RETRACTED: Evaluating Multi-Sensors Spectral and Spatial Resolutions for Tree Species Diversity Prediction
Abstract
1. Introduction
2. Materials and Methods
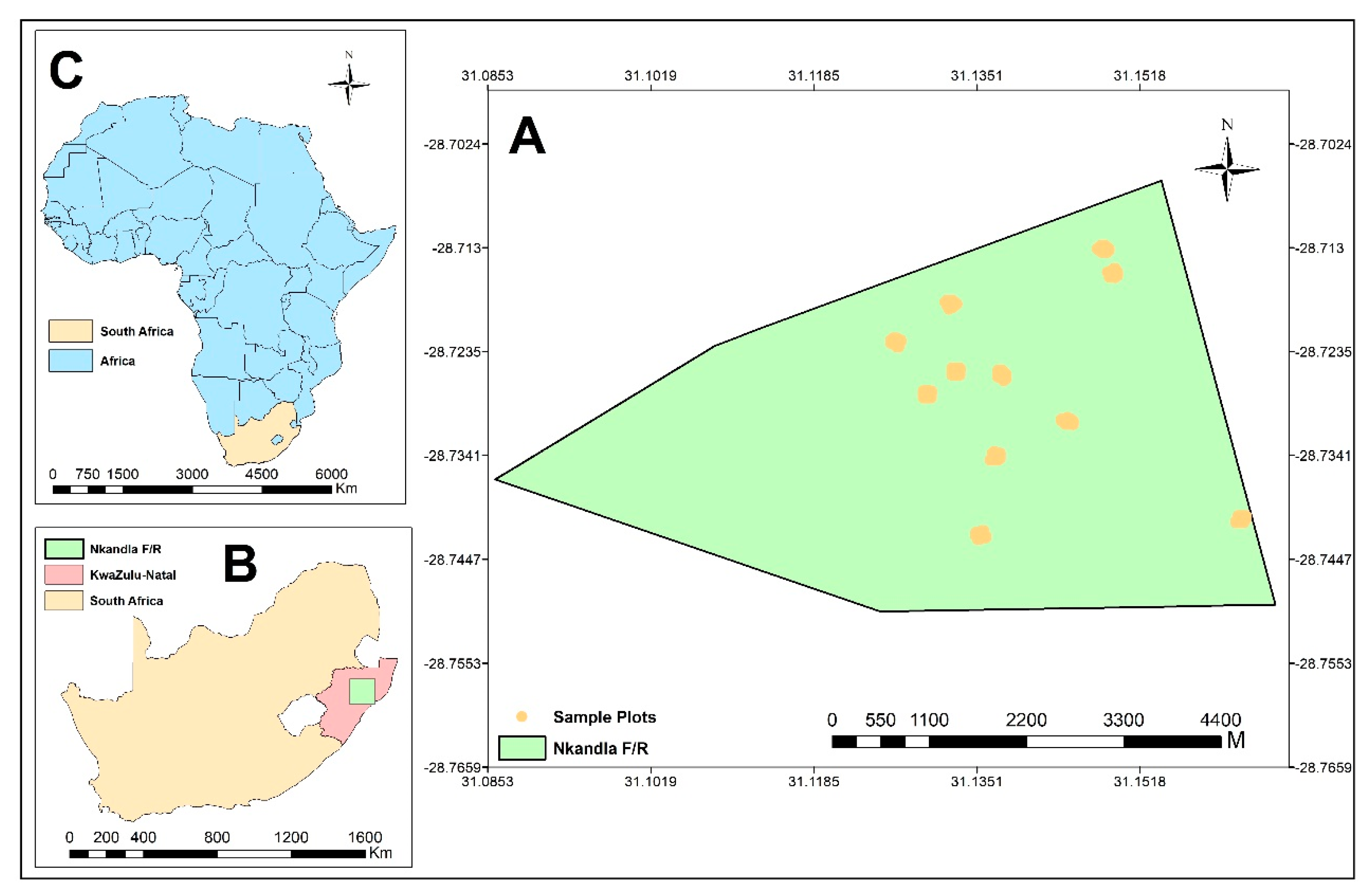
2.1. Study Area
2.2. Field Inventory and Diversity Indices Estimation
2.3. Remote Sensing Data
2.4. Important Variables Selection
2.5. Random Forest Regression Modelling
2.6. Models Evaluation
2.7. Variable Importance
3. Results
3.1. Field Inventory Data Analysis
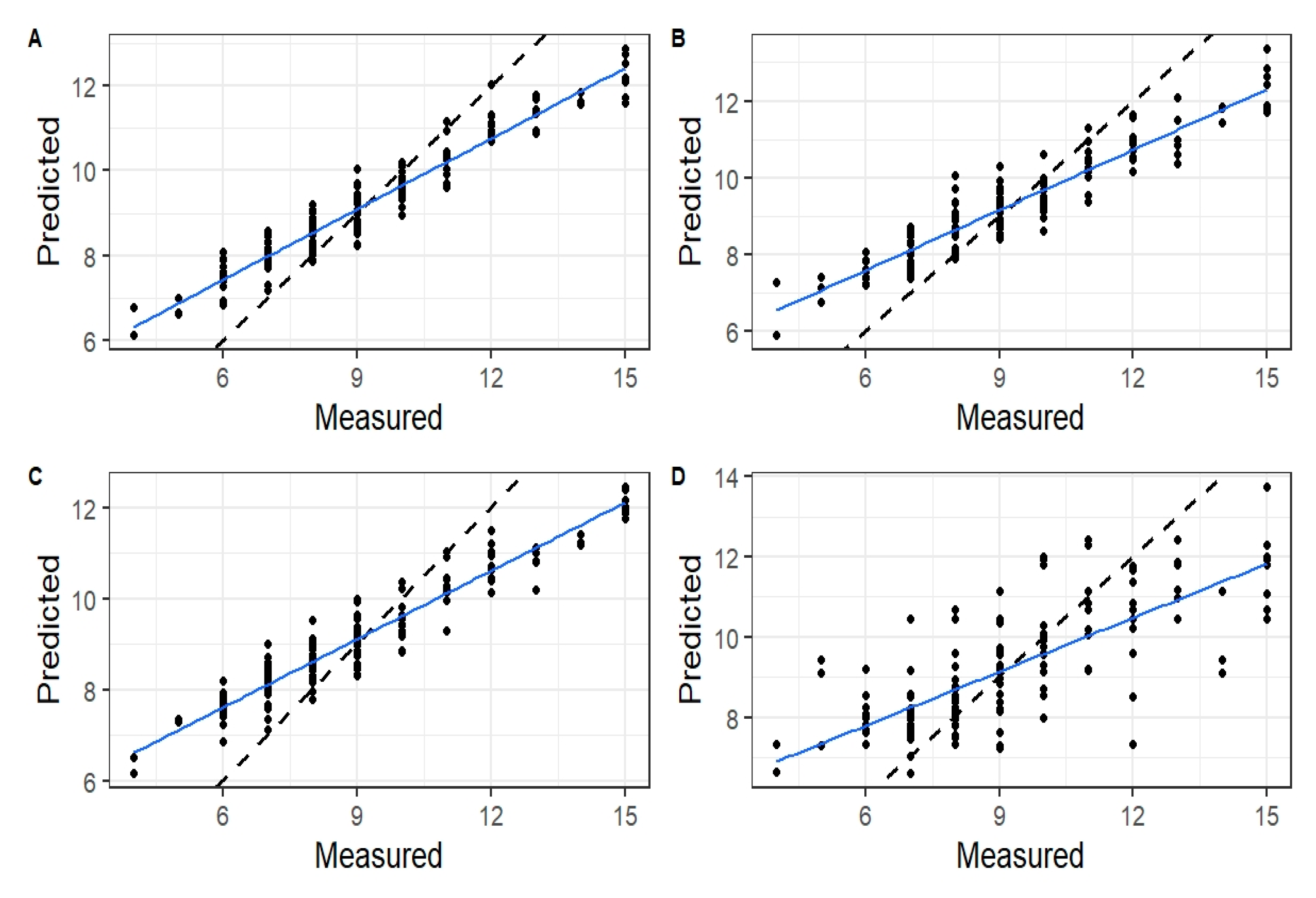
3.2. Sensor Performance Evaluation
3.3. Predicting Important Variables
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gamfeldt, L.; Snäll, T.; Bagchi, R.; Jonsson, M.; Gustafsson, L.; Kjellander, P.; Ruiz-Jaen, M.C.; Fröberg, M.; Stendahl, J.; Philipson, C.D. Higher levels of multiple ecosystem services are found in forests with more tree species. Nat. Commun. 2013, 4, 1340. [Google Scholar] [CrossRef]
- Aerts, R.; Honnay, O. Forest restoration, biodiversity and ecosystem functioning. J. BMC Ecol. 2011, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Gamon, J.A. Remote sensing of terrestrial plant biodiversity. Remote Sens. Environ. 2019, 231, 111218. [Google Scholar] [CrossRef]
- Iverson, L.R.; McKenzie, D. Tree-species range shifts in a changing climate: Detecting, modeling, assisting. Landsc. Ecol. 2013, 28, 879–889. [Google Scholar] [CrossRef]
- Millennium Ecosystem Assessment. Ecosystems and Human Well-Being; Island Press: Washington, DC, USA, 2005. [Google Scholar]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Luo, W.; Liang, J.; Gatti, R.C.; Zhao, X.; Zhan, C. Parameterization of biodiversity–productivity relationship and its scale dependency using georeferenced tree-level data. J. Ecol. 2019, 107, 1106–1119. [Google Scholar] [CrossRef]
- Fundisi, E.; Musakwa, W.; Ahmed, F.B.; Tesfamichael, S.G. Estimation of woody plant species diversity during a dry season in a savanna environment using the spectral and textural information derived from WorldView-2 imagery. PLoS ONE 2020, 15, e0234158. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest stand species mapping using the Sentinel-2 time series. J. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Adelabu, S.; Mutanga, O.; Adam, E.E.; Cho, M.A. Exploiting machine learning algorithms for tree species classification in a semiarid woodland using RapidEye image. J. Appl. Remote Sens. 2013, 7, e073480. [Google Scholar] [CrossRef]
