Detection of Standing Dead Trees after Pine Wilt Disease Outbreak with Airborne Remote Sensing Imagery by Multi-Scale Spatial Attention Deep Learning and Gaussian Kernel Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Areas and Datasets
2.2. Methods
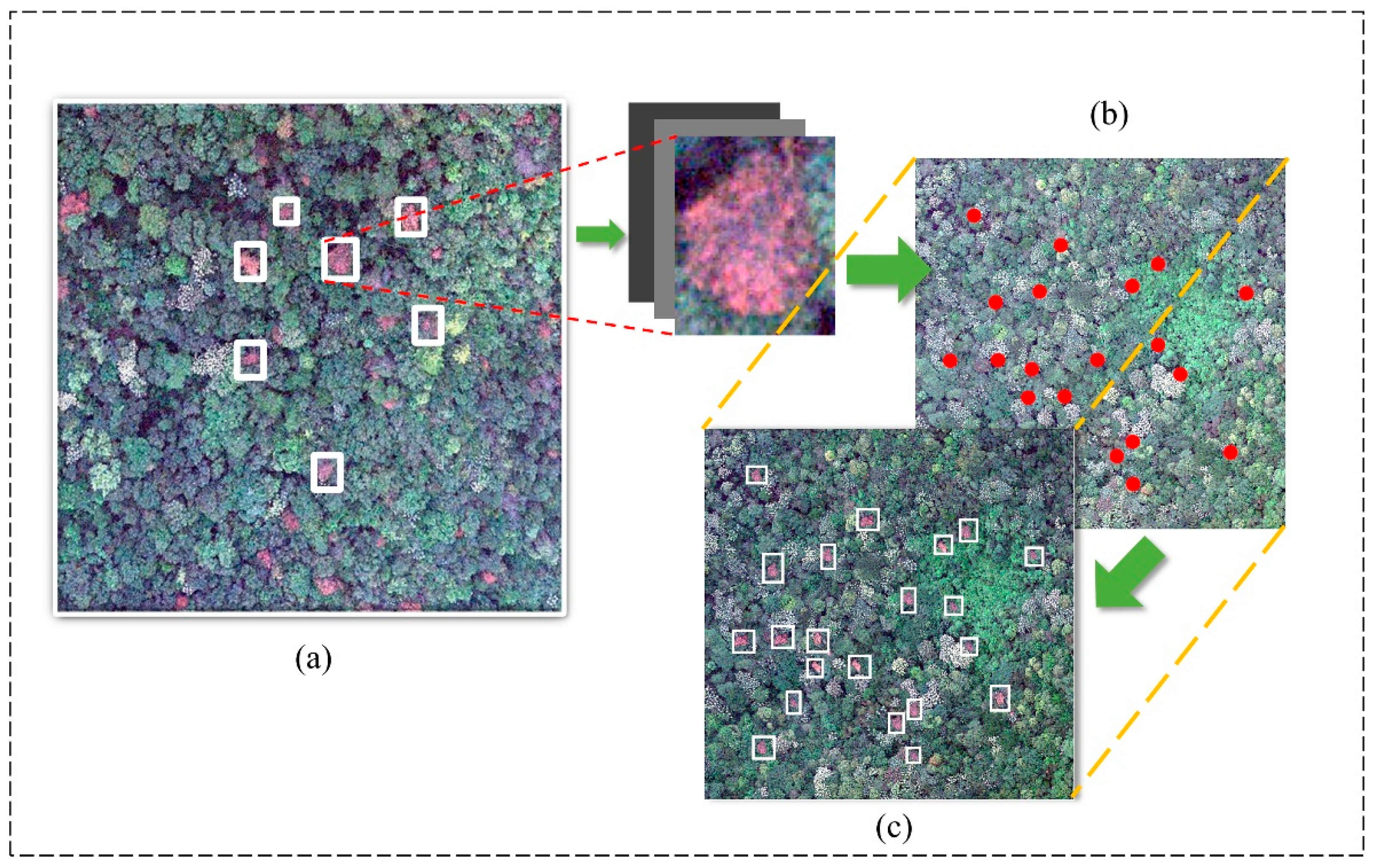
2.2.1. Confidence Map of SDT Generated by Gaussian Kernel Function
2.2.2. Augmentation Strategy for Small-SDT Detection
2.2.3. Multi-Scale Spatial Supervision Convolutional Network
2.2.4. SDT Localization from the Confidence Map
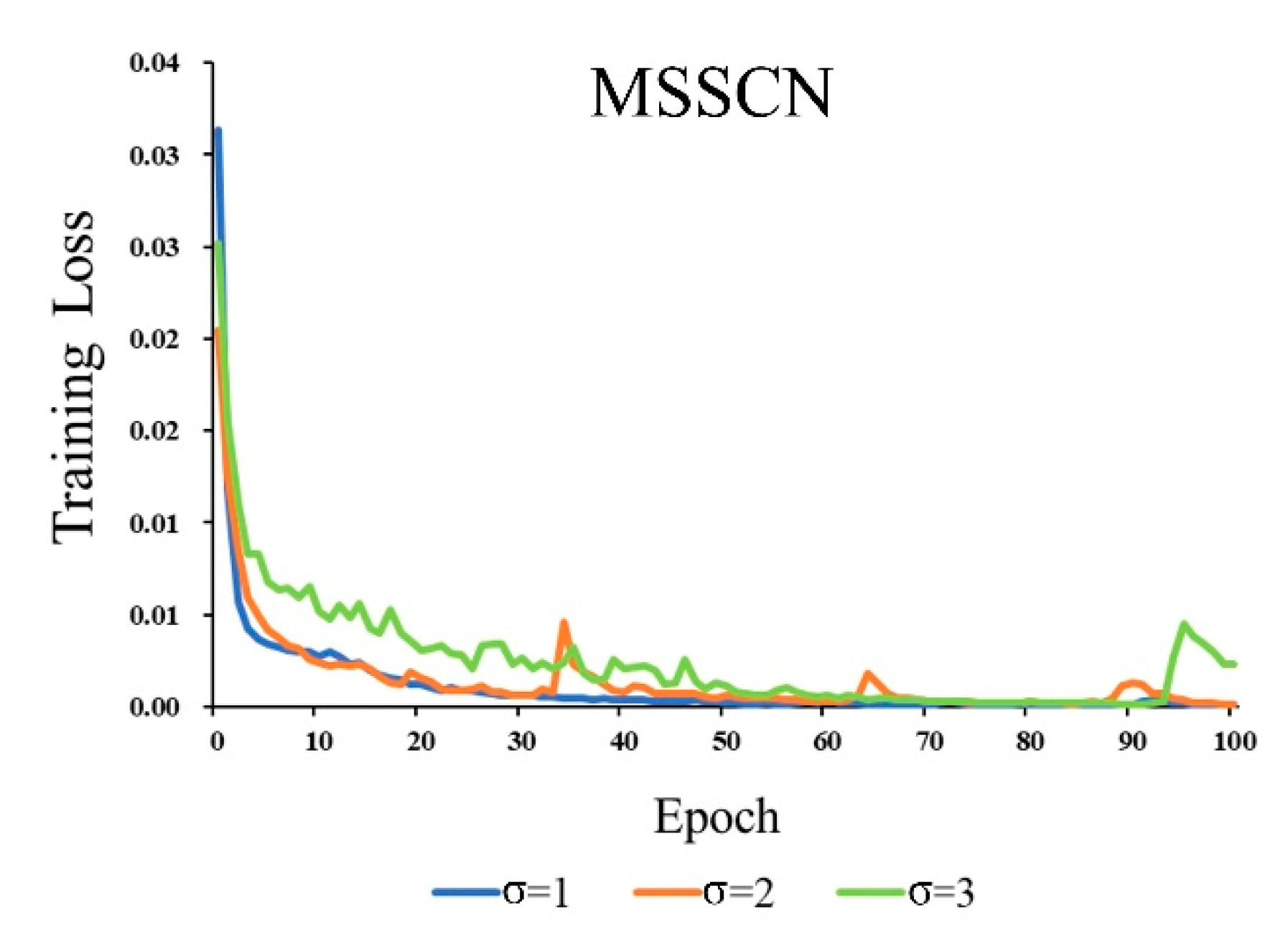
2.2.5. Experiment Setup
2.2.6. Assessment of Model Accuracy
3. Results
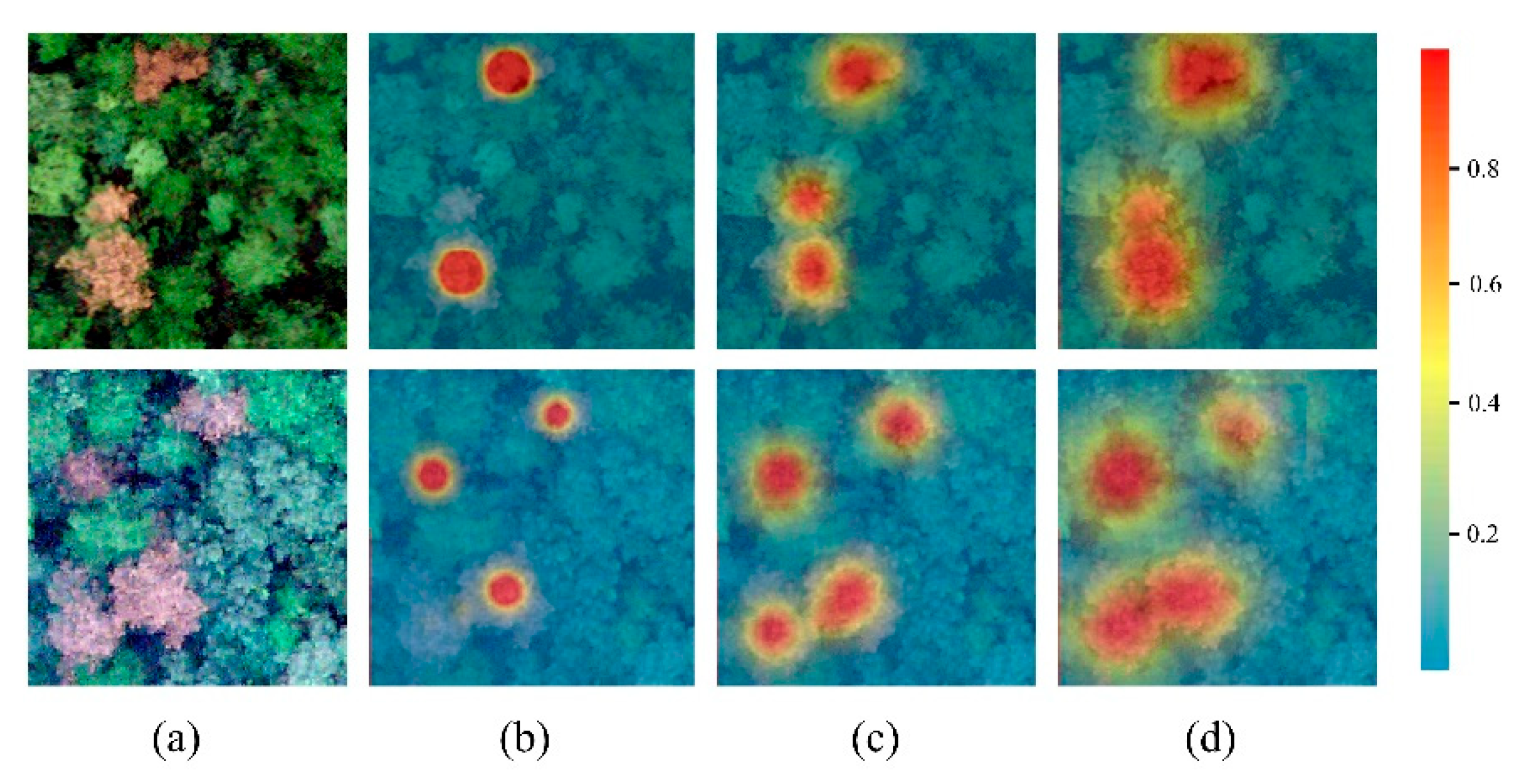
3.1. Analysis of Gaussian Kernel Parameter
3.2. Analysis of Oversampling Method
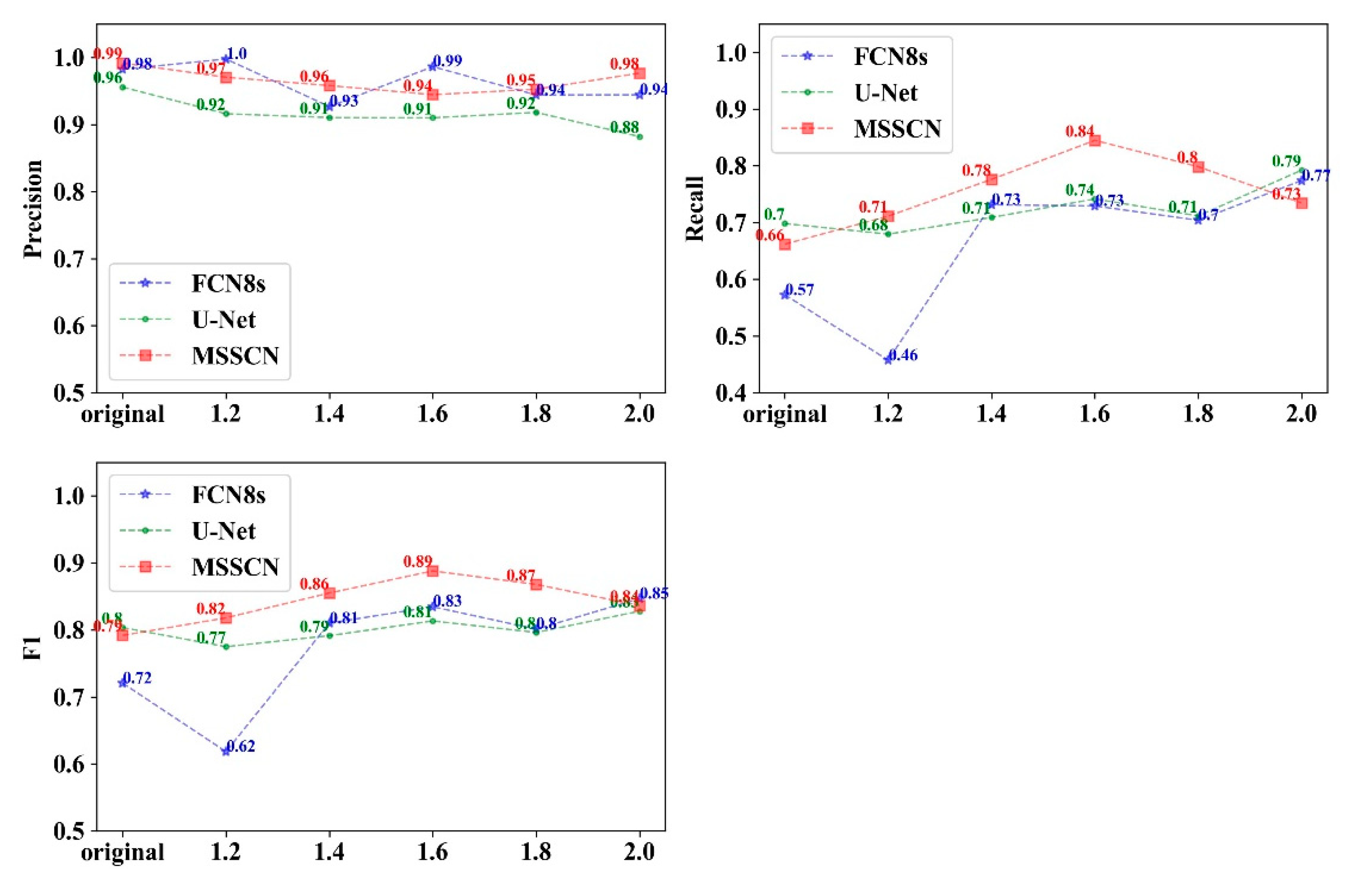
3.3. Comparison of the Accuracy of Different Models
4. Discussion
4.1. The Effect of the Gaussian Kernel Function
4.2. Oversampling Strategy in Promoting Detection Accuracy
4.3. MSSCN Model on Detection Accuracy
4.4. The Influence of Forest Type and Disease Outbreak Intensity on Detection Accuracy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tóth, Á. Bursaphelenchus Xylophilus, the Pinewood Nematode: Its Significance and a Historical Review. Acta Biol. Szeged. 2011, 55, 213–217. [Google Scholar]
- Carnegie, A.J.; Venn, T.; Lawson, S.; Nagel, M.; Wardlaw, T.; Cameron, N.; Last, I. An Analysis of Pest Risk and Potential Economic Impact of Pine Wilt Disease to Pinus Plantations in Australia. Aust. For. 2018, 81, 24–36. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, J.; Yan, J.; Fang, G. Economic Loss of Pine Wood Nematode Disease in Mainland China from 1998 to 2017. Forests 2020, 11, 1042. [Google Scholar] [CrossRef]
