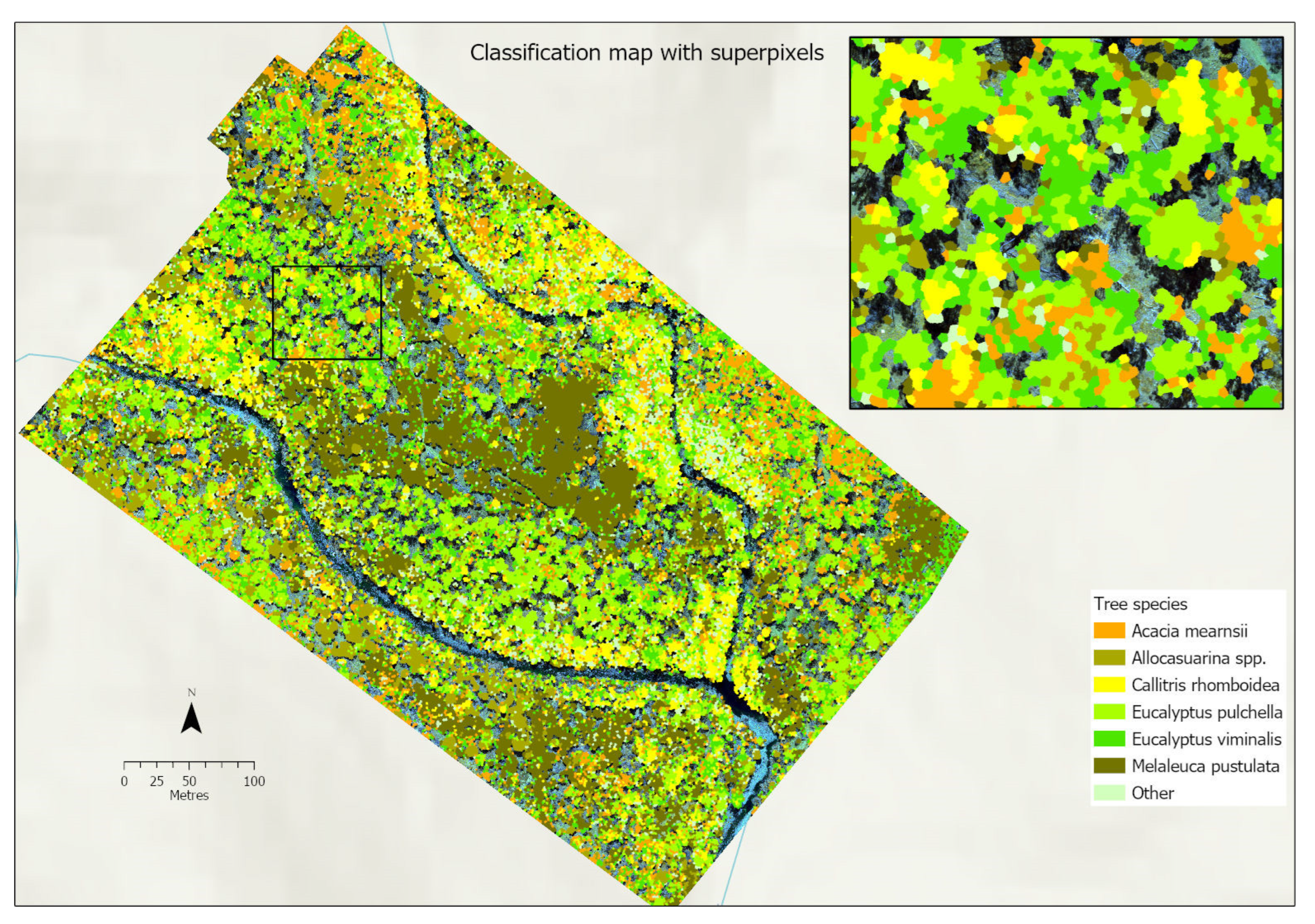
3.4.4. Classification Maps
4. Discussion
4.1. Tree Detection with DeepForest
4.2. Scale of Analysis and Classification Accuracy
4.3. Feature Selection and Importance
4.4. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matusick, G.; Ruthrof, K.X.; Fontaine, J.B.; Hardy, G.E.S.J. Eucalyptus forest shows low structural resistance and resilience to climate change-type drought. J. Veg. Sci. 2016, 27, 493–503. [Google Scholar] [CrossRef]
- Jiao, T.; Williams, C.A.; Rogan, J.; De Kauwe, M.G.; Medlyn, B.E. Drought Impacts on Australian Vegetation During the Millennium Drought Measured with Multisource Spaceborne Remote Sensing. J. Geophys. Res. Biogeosci. 2020, 125. [Google Scholar] [CrossRef]
- Brodribb, T.J.; Powers, J.; Cochard, H.; Choat, B. Hanging by a thread? Forests and drought. Science 2020, 368, 261–266. [Google Scholar] [CrossRef]
- Bunting, P.; Lucas, R. The delineation of tree crowns in Australian mixed species forests using hyperspectral Compact Airborne Spectrographic Imager (CASI) data. Remote Sens. Environ. 2006, 101, 230–248. [Google Scholar] [CrossRef]
- Williams, J.; Woinarski, J.C.Z. Eucalypt Ecology: Individuals to Ecosystems; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Coops, N.C.; Stone, C.; Culvenor, D.S.; Chisholm, L. Assessment of Crown Condition in Eucalypt Vegetation by Remotely Sensed Optical Indices. J. Environ. Qual. 2004, 33, 956–964. [Google Scholar] [CrossRef]
- Lucas, R.; Bunting, P.; Paterson, M.; Chisholm, L. Classification of Australian forest communities using aerial photography, CASI and HyMap data. Remote Sens. Environ. 2008, 112, 2088–2103. [Google Scholar] [CrossRef]
- Goodwin, N.; Turner, R.; Merton, R. Classifying Eucalyptus forests with high spatial and spectral resolution imagery: An investigation of individual species and vegetation communities. Aust. J. Bot. 2005, 53, 337–345. [Google Scholar] [CrossRef]
- Youngentob, K.N.; Roberts, D.A.; Held, A.A.; Dennison, P.E.; Jia, X.; Lindenmayer, D.B. Mapping two Eucalyptus subgenera using multiple endmember spectral mixture analysis and continuum-removed imaging spectrometry data. Remote Sens. Environ. 2011, 115, 1115–1128. [Google Scholar] [CrossRef]
- Shang, X.; Chisholm, L.A. Classification of Australian Native Forest Species Using Hyperspectral Remote Sensing and Machine-Learning Classification Algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2481–2489. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Gamon, J.A.; Townsend, P.A. Remote Sensing of Plant Biodiversity. [Electronic Resource]; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2001, 22, 2377–2400. [Google Scholar] [CrossRef]