- Ustuner, M.; Sanli, F.B.; Abdikan, S. Balanced Vs Imbalanced Training Data: Classifying Rapideye Data with Support Vector Machines. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 379–384. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Mather, P. The use of backpropagating artificial neural networks in land cover classification. Int. J. Remote Sens. 2003, 24, 4907–4938. [Google Scholar] [CrossRef]
- Chrysafis, I.; Korakis, G.; Kyriazopoulos, A.P.; Mallinis, G. Predicting Tree Species Diversity Using Geodiversity and Sentinel-2 Multi-Seasonal Spectral Information. Sustainability 2020, 12, 9250. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Ramoelo, A.; Mutanga, O.; Naidoo, L. Estimating tree species diversity in the savannah using NDVI and woody canopy cover. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 106–115. [Google Scholar] [CrossRef]
- Mutowo, G.; Murwira, A. Relationship between remotely sensed variables and tree species diversity in savanna woodlands of Southern Africa. Int. J. Remote Sens. 2012, 33, 6378–6402. [Google Scholar] [CrossRef]
- Arekhi, M.; Yilmaz, O.Y.; Yilmaz, H.; Akyuz, Y.F. Can tree species diversity be assessed with Landsat data in a temperate forest? Environ. Monit. Assess. 2017, 189, 586. [Google Scholar] [CrossRef] [PubMed]
- Foody, G.M.; Cutler, M.E.J. Mapping the species richness and composition of tropical forests from remotely sensed data with neural networks. Ecol. Model. 2006, 195, 37–42. [Google Scholar] [CrossRef]
- Gillespie, T.W.; de Geode, J.; Aguilar, L.; Jenerette, G.D.; Fricker, G.A.; Avolio, M.L.; Pincetl, S.; Johnston, T.; Clarke, L.W.; Pataki, D.E. Predicting tree species richness in urban forests. Urban Ecosyst. 2016, 20, 839–849. [Google Scholar] [CrossRef]
- Gould, W. Remote Sensing of Vegetation, Plant Species Richness, and Regional Biodiversity Hotspots. Ecol. Appl. 2000, 10, 1861–1870. [Google Scholar] [CrossRef]
- Carlson, K.M.; Asner, G.P.; Hughes, R.F.; Ostertag, R.; Martin, R.E. Hyperspectral Remote Sensing of Canopy Biodiversity in Hawaiian Lowland Rainforests. Ecosystems 2007, 10, 536–549. [Google Scholar] [CrossRef]
- Colgan, M.S.; Baldeck, C.A.; Feret, J.B.; Asner, G.P. Mapping Savanna Tree Species at Ecosystem Scales Using Support Vector Machine Classification and BRDF Correction on Airborne Hyperspectral and LiDAR Data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef]