- Cha, D.; Kim, D.; Choi, W.; Park, S.; Han, H. Point-of-care diagnostic (POCD) method for detecting Bursaphelenchus xylophilus in pinewood using recombinase polymerase amplification (RPA) with the portable optical isothermal device (POID). PLoS ONE 2020, 15, e0227476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulridha, J.; Ehsani, R.; Abd-Elrahman, A.; Ampatzidis, Y. A Remote Sensing Technique for Detecting Laurel Wilt Disease in Avocado in Presence of Other Biotic and Abiotic Stresses. Comput. Electron. Agric. 2019, 156, 549–557. [Google Scholar] [CrossRef]
- Proença, D.N.; Grass, G.; Morais, P.V. Understanding pine wilt disease: Roles of the pine endophytic bacteria and of the bacteria carried by the disease-causing pinewood nematode. MicrobiologyOpen 2017, 6, e00415. [Google Scholar] [CrossRef] [PubMed]
- Stone, C.; Mohammed, C. Application of Remote Sensing Technologies for Assessing Planted Forests Damaged by Insect Pests and Fungal Pathogens: A Review. Curr. For. Rep. 2017, 3, 75–92. [Google Scholar] [CrossRef]
- Kang, J.S.; Kim, A.-Y.; Han, H.R.; Moon, Y.S.; Koh, Y.H. Development of Two Alternative Loop-Mediated Isothermal Amplification Tools for Detecting Pathogenic Pine Wood Nematodes. For. Pathol. 2015, 45, 127–133. [Google Scholar] [CrossRef]
- Li, X.; Tong, T.; Luo, T.; Wang, J.; Rao, Y.; Li, L.; Jin, D.; Wu, D.; Huang, H. Retrieving the Infected Area of Pine Wilt Disease-Disturbed Pine Forests from Medium-Resolution Satellite Images Using the Stochastic Radiative Transfer Theory. Remote Sens. 2022, 14, 1526. [Google Scholar] [CrossRef]