- Shendryk, I.; Broich, M.; Tulbure, M.G.; McGrath, A.; Keith, D.; Alexandrov, S.V. Mapping individual tree health using full-waveform airborne laser scans and imaging spectroscopy: A case study for a floodplain eucalypt forest. Remote Sens. Environ. 2016, 187, 202–217. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Wulder, M.; Niemann, K.O.; Goodenough, D.G. Local Maximum Filtering for the Extraction of Tree Locations and Basal Area from High Spatial Resolution Imagery. Remote Sens. Environ. 2000, 73, 103–114. [Google Scholar] [CrossRef]
- Pouliot, D.A.; King, D.J.; Bell, F.W.; Pitt, D.G. Automated tree crown detection and delineation in high-resolution digital camera imagery of coniferous forest regeneration. Remote Sens. Environ. 2002, 82, 322–334. [Google Scholar] [CrossRef]
- Sankey, T.T.; McVay, J.; Swetnam, T.L.; McClaran, M.P.; Heilman, P.; Nichols, M. UAV hyperspectral and lidar data and their fusion for arid and semi-arid land vegetation monitoring. Remote Sens. Ecol. Conserv. 2018, 4, 20–33. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef]
- Marques, P.; Pádua, L.; Adão, T.; Hruška, J.; Peres, E.; Sousa, A.; Sousa, J.J. UAV-Based Automatic Detection and Monitoring of Chestnut Trees. Remote Sens. 2019, 11, 855. [Google Scholar] [CrossRef]
- Michez, A.; Piégay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Environ. Monit. Assess. 2016, 188, 1–19. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multispectral data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Gougeon, F. Comparison of Possible Multispectral Classification Schemes for Tree Crowns Individually Delineated on High Spatial Resolution MEIS Images. Can. J. Remote Sens. 1995, 21, 1–9. [Google Scholar] [CrossRef]
- Mishra, N.B.; Mainali, K.P.; Shrestha, B.B.; Radenz, J.; Karki, D. Species-Level Vegetation Mapping in a Himalayan Treeline Ecotone Using Unmanned Aerial System (UAS) Imagery. ISPRS Int. J. Geo-Inf. 2018, 7, 445. [Google Scholar] [CrossRef]
- Apostol, B.; Petrila, M.; Lorenţ, A.; Ciceu, A.; Gancz, V.; Badea, O. Species discrimination and individual tree detection for predicting main dendrometric characteristics in mixed temperate forests by use of airborne laser scanning and ultra-high-resolution imagery. Sci. Total Environ. 2020, 698, 134074. [Google Scholar] [CrossRef] [PubMed]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.; Junior, J.M.; Menezes, G.; Pistori, H.; SantaAna, D.; Goncalves, W. Image Segmentation and Classification with SLIC Superpixel and Convolutional Neural Network in Forest Context. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 6543–6546. [Google Scholar] [CrossRef]