- Oldeland, J.; Wesuls, D.; Rocchini, D.; Schmidt, M.; Jürgens, N. Does using species abundance data improve estimates of species diversity from remotely sensed spectral heterogeneity? Ecol. Indic. 2010, 10, 390–396. [Google Scholar] [CrossRef]
- Kalacska, M.; Sanchez-Azofeifa, G.A.; Rivard, B.; Caelli, T.; White, H.P.; Calvo-Alvarado, J.C. Ecological fingerprinting of ecosystem succession: Estimating secondary tropical dry forest structure and diversity using imaging spectroscopy. Remote Sens. Environ. 2007, 108, 82–96. [Google Scholar] [CrossRef]
- Laurin, V.G.; Chan, J.C.-W.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Guerriero, L.; Frate, F.D.; Miglietta, F.; Valentini, R. Biodiversity mapping in a tropical West African forest with airborne hyperspectral data. PLoS ONE 2014, 9, e97910. [Google Scholar]
- Schäfer, E.; Heiskanen, J.; Heikinheimo, V.; Pellikka, P. Mapping tree species diversity of a tropical montane forest by unsupervised clustering of airborne imaging spectroscopy data. Ecol. Indic. 2016, 64, 49–58. [Google Scholar] [CrossRef]
- Nagendra, H.; Lucas, R.; Honrado, J.P.; Jongman, R.H.G.; Tarantino, C.; Adamo, M.; Mairota, P. Remote sensing for conservation monitoring: Assessing protected areas, habitat extent, habitat condition, species diversity, and threats. Ecol. Indic. 2013, 33, 45–59. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Zhao, Y.; Zeng, Y.; Zheng, Z.; Dong, W.; Zhao, D.; Wu, B.; Zhao, Q. Forest species diversity mapping using airborne LiDAR and hyperspectral data in a subtropical forest in China. Remote Sens. Environ. 2018, 213, 104–114. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Ao, Z.; Lao, D.; Xin, Q. Deep Learning Approaches for the Mapping of Tree Species Diversity in a Tropical Wetland Using Airborne LiDAR and High-Spatial-Resolution Remote Sensing Images. Forests 2019, 10, 1047. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Ramoelo, A.; Mutanga, O. Remote sensing of species diversity using Landsat 8 spectral variables. ISPRS J. Photogramm. Remote Sens. 2017, 133, 116–127. [Google Scholar] [CrossRef]