- Zhang, Y.; Dian, Y.; Zhou, J.; Peng, S.; Hu, Y.; Hu, L.; Han, Z.; Fang, X.; Cui, H. Characterizing Spatial Patterns of Pine Wood Nematode Outbreaks in Subtropical Zone in China. Remote Sens. 2021, 13, 4682. [Google Scholar] [CrossRef]
- Zhang, B.; Ye, H.; Lu, W.; Huang, W.; Wu, B.; Hao, Z.; Sun, H. A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 2083. [Google Scholar] [CrossRef]
- Hart, S.J.; Veblen, T.T. Detection of Spruce Beetle-Induced Tree Mortality Using High- and Medium-Resolution Remotely Sensed Imagery. Remote Sens. Environ. 2015, 168, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Kelly, M.; Gong, P.; Liu, D. An Object-Based Classification Approach in Mapping Tree Mortality Using High Spatial Resolution Imagery. GIScience Remote Sens. 2007, 44, 24–47. [Google Scholar] [CrossRef]
- Iordache, M.-D.; Mantas, V.; Baltazar, E.; Pauly, K.; Lewyckyj, N. A Machine Learning Approach to Detecting Pine Wilt Disease Using Airborne Spectral Imagery. Remote Sens. 2020, 12, 2280. [Google Scholar] [CrossRef]
- Meddens, A.J.H.; Hicke, J.A.; Vierling, L.A.; Hudak, A.T. Evaluating Methods to Detect Bark Beetle-Caused Tree Mortality Using Single-Date and Multi-Date Landsat Imagery. Remote Sens. Environ. 2013, 132, 49–58. [Google Scholar] [CrossRef]
- Skakun, R.S.; Wulder, M.A.; Franklin, S.E. Sensitivity of the Thematic Mapper Enhanced Wetness Difference Index to Detect Mountain Pine Beetle Red-Attack Damage. Remote Sens. Environ. 2003, 86, 433–443. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Ghosh, A.; Joshi, P.K.; Koch, B. Assessing the Potential of Hyperspectral Imagery to Map Bark Beetle-Induced Tree Mortality. Remote Sens. Environ. 2014, 140, 533–548. [Google Scholar] [CrossRef]