- Adhikari, A.; Kumar, M.; Agrawal, S. An Integrated Object and Machine Learning Approach for Tree Canopy Extraction from UAV Datasets. J. Indian Soc. Remote Sens. 2020, 49, 471–478. [Google Scholar] [CrossRef]
- Csillik, O. Fast Segmentation and Classification of Very High Resolution Remote Sensing Data Using SLIC Superpixels. Remote Sens. 2017, 9, 243. [Google Scholar] [CrossRef]
- Abdollahnejad, A.; Panagiotidis, D. Tree Species Classification and Health Status Assessment for a Mixed Broadleaf-Conifer Forest with UAS Multispectral Imaging. Remote Sens. 2020, 12, 3722. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Miyoshi, G.T.; Imai, N.N.; Tommaselli, A.M.G.; de Moraes, M.V.A.; Honkavaara, E. Evaluation of Hyperspectral Multitemporal Information to Improve Tree Species Identification in the Highly Diverse Atlantic Forest. Remote Sens. 2020, 12, 244. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Miyoshi, G.T.; Arruda, M.d.S.; Osco, L.P.; Marcato Junior, J.; Gonçalves, D.N.; Imai, N.N.; Tommaselli, A.M.G.; Honkavaara, E.; Gonçalves, W.N. A Novel Deep Learning Method to Identify Single Tree Species in UAV-Based Hyperspectral Images. Remote Sens. 2020, 12, 1294. [Google Scholar] [CrossRef]
- Dos Santos, A.A.; Marcato Junior, J.; Araújo, M.S.; Di Martini, D.R.; Tetila, E.C.; Siqueira, H.L.; Aoki, C.; Eltner, A.; Matsubara, E.T.; Pistori, H.; et al. Assessment of CNN-Based Methods for Individual Tree Detection on Images Captured by RGB Cameras Attached to UAVs. Sensors 2019, 19, 3595. [Google Scholar] [CrossRef] [PubMed]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree Species Classification of Drone Hyperspectral and RGB Imagery with Deep Learning Convolutional Neural Networks. Remote. Sens. 2020, 12, 1070. [Google Scholar] [CrossRef]
- Zhang, C.; Xia, K.; Feng, H.; Yang, Y.; Du, X. Tree species classification using deep learning and RGB optical images obtained by an unmanned aerial vehicle. J. For. Res. 2020, 32, 1879–1888. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of Citrus Trees from Unmanned Aerial Vehicle Imagery Using Convolutional Neural Networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef]