- Mohammadi, J.; Shataee, S. Possibility investigation of tree diversity mapping using Landsat ETM+ data in the Hyrcanian forests of Iran. Remote Sens. Environ. 2010, 114, 1504–1512. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Evaluating the utility of the medium-spatial resolution Landsat 8 multispectral sensor in quantifying aboveground biomass in uMgeni catchment, South Africa. ISPRS J. Photogramm. Remote Sens. 2015, 101, 36–46. [Google Scholar] [CrossRef]
- Feilhauer, H.; Schmidtlein, S. Mapping continuous fields of forest alpha and beta diversity. Appl. Veg. Sci. 2009, 12, 429–439. [Google Scholar] [CrossRef]
- Mutowo, G.; Murwira, A. The spatial prediction of tree species diversity in savanna woodlands of Southern Africa. Geocarto Int. 2012, 27, 627–645. [Google Scholar] [CrossRef]
- Persson, M.; Lindberg, E.; Reese, H. Tree species classification with multi-temporal Sentinel-2 data. J. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef]
- Mutowo, G.; Mutanga, O.; Masocha, M. Evaluating the Applications of the Near-Infrared Region in Mapping Foliar N in the Miombo Woodlands. Remote Sens. 2018, 10, 505. [Google Scholar] [CrossRef]
- Pau, S.; Gillespie, T.W.; Wolkovich, E.M. Dissecting NDVI–species richness relationships in Hawaiian dry forests. J. Biogeogr. 2012, 39, 1678–1686. [Google Scholar] [CrossRef]
- Oindo, B.O.; Skidmore, A.K. Interannual variability of NDVI and species richness in Kenya. Int. J. Remote Sens. 2010, 23, 285–298. [Google Scholar] [CrossRef]
- Krishnaswamy, J.; Bawa, K.S.; Ganeshaiah, K.N.; Kiran, M.C. Quantifying and mapping biodiversity and ecosystem services: Utility of a multi-season NDVI based Mahalanobis distance surrogate. Remote Sens. Environ. 2009, 113, 857–867. [Google Scholar] [CrossRef]
- George-Chacon, S.P.; Dupuy, J.M.; Peduzzi, A.; Hernandez-Stefanoni, L. Combining high resolution satellite imagery and lidar data to model woody species diversity of tropical dry forests. Ecol. Indic. 2019, 101, 975–984. [Google Scholar] [CrossRef]
- Ozdemir, I.; Norton, D.A.; Ozkan, U.Y.; Mert, A.; Senturk, O. Estimation of tree size diversity using object oriented texture analysis and aster imagery. Sensors 2008, 8, 4709–4724. [Google Scholar] [CrossRef]
- Otunga, C.; Odindi, J.; Mutanga, O.; Adjorlolo, C. Evaluating the potential of the red edge channel for C3 (Festuca spp.) grass discrimination using Sentinel-2 and Rapid Eye satellite image data. Geocarto Int. 2018, 34, 1123–1143. [Google Scholar] [CrossRef]
- Tigges, J.; Lakes, T.; Hostert, P. Urban vegetation classification: Benefits of multitemporal RapidEye satellite data. Remote Sens. Environ. 2013, 136, 66–75. [Google Scholar] [CrossRef]