- Hall, R.J.; Castilla, G.; White, J.C.; Cooke, B.J.; Skakun, R.S. Remote Sensing of Forest Pest Damage: A Review and Lessons Learned from a Canadian Perspective. Can. Entomol. 2016, 148, S296–S356. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Carroll, A.L.; Coops, N.C. Challenges for the Operational Detection of Mountain Pine Beetle Green Attack with Remote Sensing. For. Chron. 2009, 85, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Hicke, J.A.; Logan, J. Mapping Whitebark Pine Mortality Caused by a Mountain Pine Beetle Outbreak with High Spatial Resolution Satellite Imagery. Int. J. Remote Sens. 2009, 30, 4427–4441. [Google Scholar] [CrossRef]
- Coops, N.C.; Johnson, M.; Wulder, M.A.; White, J.C. Assessment of QuickBird High Spatial Resolution Imagery to Detect Red Attack Damage Due to Mountain Pine Beetle Infestation. Remote Sens. Environ. 2006, 103, 67–80. [Google Scholar] [CrossRef]
- Oumar, Z.; Mutanga, O. Using WorldView-2 Bands and Indices to Predict Bronze Bug (Thaumastocoris Peregrinus) Damage in Plantation Forests. Int. J. Remote Sens. 2013, 34, 2236–2249. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of Studies on Tree Species Classification from Remotely Sensed Data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Qiao, R.; Ghodsi, A.; Wu, H.; Chang, Y.; Wang, C. Simple Weakly Supervised Deep Learning Pipeline for Detecting Individual Red-Attacked Trees in VHR Remote Sensing Images. Remote Sens. Lett. 2020, 11, 650–658. [Google Scholar] [CrossRef]
- Xiao, C.; Qin, R.; Huang, X.; Li, J. A study of using fully convolutional network for treetop detection on remote sensing data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, IV–1, 163–169. [Google Scholar]