- Ferreira, M.P.; de Almeida, D.R.A.; Papa, D.D.A.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Natesan, S.; Armenakis, C.; Vepakomma, U. Individual tree species identification using Dense Convolutional Network (DenseNet) on multitemporal RGB images from UAV. J. Unmanned Veh. Syst. 2020, 8, 310–333. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Rezaee, M.; Mohammadimanesh, F.; Zhang, Y. Very Deep Convolutional Neural Networks for Complex Land Cover Mapping Using Multispectral Remote Sensing Imagery. Remote Sens. 2018, 10, 1119. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Aubry-Kientz, M.; Vincent, G.; Senyondo, H.; White, E.P. DeepForest: A Python package for RGB deep learning tree crown delineation. Methods Ecol. Evol. 2020, 11, 1743–1751. [Google Scholar] [CrossRef]
- Skelton, R.P.; Brodribb, T.J.; McAdam, S.; Mitchell, P. Gas exchange recovery following natural drought is rapid unless limited by loss of leaf hydraulic conductance: Evidence from an evergreen woodland. New Phytol. 2017, 215, 1399–1412. [Google Scholar] [CrossRef]
- Bell, R.-A.; Callow, J.N. Investigating Banksia Coastal Woodland Decline Using Multi-Temporal Remote Sensing and Field-Based Monitoring Techniques. Remote Sens. 2020, 12, 669. [Google Scholar] [CrossRef]
- Ren, X.; Malik, J. Learning a classification model for segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Nice, France, 3–16 October 2003; pp. 10–17. [Google Scholar]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; White, E.P. Cross-site learning in deep learning RGB tree crown detection. Ecol. Inform. 2020, 56, 101061. [Google Scholar] [CrossRef]
- Anaconda; Anaconda Software Distribution; Anaconda Inc.: Austin, TX, USA, 2021; Available online: https://anaconda.com (accessed on 8 June 2021).
- Haralick, R.M. Statistical and structural approaches to texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Trimble eCognition 2021, Trimble eCognition Developer Reference Book, Trimble Inc. Available online: https://docs.ecognition.com/v10.0.2/Default.htm (accessed on 17 September 2022).
- LAStools, Version 200304, Academic; Efficient LiDAR Processing Software. 2021. Available online: http://rapidlasso.com/LAStools (accessed on 8 June 2021).
- Duff, G.A.; Reid, J.B.; Jackson, W.D. The occurrence of mixed stands of the Eucalyptus subgenera Monocalyptus and Symphyomyrtus in south-eastern Tasmania. Austral Ecol. 1983, 8, 405–414. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2021, 12, 2825–2830. [Google Scholar]
- McNicoll, G.; Burrough, P.A.; Frank, A.U. Geographic Objects with Indeterminate Boundaries; Taylor and Francis: Philadelphia, PA, USA.
- Camarretta, N.; A. Harrison, P.; Lucieer, A.; Potts, B.M.; Davidson, N.; Hunt, M. From Drones to Phenotype: Using UAV-LiDAR to Detect Species and Provenance Variation in Tree Productivity and Structure. Remote Sens. 2020, 12, 3184. [Google Scholar] [CrossRef]
