An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches
Abstract
1. Introduction
2. Materials and Methods
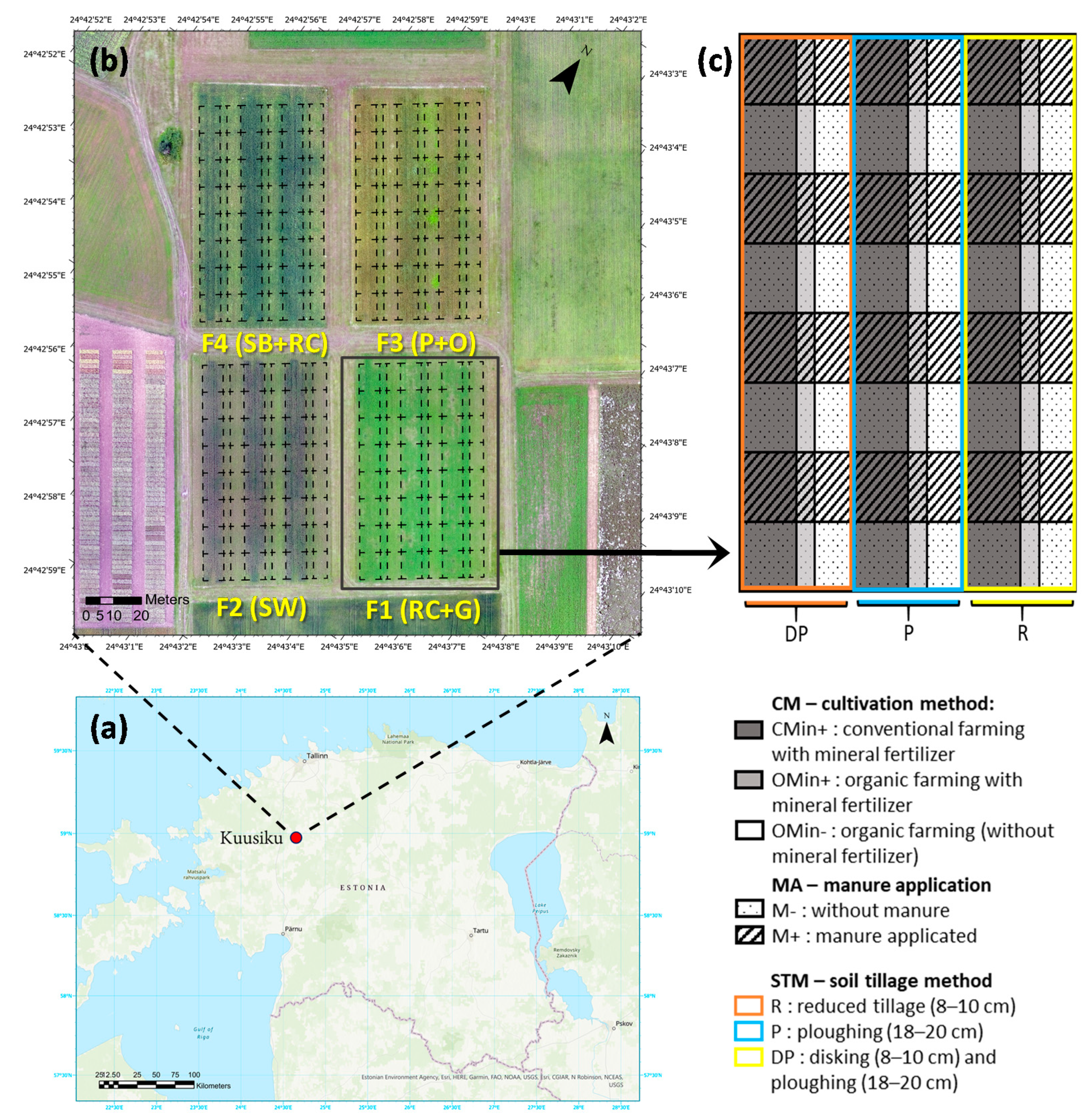
2.1. Study Area and Experiment Layout
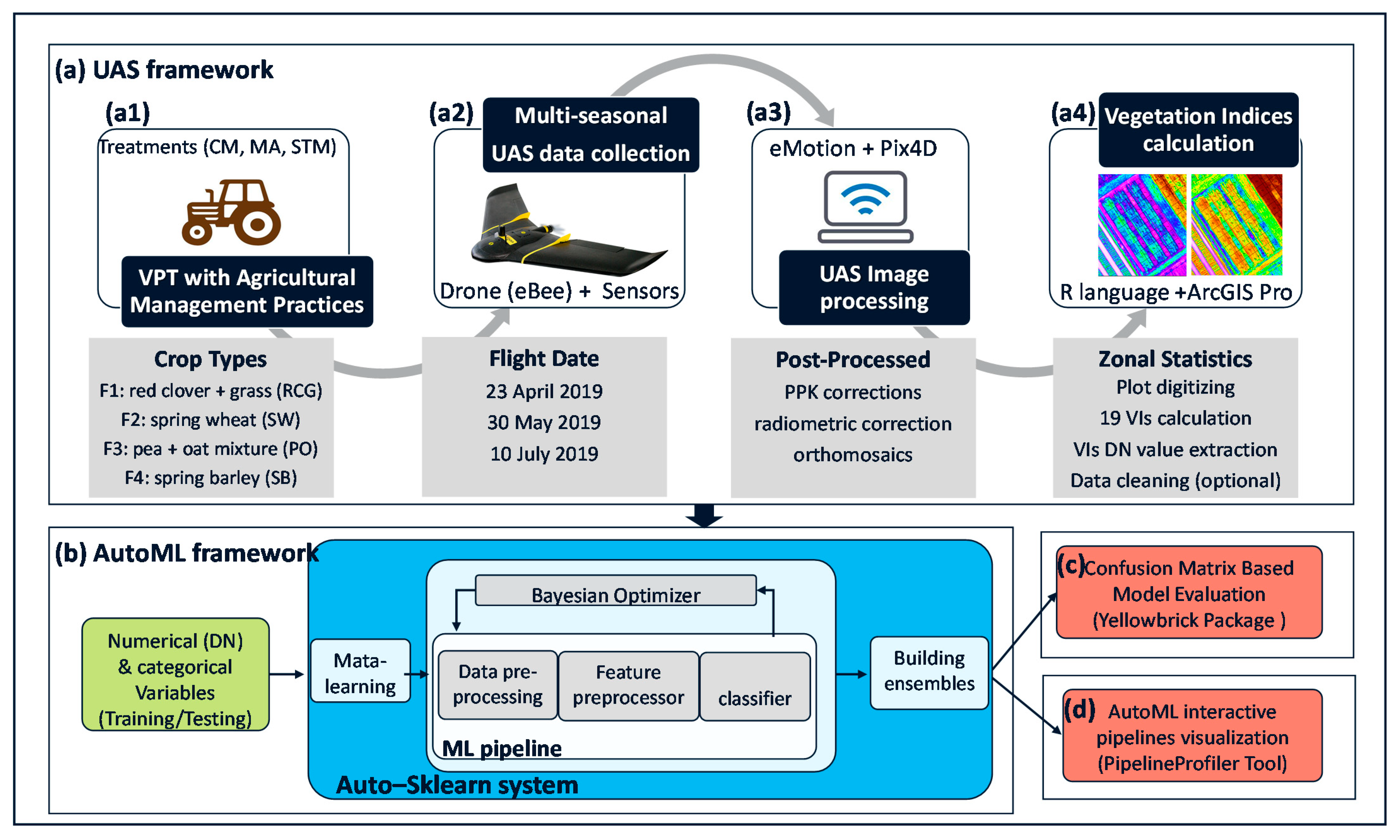
2.2. UAS Image Acquisition
2.3. UAS Image Processing
2.4. Vegetation Indices Calculation
2.5. Principal Component Analysis and VI Extraction
2.6. AutoML Modeling with Auto-Sklearn
2.7. AutoML Model Evaluation and Visualization
3. Results
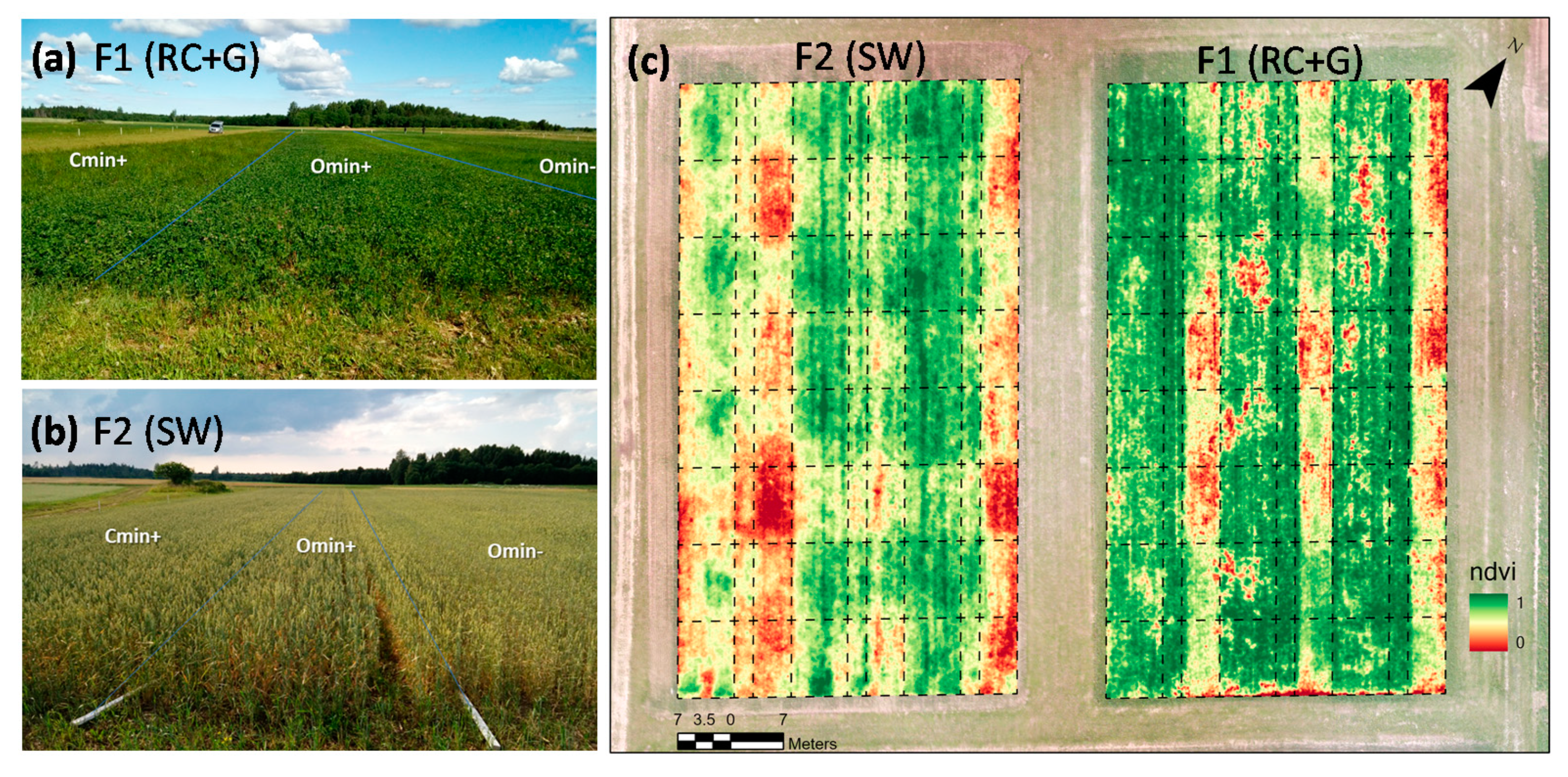
3.1. The AMPs Observation in VPTs and VIs Calculation
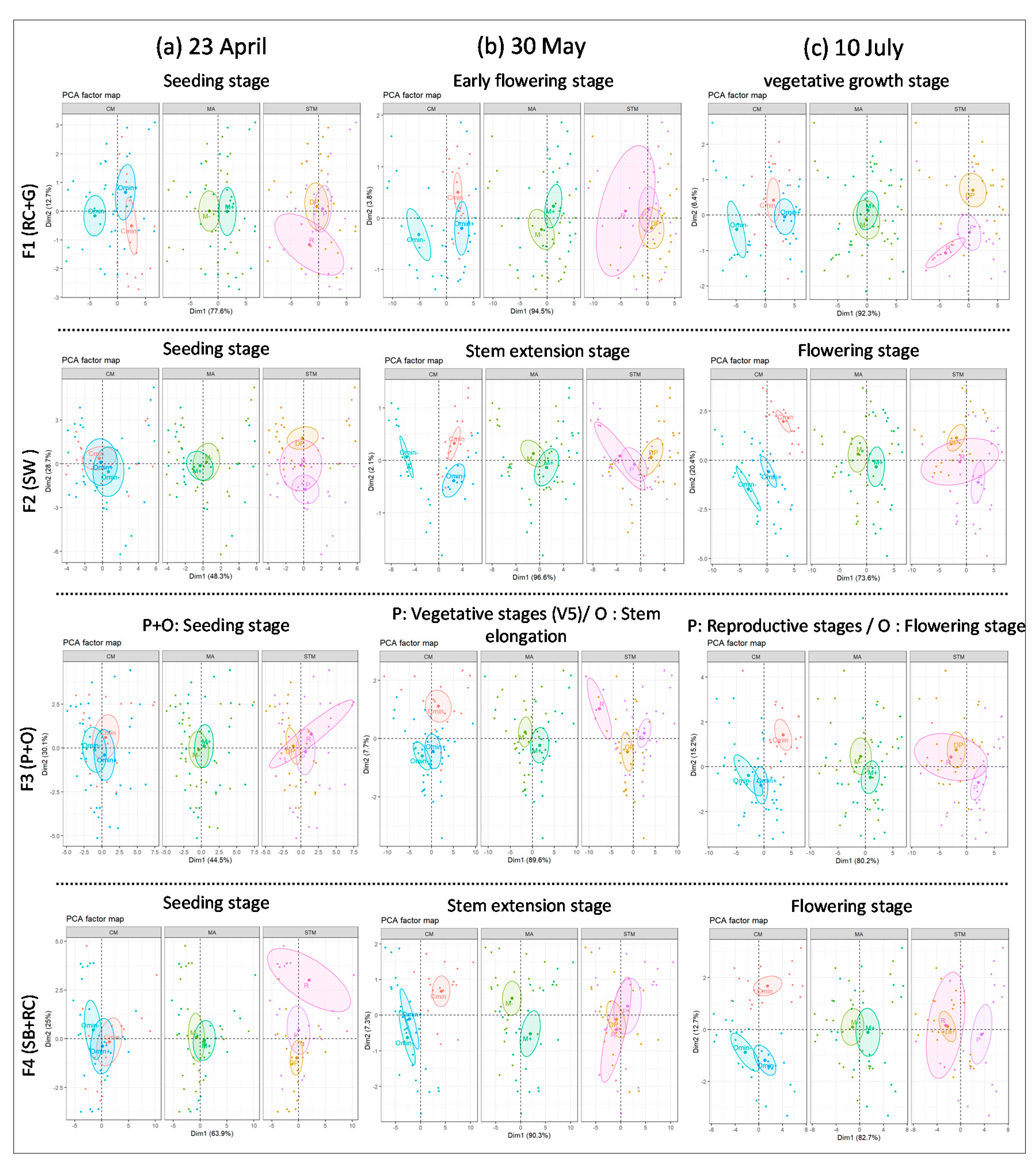
3.2. Monthly PCA Analysis in Various Crop Growth Periods
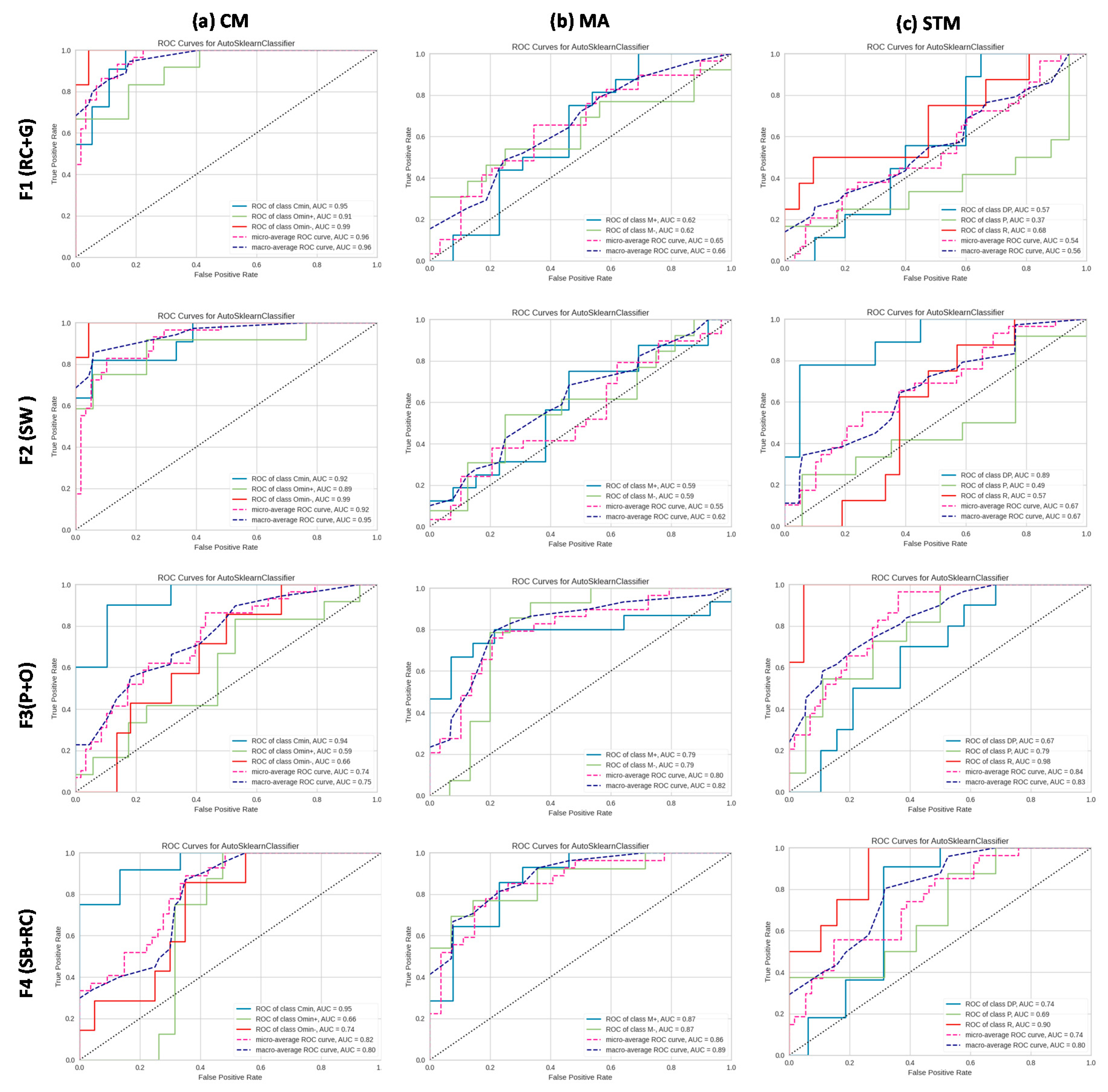
3.3. AutoML ROC and AUC Evaluation of AMP Recognition in May
3.4. AutoML Precision–Recall, Prediction Error, and Classification Report of CM Recognition
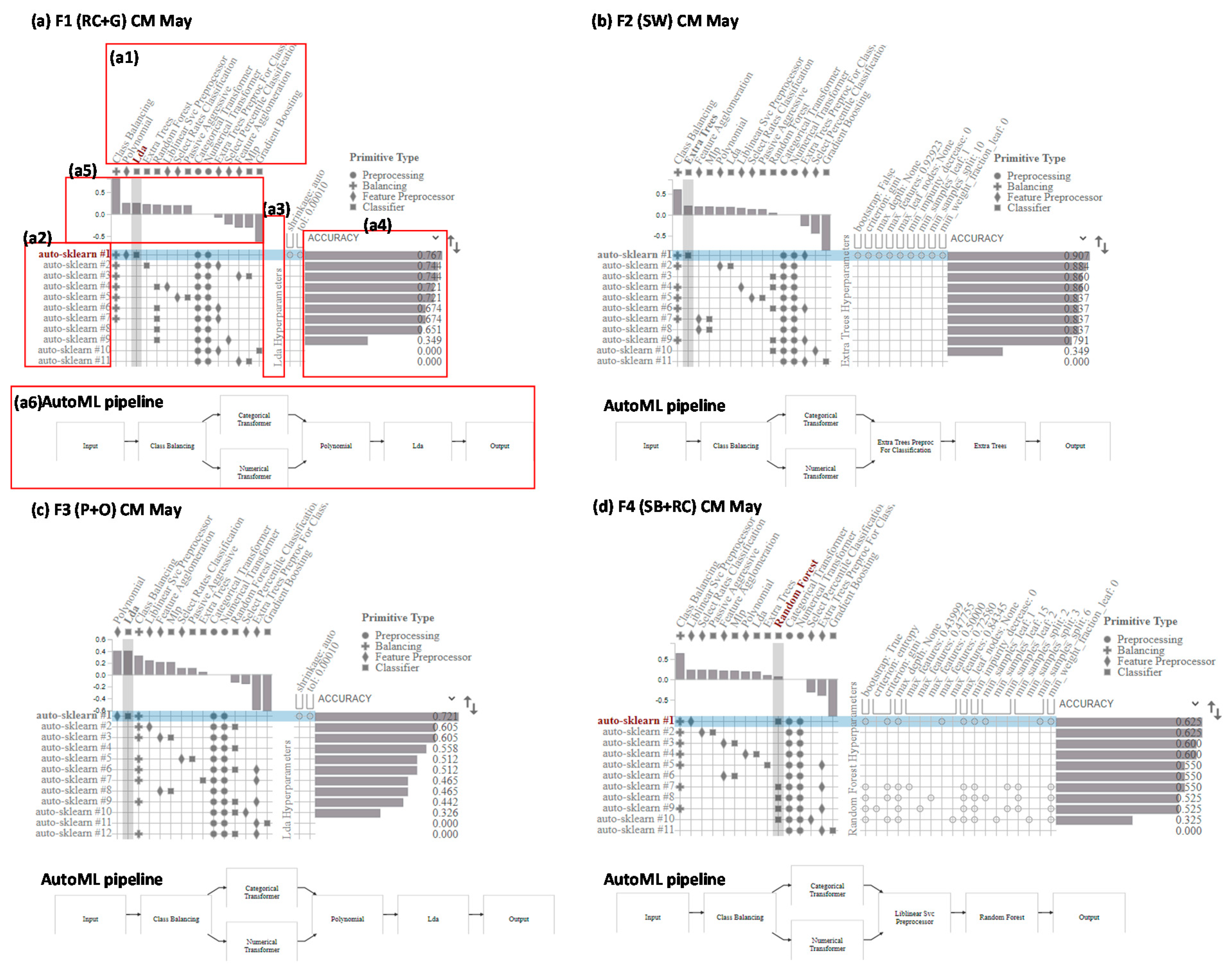
3.5. AutoML Pipeline Visualization
3.6. Comparison of Performance between AutoML and Other Machine Learning Technologies
4. Discussion
4.1. Applicability and the Impact of the AutoML Method in UAS
4.2. The Impact of Algorithm Selection, Cultivated Period, and Crop Types in AutoML AMP Recognization
4.3. The Limitations of Our Method
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Tripicchio, P.; Satler, M.; Dabisias, G.; Ruffaldi, E.; Avizzano, C.A. Towards Smart Farming and Sustainable Agriculture with Drones. In Proceedings of the 2015 International Conference on Intelligent Environments, IEEE, Rabat, Morocco, 15–17 July 2015; pp. 140–143. [Google Scholar] [CrossRef]