- Qin, J.; Wang, B.; Wu, Y.; Lu, Q.; Zhu, H. Identifying Pine Wood Nematode Disease Using Uav Images and Deep Learning Algorithms. Remote Sens. 2021, 13, 162. [Google Scholar] [CrossRef]
- Buda, M.; Maki, A.; Mazurowski, M.A. A Systematic Study of the Class Imbalance Problem in Convolutional Neural Networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kisantal, M.; Wojna, Z.; Murawski, J.; Naruniec, J.; Cho, K. Augmentation for small object detection. arXiv 2019, arXiv:1902.07296. [Google Scholar]
- Lopatin, J.; Dolos, K.; Kattenborn, T.; Fassnacht, F.E. How Canopy Shadow Affects Invasive Plant Species Classification in High Spatial Resolution Remote Sensing. Remote Sens. Ecol. Conserv. 2019, 5, 302–317. [Google Scholar] [CrossRef]
- Liu, X.; Frey, J.; Denter, M.; Zielewska-Büttner, K.; Still, N.; Koch, B. Mapping Standing Dead Trees in Temperate Montane Forests Using a Pixel- and Object-Based Image Fusion Method and Stereo WorldView-3 Imagery. Ecol. Indic. 2021, 133, 108438. [Google Scholar] [CrossRef]
- Osco, L.P.; de Arruda, M.D.S.; Marcato Junior, J.; da Silva, N.B.; Ramos, A.P.M.; Moryia, É.A.S.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A Convolutional Neural Network Approach for Counting and Geolocating Citrus-Trees in UAV Multispectral Imagery. ISPRS J. Photogramm. Remote Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfister, T.; Charles, J.; Zisserman, A. Flowing Convnets for Human Pose Estimation in Videos. In Proceedings of the IEEE international conference on computer vision, Santiago, Chile, 7–13 December 2015; pp. 1913–1921. [Google Scholar]