- van Deventer, H.; Cho, M.A.; Mutanga, O. Improving the classification of six evergreen subtropical tree species with multi-season data from leaf spectra simulated to WorldView-2 and RapidEye. Int. J. Remote Sens. 2017, 38, 4804–4830. [Google Scholar] [CrossRef]
- Tuominen, S.; Näsi, R.; Honkavaara, E.; Balazs, A.; Hakala, T.; Viljanen, N.; Pölönen, I.; Saari, H.; Ojanen, H. Assessment of classifiers and remote sensing features of hyperspectral imagery and stereo-photogrammetric point clouds for recognition of tree species in a forest area of high species diversity. Remote Sens. 2018, 10, 714. [Google Scholar] [CrossRef]
- Pedro, M.S.; Rammer, W.; Seidl, R. Tree species diversity mitigates disturbance impacts on the forest carbon cycle. Oecologia 2015, 177, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Mallinis, G.; Chrysafis, I.; Korakis, G.; Pana, E.; Kyriazopoulos, A.P. A Random Forest Modelling Procedure for a Multi-Sensor Assessment of Tree Species Diversity. Remote Sens. 2020, 12, 1210. [Google Scholar] [CrossRef]
- John, R.; Chen, J.; Lu, N.; Guo, K.; Liang, C.; Wei, Y.; Noormets, A.; Ma, K.; Han, X. Predicting plant diversity based on remote sensing products in the semi-arid region of Inner Mongolia. Remote Sens. Environ. 2008, 112, 2018–2032. [Google Scholar] [CrossRef]
- Ouyang, S.; Xiang, W.; Wang, X.; Zeng, Y.; Lei, P.; Deng, X.; Peng, C. Significant effects of biodiversity on forest biomass during the succession of subtropical forest in south China. For. Ecol. Manag. 2016, 372, 291–302. [Google Scholar] [CrossRef]
- Huang, Y.; Ma, Y.; Zhao, K.; Niklaus, P.A.; Schmid, B.; He, J.-S. Positive effects of tree species diversity on litterfall quantity and quality along a secondary successional chronosequence in a subtropical forest. J. Plant Ecol. 2017, 10, 28–35. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Schmid, B.; Bruelheide, H.; Bu, W.; Ma, K. Positive effects of tree species richness on fine-root production in a subtropical forest in SE-China. J. Plant Ecol. 2017, 10, 146–157. [Google Scholar] [CrossRef]
- Ezemvelo KwaZulu-Natal Wildlife. Nkandla Forest Complex: Protected Area Management Plan. 2015, pp. 1–156. Available online: www.kznwildlife.com/Documents/Nkandla%20Forest%20Complex%20Management%20Plan%20Final%2012102016%20compressed.pdf (accessed on 1 January 2021).