- Herwitz, S.R.; Johnson, L.F.; Dunagan, S.E.; Higgins, R.G.; Sullivan, D.V.; Zheng, J.; Lobitz, B.M.; Leung, J.G.; Gallmeyer, B.A.; Aoyagi, M.; et al. Imaging from an unmanned aerial vehicle: Agricultural surveillance and decision support. Comput. Electron. Agric. 2004, 44, 49–61. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Xiang, H.; Tian, L. Development of a low-cost agricultural remote sensing system based on an autonomous unmanned aerial vehicle (UAV). Biosyst. Eng. 2011, 108, 174–190. [Google Scholar] [CrossRef]
- Raeva, P.L.; Šedina, J.; Dlesk, A. Monitoring of crop fields using multispectral and thermal imagery from UAV. Eur. J. Remote Sens. 2018, 52, 192–201. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Carter, A.H. Field-based crop phenotyping: Multispectral aerial imaging for evaluation of winter wheat emergence and spring stand. Comput. Electron. Agric. 2015, 118, 372–379. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Andrade, J.; Edreira, J.I.R.; Mourtzinis, S.; Conley, S.; Ciampitti, I.; Dunphy, J.E.; Gaska, J.M.; Glewen, K.; Holshouser, D.L.; Kandel, H.J.; et al. Assessing the influence of row spacing on soybean yield using experimental and producer survey data. Field Crop. Res. 2018, 230, 98–106. [Google Scholar] [CrossRef]
- Laidig, F.; Piepho, H.-P.; Drobek, T.; Meyer, U. Genetic and non-genetic long-term trends of 12 different crops in German official variety performance trials and on-farm yield trends. Theor. Appl. Genet. 2014, 127, 2599–2617. [Google Scholar] [CrossRef]