- Yun, T.; Jiang, K.; Li, G.; Eichhorn, M.P.; Fan, J.; Liu, F.; Chen, B.; An, F.; Cao, L. Individual Tree Crown Segmentation from Airborne LiDAR Data Using a Novel Gaussian Filter and Energy Function Minimization-Based Approach. Remote Sens. Environ. 2021, 256, 112307. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Brooks, D.; Reich, R.; Wheate, R.D. Detection of Red Attack Stage Mountain Pine Beetle Infestation with High Spatial Resolution Satellite Imagery. Remote Sens. Environ. 2005, 96, 340–351. [Google Scholar] [CrossRef]
- Mai, Z.; Hu, X.; Peng, S.; Wei, Y. Human Pose Estimation via Multi-Scale Intermediate Supervision Convolution Network. In Proceedings of the 2019 12th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Suzhou, China, 19–21 October 2019; IEEE: Suzhou, China, 2019; pp. 1–6. [Google Scholar]
- Han, Z.; Dian, Y.; Xia, H.; Zhou, J.; Jian, Y.; Yao, C.; Wang, X.; Li, Y. Comparing Fully Deep Convolutional Neural Networks for Land Cover Classification with High-Spatial-Resolution Gaofen-2 Images. ISPRS Int. J. Geo-Inf. 2020, 9, 478. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE conference on computer vision and pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Yu, F.; Koltun, V.; Funkhouser, T. Dilated Residual Networks. In Proceedings of the Proceedings, 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 636–644. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. arXiv 2018, arXiv:1807.06521. [Google Scholar]
- Li, X.; Xu, F.; Lyu, X.; Gao, H.; Tong, Y.; Cai, S.; Li, S.; Liu, D. Dual Attention Deep Fusion Semantic Segmentation Networks of Large-Scale Satellite Remote-Sensing Images. Int. J. Remote Sens. 2021, 42, 3583–3610. [Google Scholar] [CrossRef]