- Gyamfi-Ampadu, E.; Gebreslasie, M.; Mendoza-Ponce, A. Mapping natural forest cover using satellite imagery of Nkandla forest reserve, KwaZulu-Natal, South Africa. Remote Sens. Appl. Soc. Environ. 2020, 18, 100302. [Google Scholar] [CrossRef]
- Shannon, C. A mathematical theory of communication. J. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E. Measurement of diversity. J. Nat. 1949, 163, 688. [Google Scholar] [CrossRef]
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D. Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef]
- Daly, A.J.; Baetens, J.M.; de Baets, B.J.M. Ecological diversity: Measuring the unmeasurable. Mathematics 2018, 6, 119. [Google Scholar] [CrossRef]
- Ifo, S.A.; Moutsambote, J.-M.; Koubouana, F.; Yoka, J.; Ndzai, S.F.; Bouetou-Kadilamio, L.N.O.; Mampouya, H.; Jourdain, C.; Bocko, Y.; Mantota, A.B. Tree species diversity, richness, and similarity in intact and degraded forest in the tropical rainforest of the Congo Basin: Case of the forest of Likouala in the Republic of Congo. Int. J. For. Res. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Wang, D.; Wan, B.; Qiu, P.; Su, Y.; Guo, Q.; Wang, R.; Sun, F.; Wu, X. Evaluating the Performance of Sentinel-2, Landsat 8 and Pléiades-1 in Mapping Mangrove Extent and Species. Remote Sens. 2018, 10, 1468. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rodríguez-Galiano, V.F.; Abarca-Hernández, F.; Ghimire, B.; Chica-Olmo, M.; Atkinson, P.M.; Jeganathan, C. Incorporating Spatial Variability Measures in Land-cover Classification using Random Forest. Procedia Environ. Sci. 2011, 3, 44–49. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.M.; Ahmed, F.B.; Ismail, R. Random forest regression and spectral band selection for estimating sugarcane leaf nitrogen concentration using EO-1 Hyperion hyperspectral data. Int. J. Remote 2013, 34, 712–728. [Google Scholar] [CrossRef]
- Ramoelo, A.; Cho, M.A.; Mathieu, R.; Madonsela, S.; Van De Kerchove, R.; Kaszta, Z.; Wolff, E. Monitoring grass nutrients and biomass as indicators of rangeland quality and quantity using random forest modelling and WorldView-2 data. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 43–54. [Google Scholar] [CrossRef]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Nitze, I.; Barrett, B.; Cawkwell, F. Temporal optimisation of image acquisition for land cover classification with Random Forest and MODIS time-series. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 136–146. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Ganivet, E.; Bloomberg, M. Towards rapid assessments of tree species diversity and structure in fragmented tropical forests: A review of perspectives offered by remotely-sensed and field-based data. Forest Ecol. Manag. 2019, 432, 40–53. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, P.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Todd, S.W.; Hoffer, R.M.; Milchunas, D.G. Biomass estimation on grazed and ungrazed rangelands using spectral indices. Int. J. Remote Sens. 1998, 19, 427–438. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O.; Kiala, Z. The utility of Sentinel-2 Vegetation Indices (VIs) and Sentinel-1 Synthetic Aperture Radar (SAR) for invasive alien species detection and mapping. Nat. Conserv. 2019, 35, 41–61. [Google Scholar] [CrossRef]
- Rocchini, D.; Ricotta, C.; Chiarucci, A. Using satellite imagery to assess plant species richness: The role of multispectral systems. J. Appl. Veg. Sci. 2007, 10, 325–331. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A.; Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Zygielbaum, A.I.; Kothari, S. Influence of species richness, evenness, and composition on optical diversity: A simulation study. J. Remote Sens. Environ. 2018, 211, 218–228. [Google Scholar] [CrossRef]