- Lollato, R.P.; Roozeboom, K.; Lingenfelser, J.F.; Da Silva, C.L.; Sassenrath, G. Soft winter wheat outyields hard winter wheat in a subhumid environment: Weather drivers, yield plasticity, and rates of yield gain. Crop. Sci. 2020, 60, 1617–1633. [Google Scholar] [CrossRef]
- Zhu-Barker, X.; Steenwerth, K.L. Nitrous Oxide Production from Soils in the Future: Processes, Controls, and Responses to Climate Change, Chapter Six. In Climate Change Impacts on Soil Processes and Ecosystem Properties; Elsevier: Amsterdam, The Netherlands, 2018; pp. 131–183. [Google Scholar] [CrossRef]
- De Longe, M.S.; Owen, J.J.; Silver, W.L. Greenhouse Gas Mitigation Opportunities in California Agriculture: Review of California Rangeland Emissions and Mitigation Potential; Nicholas Institute for Environ, Policy Solutions Report; Duke University: Durham, NC, USA, 2014. [Google Scholar]
- Zhu-Barker, X.; Steenwerth, K.L. Nitrous Oxide Production from Soils in the Future. In Developments in Soil Science; Elsevier: Amsterdam, The Netherlands, 2018; Volume 35, pp. 131–183. [Google Scholar] [CrossRef]
- Crews, T.; Peoples, M. Legume versus fertilizer sources of nitrogen: Ecological tradeoffs and human needs. Agric. Ecosyst. Environ. 2004, 102, 279–297. [Google Scholar] [CrossRef]
- Munaro, L.B.; Hefley, T.J.; DeWolf, E.; Haley, S.; Fritz, A.K.; Zhang, G.; Haag, L.A.; Schlegel, A.J.; Edwards, J.T.; Marburger, D.; et al. Exploring long-term variety performance trials to improve environment-specific genotype × management recommendations: A case-study for winter wheat. Field Crop. Res. 2020, 255, 107848. [Google Scholar] [CrossRef]