- Chadwick, A.J.; Goodbody, T.R.H.; Coops, N.C.; Hervieux, A.; Bater, C.W.; Martens, L.A.; White, B.; Röeser, D. Automatic Delineation and Height Measurement of Regenerating Conifer Crowns under Leaf-off Conditions Using UAV Imagery. Remote Sens. 2020, 12, 4104. [Google Scholar] [CrossRef]
- Meddens, A.J.H.; Hicke, J.A. Spatial and Temporal Patterns of Landsat-Based Detection of Tree Mortality Caused by a Mountain Pine Beetle Outbreak in Colorado, USA. For. Ecol. Manag. 2014, 322, 78–88. [Google Scholar] [CrossRef]




| Low-Intensity Area | High-Intensity Area | |||
|---|---|---|---|---|
| Area | A-1 | A-3 | A-2 | A-4 |
| Number | 116 | 67 | 118 | 396 |
| Density | 9 ha−1 | 5 ha−1 | 68 ha−1 | 58 ha−1 |
| Low PWD Dead-Tree Intensity Area | High PWD Dead-Tree Intensity Area | |||||||
|---|---|---|---|---|---|---|---|---|
| Area | B-1 | B-2 | B-4 | B-7 | B-3 | B-5 | B-6 | B-8 |
| Number | 42 | 43 | 85 | 25 | 136 | 62 | 56 | 123 |
| Density | 12 ha−1 | 18 ha−1 | 18 ha−1 | 11 ha−1 | 25 ha−1 | 28 ha−1 | 21 ha−1 | 34 ha−1 |
| Precision | Recall | F1-Score | |
|---|---|---|---|
| 1.0 | 0.95 | 0.62 | 0.74 |
| 2.0 | 0.92 | 0.69 | 0.79 |
| 3.0 | 0.83 | 0.62 | 0.71 |
| Model | Site | TP | FN | FP | P | R | F1 |
|---|---|---|---|---|---|---|---|
| FCN8s | A-1 | 99 | 17 | 2 | 0.98 | 0.85 | 0.91 |
| A-2 | 71 | 47 | 0 | 1.00 | 0.60 | 0.75 | |
| A-3 | 45 | 22 | 1 | 0.98 | 0.67 | 0.80 | |
| A-4 | 313 | 83 | 4 | 0.99 | 0.79 | 0.88 | |
| Avg | 0.99 | 0.73 | 0.83 | ||||
| U-Net | A-1 | 100 | 16 | 8 | 0.93 | 0.86 | 0.89 |
| A-2 | 70 | 48 | 5 | 0.93 | 0.59 | 0.73 | |
| A-3 | 51 | 16 | 6 | 0.89 | 0.76 | 0.82 | |
| A-4 | 297 | 99 | 38 | 0.89 | 0.75 | 0.81 | |