- Immitzer, M.; Neuwirth, M.; Böck, S.; Brenner, H.; Vuolo, F.; Atzberger, C. Optimal Input Features for Tree Species Classification in Central Europe Based on Multi-Temporal Sentinel-2 Data. J. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef]
- Martin-Gallego, P.; Aplin, P.; Marston, C.; Altamirano, A.; Pauchard, A. Detecting and modelling alien tree presence using Sentinel-2 satellite imagery in Chile’s temperate forests. For. Ecol. Manag. 2020, 474, 118353. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M. On the Importance of Training Data Sample Selection in Random Forest Image Classification: A Case Study in Peatland Ecosystem Mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef]
- Bolyn, C.; Michez, A.; Gaucher, P.; Lejeune, P.; Bonnet, S. Forest mapping and species composition using supervised per pixel classification of Sentinel-2 imagery. Biotechnol. Agron. Société Environ. 2018, 22, 16. [Google Scholar]
- Parson, E.A. Climate Engineering in Global Climate Governance: Implications for Participation and Linkage. Transnatl. Environ. Law 2013, 3, 89–110. [Google Scholar] [CrossRef]
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2001, 22, 2377–2400. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O.; Elhadi, A.; Ismail, R. Intra-and-inter species biomass prediction in a plantation forest: Testing the utility of high spatial resolution spaceborne multispectral RapidEye sensor and advanced machine learning algorithms. Sensors 2014, 14, 15348–15370. [Google Scholar] [CrossRef]
- Wallner, A.; Elatawneh, A.; Schneider, T.; Knoke, T. Estimation of forest structural information using RapidEye satellite data. Forestry 2014, 88, 96–107. [Google Scholar] [CrossRef]
- Maeda, E.E.; Heiskanen, J.; Thijs, K.W.; Pellikka, P.K.E. Season-dependence of remote sensing indicators of tree species diversity. Remote Sens. Lett. 2014, 5, 404–412. [Google Scholar] [CrossRef]
- El-Askary, H.; Abd El-Mawla, S.; Li, J.; El-Hattab, M.; El-Raey, M. Change detection of coral reef habitat using Landsat-5 TM, Landsat 7 ETM+ and Landsat 8 OLI data in the Red Sea (Hurghada, Egypt). Int. J. Remote Sens. 2014, 35, 2327–2346. [Google Scholar] [CrossRef]
- Pahlevan, N.; Lee, Z.; Wei, J.; Schaaf, C.B.; Schott, J.R.; Berk, A. On-orbit radiometric characterization of OLI (Landsat-8) for applications in aquatic remote sensing. Remote Sens. Environ. 2014, 154, 272–284. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]


| Sentinel 2 | Landsat 8 | RapidEye | PlanetScope | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bands | Bandwidth (nm) | Spatial Resolution (m) | Bands | Bandwidth | Spatial Resolution (m) | Bands | Bandwidth (nm) | Spatial Resolution (m) | Bands | Bandwidth (nm) | Spatial Resolution (m) |
| Blue | 458-523 | 10 | Blue | 452-512 | 30 | Blue | 440-510 | 5 | Blue | 455-515 | 3 |
| Green | 543-573 | 10 | Green | 533-590 | 30 | Green | 520-590 | 5 | Green | 500-590 | 3 |
| Red | 650-680 | 10 | Red | 636-673 | 30 | Red | 630-685 | 5 | Red | 590-670 | 3 |
| RE1 | 698-713 | 20 | NIR | 851-879 | 30 | RE | 690-730 | 5 | NIR | 780-860 | 3 |
| RE2 | 733-748 | 20 | SWIR1 | 1566-1651 | 30 | NIR | 760-850 | 5 | |||
| RE3 | 773-793 | 20 | SWIR2 | 2107-2294 | 30 | ||||||
| NIR | 785-899 | 10 | |||||||||
| NNIR | 855-875 | 20 | |||||||||
| SWIR2 | 1565-1655 | 20 | |||||||||
| SWIR2 | 2100-2280 | 20 | |||||||||
| Parameter | Shannon Index | Simpson Index | Species Richness |
|---|---|---|---|
| Mean | 2.055 | 0.891 | 9 |
| Minimum | 0.949 | 0.155 | 4 |