- Gardner, J.B.; Drinkwater, L.E. The fate of nitrogen in grain cropping systems: A meta-analysis of 15N field experiments. Ecol. Appl. 2009, 19, 2167–2184. [Google Scholar] [CrossRef]
- Drinkwater, L.; Snapp, S. Nutrients in Agroecosystems: Rethinking the Management Paradigm. Adv. Agron. 2007, 92, 163–186. [Google Scholar] [CrossRef]
- van Ittersum, M.K.; Cassman, K.G.; Grassini, P.; Wolf, J.; Tittonell, P.; Hochman, Z. Yield gap analysis with local to global relevance—A review. Field Crop. Res. 2013, 143, 4–17. [Google Scholar] [CrossRef]
- Nawar, S.; Corstanje, R.; Halcro, G.; Mulla, D.; Mouazen, A.M. Delineation of Soil Management Zones for Variable-Rate Fertilization. Adv. Agron. 2017, 143, 175–245. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Z.; Chao, H.-C.; Guizani, M. Toward Intelligent Network Optimization in Wireless Networking: An Auto-Learning Framework. IEEE Wirel. Commun. 2019, 26, 76–82. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A survey of the state-of-the-art. Knowl. Based Syst. 2020, 212, 106622. [Google Scholar] [CrossRef]
- Mendoza, H.; Klein, A.; Feurer, M.; Springenberg, J.T.; Hutter, F. Towards Automatically-Tuned Neural Networks. In Proceedings of the Workshop on Automatic Machine Learning, New York, NY, USA, 24 June 2016. [Google Scholar]