- Sothe, C.; Dalponte, M.; de Almeida, C.M.; Schimalski, M.B.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; Tommaselli, A.M.G. Tree Species Classification in a Highly Diverse Subtropical Forest Integrating UAV-Based Photogrammetric Point Cloud and Hyperspectral Data. Remote Sens. 2019, 11, 1338. [Google Scholar] [CrossRef]
- Braga, J.R.G.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragão, L.E.O.C.; Velho, H.F.D.C.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef]
- Chadwick, A.; Goodbody, T.; Coops, N.; Hervieux, A.; Bater, C.; Martens, L.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-Off Conditions Using UAV Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Hao, Z.; Lin, L.; Post, C.J.; Mikhailova, E.A.; Li, M.; Chen, Y.; Yu, K.; Liu, J. Automated tree-crown and height detection in a young forest plantation using mask region-based convolutional neural network (Mask R-CNN). ISPRS J. Photogramm. Remote Sens. 2021, 178, 112–123. [Google Scholar] [CrossRef]
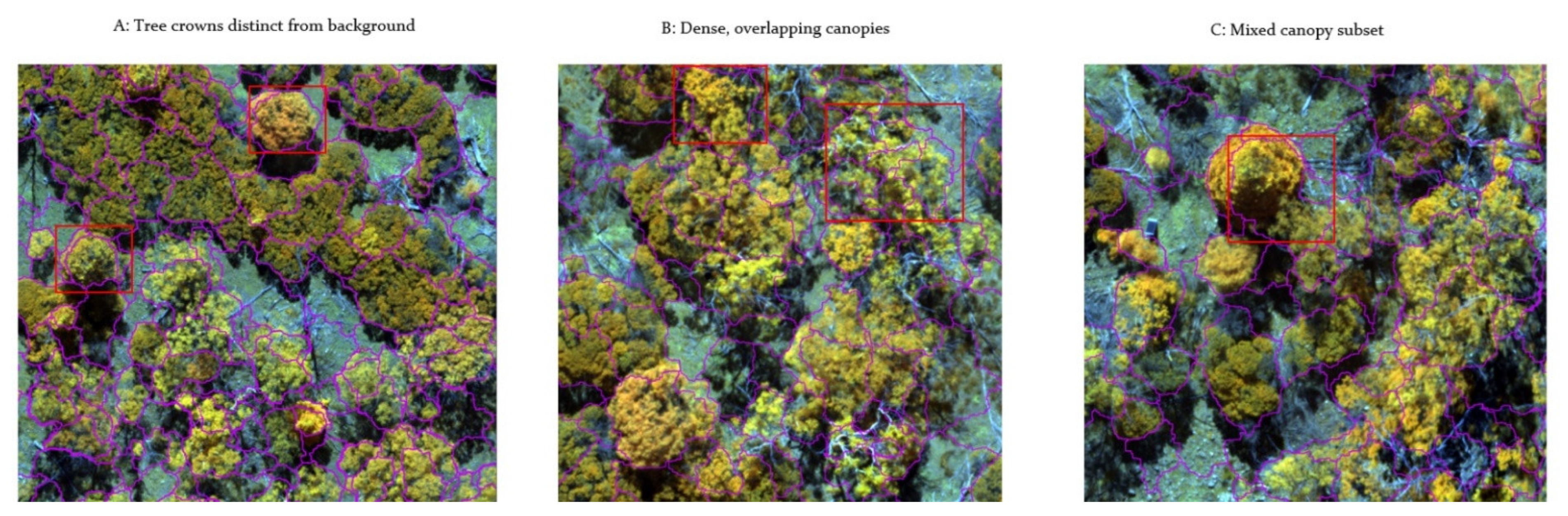
| DeepForest Model | Patch Size (Pixels) | Training Accuracy mAP | Test Accuracy mAP | ||
|---|---|---|---|---|---|
| Closed Canopy and Trees Distinct from Background | Dense, Closed Canopy Subset | Overlapping Canopy Subset | |||
| Pre-built | 400 | N/A | 0.04 | 0.03 | 0.02 |
| Trained | 1000 | 0.82 | 0.65 | 0.51 | 0.5 |
| Scale of Analysis | Input Features | OOB Training Accuracy | Test Accuracy | ||
|---|---|---|---|---|---|
| Spectra from All Pixels within Tree Canopy | Spectra from Pixels with SR Greater than Average | Spectra from All Pixels within Tree Canopy | Spectra from Pixels with SR Greater than Average | ||
| Superpixel | Spectral, CHM | 0.77 | 0.81 | 0.79 | 0.83 |
| Object | 0.70 | 0.73 | 0.71 | 0.74 | |
| Bounding box | 0.63 | 0.71 | 0.63 | 0.65 | |
| Scale of Analysis | Input Features | Classes | OOB Training Accuracy | Test Accuracy |
|---|---|---|---|---|
| Superpixel | Spectral, CHM | All | 0.81 | 0.83 |
| Spectral, CHM, Texture | All | 0.8 | 0.84 | |
| Spectral, CHM, Texture | Euc spp. grouped | 0.88 | 0.93 | |
| Object | Spectral, CHM | All | 0.73 | 0.74 |
| Spectral, CHM, Texture | All | 0.74 | 0.77 | |
| Spectral, CHM, PPC | All | 0.71 | 0.71 | |
| Spectral, CHM, Shape | All | 0.71 | 0.75 | |
| Spectral, CHM, Texture, PPC, Shape | All | 0.71 | 0.75 | |
| Spectral, CHM, Texture | Euc spp. grouped | 0.8 | 0.77 | |
| Bounding box | Spectral, CHM | All | 0.71 | 0.65 |
| Spectral, CHM, Texture | All | 0.69 | 0.67 | |
| Spectral, CHM, PPC | All | 0.68 | 0.65 | |
| Spectral, CHM, Texture, PPC | All | 0.69 | 0.69 | |