| Maximum | 2.718 | 0.993 | 15 |
| Standard Deviation | 0.290 | 0.068 | 2.47 |
| Image | Shannon Index | Simpson Index | Species Richness | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | p Value | R2 | RMSE | p Value | R2 | RMSE | p Value | |
| Sentinel 2 | 0.926 | 0.148 | <2.2 × | 0.917 | 0.043 | <2.2 × | 0.923 | 1.183 | <2.2 × |
| RapidEye | 0.902 | 0.147 | <2.2 × | 0.915 | 0.044 | <2.2 × | 0.833 | 1.287 | <2.2 × |
| PlanetScope | 0.898 | 0.156 | <2.2 × | 0.899 | 0.045 | <2.2 × | 0.900 | 1.293 | <2.2 × |
| Landsat 8 | 0.529 | 1.748 | <2.2 × | 0.410 | 0.063 | <2.2 × | 0.532 | 1.746 | <2.2 × |
| Satellite Image | Parameter | Shannon Index | Simpson Index | Species Richness |
|---|---|---|---|---|
| Sentinel 2 | Mean | 2.05 | 0.89 | 9.24 |
| Minimum | 1.51 | 0.55 | 6.58 | |
| Maximum | 2.34 | 0.95 | 12.7 | |
| Standard deviation | 0.15 | 0.04 | 1.33 | |
| RapidEye | Mean | 2.05 | 0.89 | 9.22 |
| Minimum | 1.40 | 0.53 | 5.73 | |
| Maximum | 2.44 | 0.94 | 13.48 | |
| Standard deviation | 0.18 | 0.04 | 1.41 | |
| PlanetScope | Mean | 2.05 | 0.89 | 9.26 |
| Minimum | 1.61 | 0.56 | 6.25 | |
| Maximum | 2.44 | 0.95 | 12.67 | |
| Standard deviation | 0.15 | 0.04 | 1.30 | |
| Landsat 8 | Mean | 2.06 | 0.89 | 9.04 |
| Minimum | 1.73 | 0.73 | 6.13 | |
| Maximum | 2.42 | 0.95 | 13.37 | |
| Standard deviation | 0.15 | 0.04 | 1.41 |
| Shannon Index | Simpson Index | Species Richness | |||
|---|---|---|---|---|---|
| Band | %IncMSE | Band | %IncMSE | Band | %IncMSE |
| SWIR1 | 17.75 | RE2 | 6.71 | SWIR1 | 18.57 |
| SWIR2 | 14.11 | SWIR1 | 6.05 | NNIR | 16.19 |
| RE2 | 13.52 | SWIR2 | 5.43 | SWIR2 | 13.27 |
| NNIR | 8.97 | NNIR | 4.50 | RE2 | 10.88 |
| NIR | 4.89 | NIR | −0.03 | NIR | 6.99 |
| Shannon Index | Simpson Index | Species Richness | |||
|---|---|---|---|---|---|
| Band | %IncMSE | Band | %IncMSE | Band | %IncMSE |
| Red | 19.64 | NIR | 6.28 | NIR | 12.58 |
| NIR | 14.24 | RE | 4.79 | RE | 9.25 |
| RE | 14.01 | Red | 1.91 | Red | 8.39 |
| Shannon Index | Simpson Index | Species Richness | |||
|---|---|---|---|---|---|
| Band | %IncMSE | Band | %IncMSE | Band | %IncMSE |
| Green | 9.93 | Green | 6.03 | NIR | 14.54 |
| Red | 9.91 | NIR | 5.83 | Green | 13.46 |
| Blue | 7.78 | Blue | 5.44 | Red | 12.78 |
| NIR | 7.04 | Red | 2.01 | Blue | 10.11 |
| Shannon Index | Simpson Index | Species Richness | |||
|---|---|---|---|---|---|
| Band | %IncMSE | Band | %IncMSE | Band | %IncMSE |
| SWIR1 | 18.01 | NIR | 8.36 | SWIR1 | 18.96 |
| NIR | 15.38 | SWIR1 | 7.87 | NIR | 16.71 |
| Green | 14.16 | Red | 5.11 | Red | 13.91 |
| Red | 11.96 | Green | 3.77 | Green | 13.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gyamfi-Ampadu, E.; Gebreslasie, M.; Mendoza-Ponce, A. RETRACTED: Evaluating Multi-Sensors Spectral and Spatial Resolutions for Tree Species Diversity Prediction. Remote Sens. 2021, 13, 1033. https://doi.org/10.3390/rs13051033
Gyamfi-Ampadu E, Gebreslasie M, Mendoza-Ponce A. RETRACTED: Evaluating Multi-Sensors Spectral and Spatial Resolutions for Tree Species Diversity Prediction. Remote Sensing. 2021; 13(5):1033. https://doi.org/10.3390/rs13051033
Chicago/Turabian StyleGyamfi-Ampadu, Enoch, Michael Gebreslasie, and Alma Mendoza-Ponce. 2021. "RETRACTED: Evaluating Multi-Sensors Spectral and Spatial Resolutions for Tree Species Diversity Prediction" Remote Sensing 13, no. 5: 1033. https://doi.org/10.3390/rs13051033
APA StyleGyamfi-Ampadu, E., Gebreslasie, M., & Mendoza-Ponce, A. (2021). RETRACTED: Evaluating Multi-Sensors Spectral and Spatial Resolutions for Tree Species Diversity Prediction. Remote Sensing, 13(5), 1033. https://doi.org/10.3390/rs13051033