- Zöller, M.-A.; Huber, M.F. Benchmark and Survey of Automated Machine Learning Frameworks. J. Artif. Intell. Res. 2021, 70, 409–472. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, M.; Chen, Y.; Dai, W.; Hu, Y.Q.; Li, Y.F.; Tu, W.W.; Yang, Q.; Yu, Y. Taking the Human out of Learning Applications: A Survey on Automated Machine Learning. arXiv 2018, arXiv:1810.13306. [Google Scholar]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined Selection and Hyperparame-ter Optimization of Classification Algorithms. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 847–855. [Google Scholar]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.T.; Blum, M.; Hutter, F. Auto-sklearn: Efficient and Robust Automated Machine Learning. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 113–134. [Google Scholar] [CrossRef]
- Remeseiro, B.; Bolon-Canedo, V. A review of feature selection methods in medical applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef]
- Babaeian, E.; Paheding, S.; Siddique, N.; Devabhaktuni, V.K.; Tuller, M. Estimation of root zone soil moisture from ground and remotely sensed soil information with multisensor data fusion and automated machine learning. Remote Sens. Environ. 2021, 260, 112434. [Google Scholar] [CrossRef]
- Ledell, E.; Poirier, S. H2O AutoML: Scalable Automatic Machine Learning. In Proceedings of the 7th ICML Workshop on Automated Machine Learning, Vienna, Austria, 17 July 2020; Available online: https://www.automl.org/wp-content/uploads/2020/07/AutoML_2020_paper_61.pdf?fbclid=IwAR2QaAJWDbgi1jIfnhK83x2g3hV6APfvTZoeUblcf4q44wxqT1z5oRTiEVo (accessed on 20 October 2020).