| Spectral, CHM, Texture, PPC | Euc spp. grouped | 0.76 | 0.77 |
| Class | Superpixel | Object | Bounding Box | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F-Score | Precision | Recall | F-Score | Precision | Recall | F-Score | |
| Acacia mearnsii | 1 | 0.83 | 0.91 | 0.5 | 0.5 | 0.5 | 0.6 | 0.6 | 0.6 |
| Allocasuarina spp. | 0.81 | 1 | 0.9 | 0.67 | 0.67 | 0.67 | 0.86 | 0.6 | 0.71 |
| Callitris rhomboidea | 0.86 | 0.96 | 0.91 | 0.89 | 0.76 | 0.82 | 0.83 | 0.83 | 0.83 |
| Eucalyptus pulchella | 0.79 | 0.73 | 0.76 | 0.77 | 0.94 | 0.85 | 0.58 | 0.85 | 0.69 |
| Eucalyptus viminalis | 0.6 | 0.55 | 0.57 | 0.79 | 0.79 | 0.79 | 0.5 | 0.25 | 0.33 |
| F-Score | 0.83 | 0.77 | 0.69 | ||||||
| Class | Superpixel | Object | Bounding Box | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F-Score | Precision | Recall | F-Score | Precision | Recall | F-Score | |
| Acacia mearnsii | 1 | 0.83 | 0.91 | 0.43 | 0.38 | 0.4 | 0.75 | 0.6 | 0.67 |
| Allocasuarina spp. | 0.92 | 1 | 0.96 | 1 | 0.5 | 0.67 | 0.88 | 0.7 | 0.78 |
| Callitris rhomboidea | 0.89 | 0.96 | 0.92 | 0.89 | 0.76 | 0.82 | 0.85 | 0.92 | 0.88 |
| Eucalyptus spp. | 0.93 | 0.97 | 0.95 | 0.73 | 0.94 | 0.82 | 0.75 | 0.83 | 0.79 |
| F-Score | 0.93 | 0.77 | 0.76 | ||||||
| Scale of Analysis | Important Features (Based on Permutating Features) | OOB Training Accuracy | Test Accuracy |
|---|---|---|---|
| Superpixel | NRBI, SR, r_rededge, CHM, NDVI | 0.88 | 0.93 |
| Object | NRBI, RBNDVI, CHM, NGBI, r_rededge, CIRE | 0.8 | 0.77 |
| Bounding box | NRBI, CHM, M3CI, r_rededge, PSRI | 0.76 | 0.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivanandam, P.; Lucieer, A. Tree Detection and Species Classification in a Mixed Species Forest Using Unoccupied Aircraft System (UAS) RGB and Multispectral Imagery. Remote Sens. 2022, 14, 4963. https://doi.org/10.3390/rs14194963
Sivanandam P, Lucieer A. Tree Detection and Species Classification in a Mixed Species Forest Using Unoccupied Aircraft System (UAS) RGB and Multispectral Imagery. Remote Sensing. 2022; 14(19):4963. https://doi.org/10.3390/rs14194963
Chicago/Turabian StyleSivanandam, Poornima, and Arko Lucieer. 2022. "Tree Detection and Species Classification in a Mixed Species Forest Using Unoccupied Aircraft System (UAS) RGB and Multispectral Imagery" Remote Sensing 14, no. 19: 4963. https://doi.org/10.3390/rs14194963
APA StyleSivanandam, P., & Lucieer, A. (2022). Tree Detection and Species Classification in a Mixed Species Forest Using Unoccupied Aircraft System (UAS) RGB and Multispectral Imagery. Remote Sensing, 14(19), 4963. https://doi.org/10.3390/rs14194963