| Avg | 0.91 | 0.74 | 0.81 | ||||
| MSSCN | A-1 | 104 | 12 | 11 | 0.90 | 0.90 | 0.90 |
| A-2 | 84 | 34 | 0 | 1.00 | 0.71 | 0.83 | |
| A-3 | 62 | 5 | 5 | 0.93 | 0.93 | 0.93 | |
| A-4 | 335 | 61 | 18 | 0.95 | 0.85 | 0.89 | |
| Avg | 0.94 | 0.84 | 0.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Hu, W.; Peng, S.; Lin, H.; Zhang, J.; Zhou, J.; Wang, P.; Dian, Y. Detection of Standing Dead Trees after Pine Wilt Disease Outbreak with Airborne Remote Sensing Imagery by Multi-Scale Spatial Attention Deep Learning and Gaussian Kernel Approach. Remote Sens. 2022, 14, 3075. https://doi.org/10.3390/rs14133075
Han Z, Hu W, Peng S, Lin H, Zhang J, Zhou J, Wang P, Dian Y. Detection of Standing Dead Trees after Pine Wilt Disease Outbreak with Airborne Remote Sensing Imagery by Multi-Scale Spatial Attention Deep Learning and Gaussian Kernel Approach. Remote Sensing. 2022; 14(13):3075. https://doi.org/10.3390/rs14133075
Chicago/Turabian StyleHan, Zemin, Wenjie Hu, Shoulian Peng, Haoran Lin, Jian Zhang, Jingjing Zhou, Pengcheng Wang, and Yuanyong Dian. 2022. "Detection of Standing Dead Trees after Pine Wilt Disease Outbreak with Airborne Remote Sensing Imagery by Multi-Scale Spatial Attention Deep Learning and Gaussian Kernel Approach" Remote Sensing 14, no. 13: 3075. https://doi.org/10.3390/rs14133075
APA StyleHan, Z., Hu, W., Peng, S., Lin, H., Zhang, J., Zhou, J., Wang, P., & Dian, Y. (2022). Detection of Standing Dead Trees after Pine Wilt Disease Outbreak with Airborne Remote Sensing Imagery by Multi-Scale Spatial Attention Deep Learning and Gaussian Kernel Approach. Remote Sensing, 14(13), 3075. https://doi.org/10.3390/rs14133075