- Koh, J.C.O.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Jin, H.; Song, Q.; Hu, X. Auto-Keras: An Efficient Neural Architecture Search System. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Anchorage, AL, USA, 4–8 August 2019. [Google Scholar] [CrossRef]
- Komer, B.; Bergstra, J.; Eliasmith, C. Hyperopt-Sklearn: Automatic Hyperparameter Configuration for Scikit-Learn. In Proceedings of the 13th Python in Science Conference, Austin, TX, USA, 6–12 July 2014; pp. 34–40. [Google Scholar] [CrossRef]
- FAO. World Reference Base for Soil Resources; World Soil Resources Report 103; FAO: Rome, Italy, 2006; ISBN 9251055114. [Google Scholar]
- Poncet, A.M.; Knappenberger, T.; Brodbeck, C.; Fogle, J.M.; Shaw, J.N.; Ortiz, B.V. Multispectral UAS Data Accuracy for Different Radiometric Calibration Methods. Remote Sens. 2019, 11, 1917. [Google Scholar] [CrossRef]
- Kelcey, J.; Lucieer, A. Sensor Correction of a 6-Band Multispectral Imaging Sensor for UAV Remote Sensing. Remote Sens. 2012, 4, 1462–1493. [Google Scholar] [CrossRef]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Practical Automated Machine Learning for the AutoML Challenge 2018. In Proceedings of the International Workshop on Automatic Machine Learning at ICML, Stockholm, Sweden, 10–15 July 2018. [Google Scholar]
- Metsar, J.; Kollo, K.; Ellmann, A. Modernization of the Estonian National GNSS Reference Station Network. Geod. Cartogr. 2018, 44, 55–62. [Google Scholar] [CrossRef][Green Version]
- de Lima, R.; Lang, M.; Burnside, N.; Peciña, M.; Arumäe, T.; Laarmann, D.; Ward, R.; Vain, A.; Sepp, K. An Evaluation of the Effects of UAS Flight Parameters on Digital Aerial Photogrammetry Processing and Dense-Cloud Production Quality in a Scots Pine Forest. Remote Sens. 2021, 13, 1121. [Google Scholar] [CrossRef]
- Tomaštík, J.; Mokroš, M.; Surový, P.; Grznárová, A.; Merganič, J. UAV RTK/PPK Method—An Optimal Solution for Mapping Inaccessible Forested Areas? Remote Sens. 2019, 11, 721. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, W.; Zhou, Q. Reflectance Variation within the In-Chlorophyll Centre Waveband for Robust Retrieval of Leaf Chlorophyll Content. PLoS ONE 2014, 9, e110812. [Google Scholar] [CrossRef]
- Dong, T.; Meng, J.; Shang, J.; Liu, J.; Wu, B. Evaluation of Chlorophyll-Related Vegetation Indices Using Simulated Sentinel-2 Data for Estimation of Crop Fraction of Absorbed Photosynthetically Active Radiation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4049–4059. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus hippocastanum L. and Acer platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Merton, R.; Huntington, J. Early Simulation Results of the Aries-1 Satellite Sensor for Multi-Temporal Vege-tation Research Derived from Aviris. In Proceedings of the Eighth Annual JPL, Orlando, FL, USA, 7–14 May 1999; Available online: http://www.eoc.csiro.au/hswww/jpl_99.htm (accessed on 22 October 2020).
- Henebry, G.; Viña, A.; Gitelson, A. The Wide Dynamic Range Vegetation Index and Its Potential Utility for Gap Analysis. Papers in Natural Resources 2004. Available online: https://digitalcommons.unl.edu/natrespapers/262 (accessed on 22 October 2020).
- Wu, W. The Generalized Difference Vegetation Index (GDVI) for Dryland Characterization. Remote Sens. 2014, 6, 1211–1233. [Google Scholar] [CrossRef]
- Strong, C.J.; Burnside, N.G.; Llewellyn, D. The potential of small-Unmanned Aircraft Systems for the rapid detection of threatened unimproved grassland communities using an Enhanced Normalized Difference Vegetation Index. PLoS ONE 2017, 12, e0186193. [Google Scholar] [CrossRef]
- Kross, A.; McNairn, H.; Lapen, D.; Sunohara, M.; Champagne, C. Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 235–248. [Google Scholar] [CrossRef]
- Chen, P.F.; Tremblay, N.; Wang, J.H.; Vigneault, P.; Huang, W.J.; Li, B.G. New Index for Crop Canopy Fresh Biomass Estimation. Spectrosc. Spectr. Anal. 2010, 30, 512–517. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Vasudevan, A.; Kumar, D.A.; Bhuvaneswari, N.S. Precision farming using unmanned aerial and ground vehicles. In Proceedings of the 2016 IEEE International Conference on Technological Innovations in ICT for Agriculture and Rural Development, TIAR, Chennai, India, 15–16 July 2016; pp. 146–150. [Google Scholar] [CrossRef]
- Ballester, C.; Brinkhoff, J.; Quayle, W.C.; Hornbuckle, J. Monitoring the Effects of Water Stress in Cotton using the Green Red Vegetation Index and Red Edge Ratio. Remote Sens. 2019, 11, 873. [Google Scholar] [CrossRef]
- Feng, H.; Tao, H.; Zhao, C.; Li, Z.; Yang, G. Comparison of UAV RGB Imagery and Hyperspectral Remote-Sensing Data for Monitoring Winter-Wheat Growth. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Cross, M.D.; Scambos, T.; Pacifici, F.; Marshall, W.E. Determining Effective Meter-Scale Image Data and Spectral Vegetation Indices for Tropical Forest Tree Species Differentiation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 2934–2943. [Google Scholar] [CrossRef]
- Datt, B. Remote Sensing of Chlorophyll a, Chlorophyll b, Chlorophyll a+b, and Total Carotenoid Content in Eucalyptus Leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Crippen, R. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Sripada, R.P.; Heiniger, R.W.; White, J.; Meijer, A.D. Aerial Color Infrared Photography for Determining Early In-Season Nitrogen Requirements in Corn. Agron. J. 2006, 98, 968–977. [Google Scholar] [CrossRef]
- Gianelle, D.; Vescovo, L. Determination of green herbage ratio in grasslands using spectral reflectance. Methods and ground measurements. Int. J. Remote Sens. 2007, 28, 931–942. [Google Scholar] [CrossRef]
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; Final Report, RSC 1978-4; Texas A&M University: College Station, TX, USA, 1974. [Google Scholar]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide Dynamic Range Vegetation Index for Remote Quantification of Biophysical Characteristics of Vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef]
- Kambhatla, N.; Leen, T.K. Dimension Reduction by Local Principal Component Analysis. Neural Comput. 1997, 9, 1493–1516. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- ESRI. ArcGIS PRO: Essential Workflows; ESRI: Redlands, CA, USA, 2016; Available online: https://community.esri.com/t5/esritraining-documents/arcgis-pro-essential-workflows-course-resources/ta-p/914710 (accessed on 1 May 2021).
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Auto-Sklearn 2.0: The Next Generation. arXiv 2020, arXiv:2007.04074. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Sequential Model-Based Optimization for General Algorithm Configuration. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Rome, Italy, 17–21 January 2011; pp. 507–523. [Google Scholar] [CrossRef]
- Suykens, J.; Vandewalle, J. Least Squares Support Vector Machine Classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Olson, R.S.; Urbanowicz, R.J.; Andrews, P.C.; Lavender, N.A.; Kidd, L.C.; Moore, J.H. Automating Biomedical Data Science Through Tree-Based Pipeline Optimization. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Porto, Portugal, 30 March–1 April 2016; pp. 123–137. [Google Scholar] [CrossRef]
- Feurer, M.; Springenberg, J.T.; Hutter, F. Initializing Bayesian Hyperparameter Optimization via Me-ta-Learning. In Proceedings of the Proceedings of the National Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 2. Available online: https://ojs.aaai.org/index.php/AAAI/article/view/9354 (accessed on 20 October 2020).
- Franceschi, L.; Frasconi, P.; Salzo, S.; Grazzi, R.; Pontil, M. Bilevel Programming for Hyperparameter Opti-mization and Meta-Learning. arXiv 2018, arXiv:1806.04910. [Google Scholar]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- Kumar, A.; McCann, R.; Naughton, J.; Patel, J.M. Model Selection Management Systems: The Next Frontier of Advanced Analytics. ACM SIGMOD Rec. 2016, 44, 17–22. [Google Scholar] [CrossRef]
- Bengfort, B.; Bilbro, R. Yellowbrick: Visualizing the Scikit-Learn Model Selection Process. J. Open Source Softw. 2019, 4, 1075. [Google Scholar] [CrossRef]
- Anwar, M.Z.; Kaleem, Z.; Jamalipour, A. Machine Learning Inspired Sound-Based Amateur Drone Detection for Public Safety Applications. IEEE Trans. Veh. Technol. 2019, 68, 2526–2534. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2005, 27, 861–874. [Google Scholar] [CrossRef]
- Pencina, M.J.; Agostino, R.B.D.; Vasan, R.S. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat. Med. 2007, 27, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Gunčar, G.; Kukar, M.; Notar, M.; Brvar, M.; Černelč, P.; Notar, M.; Notar, M. An application of machine learning to haematological diagnosis. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Boyd, K.; Eng, K.H.; Page, C.D. Area under the Precision-Recall Curve: Point Estimates and Confidence In-tervals. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Prague, Czech Republic, 23–27 September 2013; Volume 8190 LNAI. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ming, Y.; Jin, Z.; Shen, Q.; Liu, D.; Smith, M.J.; Veeramachaneni, K.; Qu, H. ATMSeer: Increasing Transparency and Controllability in Automated Machine Learning. arXiv 2019, arXiv:1902.05009. [Google Scholar]
- Ono, J.P.; Castelo, S.; Lopez, R.; Bertini, E.; Freire, J.; Silva, C. PipelineProfiler: A Visual Analytics Tool for the Exploration of AutoML Pipelines. IEEE Trans. Vis. Comput. Graph. 2021, 27, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Serpico, S.B.; D’Inca, M.; Melgani, F.; Moser, G. Comparison of Feature Reduction Techniques for Classification of Hyperspectral Remote Sensing Data. In Proceedings of the Image and Signal Processing for Remote Sensing VIII, Crete, Greece, 13 March 2003; Volume 4885. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Samaras, S.; Diamantidou, E.; Ataloglou, D.; Sakellariou, N.; Vafeiadis, A.; Magoulianitis, V.; Lalas, A.; Dimou, A.; Zarpalas, D.; Votis, K.; et al. Deep Learning on Multi Sensor Data for Counter UAV Applications—A Systematic Review. Sensors 2019, 19, 4837. [Google Scholar] [CrossRef]
- David, L.C.; Ballado, A.J. Vegetation indices and textures in object-based weed detection from UAV imagery. In Proceedings of the 6th IEEE International Conference on Control System, Computing and Engineering, ICCSCE, Penang, Malaysia, 25–27 November 2016. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Lopez-Granados, F.; De Castro, A.I.; Peña-Barragan, J.M. Configuration and Specifications of an Unmanned Aerial Vehicle (UAV) for Early Site Specific Weed Management. PLoS ONE 2013, 8, e58210. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Sathuvalli, V.R.; VanDeMark, G.J.; Miklas, P.N.; Carter, A.; Pumphrey, M.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Chawade, A.; Van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Young, S.N. A Framework for Evaluating Field-Based, High-Throughput Phenotyping Systems: A Meta-Analysis. Sensors 2019, 19, 3582. [Google Scholar] [CrossRef]
- Vivaldini, K.C.T.; Martinelli, T.H.; Guizilini, V.C.; Souza, J.; Oliveira, M.D.; Ramos, F.T.; Wolf, D.F. UAV route planning for active disease classification. Auton. Robot. 2018, 43, 1137–1153. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A Deep Learning-Based Approach for Automated Yellow Rust Disease Detection from High-Resolution Hyperspectral UAV Images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Villoslada, M.; Bergamo, T.; Ward, R.; Burnside, N.; Joyce, C.; Bunce, R.; Sepp, K. Fine scale plant community assessment in coastal meadows using UAV based multispectral data. Ecol. Indic. 2020, 111, 105979. [Google Scholar] [CrossRef]
- Burnside, N.G.; Joyce, C.B.; Puurmann, E.; Scott, D.M. Use of Vegetation Classification and Plant Indicators to Assess Grazing Abandonment in Estonian Coastal Wetlands. J. Veg. Sci. 2007, 18, 645–654. [Google Scholar] [CrossRef]
- Sona, G.; Passoni, D.; Pinto, L.; Pagliari, D.; Masseroni, D.; Ortuani, B.; Facchi, A. UAV multispectral survey to map soil and crop for precision farming applications. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B1, 1023–1029. [Google Scholar] [CrossRef]
- Kwak, G.-H.; Park, N.-W. Impact of Texture Information on Crop Classification with Machine Learning and UAV Images. Appl. Sci. 2019, 9, 643. [Google Scholar] [CrossRef]
- Yang, M.-D.; Tseng, H.-H.; Hsu, Y.-C.; Tsai, H.P. Semantic Segmentation Using Deep Learning with Vegetation Indices for Rice Lodging Identification in Multi-date UAV Visible Images. Remote Sens. 2020, 12, 633. [Google Scholar] [CrossRef]
- Yang, M.-D.; Boubin, J.G.; Tsai, H.P.; Tseng, H.-H.; Hsu, Y.-C.; Stewart, C.C. Adaptive autonomous UAV scouting for rice lodging assessment using edge computing with deep learning EDANet. Comput. Electron. Agric. 2020, 179, 105817. [Google Scholar] [CrossRef]
- Najafi, P.; Feizizadeh, B.; Navid, H. A Comparative Approach of Fuzzy Object Based Image Analysis and Machine Learning Techniques Which Are Applied to Crop Residue Cover Mapping by Using Sentinel-2 Satellite and UAV Imagery. Remote Sens. 2021, 13, 937. [Google Scholar] [CrossRef]
- Duro, D.; Franklin, S.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Kim, H.-O.; Yeom, J.-M. A Study on Object-Based Image Analysis Methods for Land Cover Classification in Agricultural Areas. J. Korean Assoc. Geogr. Inf. Stud. 2012, 15, 26–41. [Google Scholar] [CrossRef][Green Version]
- Telles, T.S.; Reydon, B.P.; Maia, A.G. Effects of no-tillage on agricultural land values in Brazil. Land Use Policy 2018, 76, 124–129. [Google Scholar] [CrossRef]
- Yeom, J.; Jung, J.; Chang, A.; Ashapure, A.; Maeda, M.; Maeda, A.; Landivar, J. Comparison of Vegetation Indices Derived from UAV Data for Differentiation of Tillage Effects in Agriculture. Remote Sens. 2019, 11, 1548. [Google Scholar] [CrossRef]
- Bisong, E. Google AutoML: Cloud Vision. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer Nature: Cham, Switzerland, 2019; pp. 581–598. [Google Scholar] [CrossRef]

| Vegetation Index | Equation | Reference |
|---|---|---|
| Datt4 | ρ R/(ρ G * ρ REG) | [61] |
| Green Infrared Percentage Vegetation Index (GIPVI) | ρ NIR/(ρ NIR + ρ G) | [62] |
| Green Normalized Difference Vegetation Index (GNDVI) | (ρ NIR − ρ G)/(NIR + ρ G) | [63] |
| Green Difference Vegetation Index (GDVI) | ρ NIR − ρ G | [64] |
| Green Ration Vegetation Index (GRVI) | ρ NIR/ρ G | [64] |
| Green Difference Index (GDI) | ρ NIR − ρ R + ρ G | [65] |
| Green Red Difference Index (GRDI) | (ρ G − ρ R)/(ρ G + ρ R) | [65] |
| Normalized Difference Vegetation Index (NDVI) | (ρ NIR − ρ R)/(ρ NIR + ρ R) | [66] |
| Red-Edge Normalized Difference Vegetation Index (NDVIre) | (ρ NIR − ρ REG)/(ρ NIR + ρ REG) | [46] |
| Red-Edge Simple Ratio (SRre) | ρ NIR/ρ REG | [46] |
| Renormalized Difference Vegetation Index (RDVI) | ((ρ NIR − ρ R)/((ρ NIR + ρ R) ** (0.5))) | [67] |
| Red-Edge Modified Simple Ratio (MSRre) | ((ρ NIR − ρ REG) − 1)/(((ρ NIR + ρ REG) ** (0.5)) + 1) | [49] |
| Red-Edge Triangular Vegetation Index (RTVIcore) | (100 * (ρ NIR − ρ REG)) − (10 * (ρ NIR − ρ G)) | [55] |
| Red-Edge Vegetation Stress Index (RVSI) | ((ρ R + ρ NIR)/2) − ρ REG | [50] |
| Red-Edge Greenness Vegetation Index (REGVI) | (ρ REG − ρ G)/(ρ REG + ρ G) | [68] |
| Simple Ratio (SR) | ρ NIR/ρ R | [69] |
| Modified Simple Ratio (MSR) | ((ρ NIR − ρ R) − 1)/(((NIR + ρ R) ** (0.5)) + 1) | [48] |
| Modified Triangular Vegetation Index (MTVI) | 1.2 * ((1.2 * (ρ NIR − ρ G)) − (2.5 * (ρ R − ρ G))) | [47] |
| Wide Dynamic Range Vegetation Index (WDRVI) | (((0.2 * ρ NIR) − ρ R)/((0.2 * ρ NIR) + ρ R)) | [70] |
| Parameter Name | Range Value | Description |
|---|---|---|
| time_left_for_this_task | 60–1200 s | The time limit for the search of appropriate models. |
| per_run_time_limit | 10 s | The time limit for a single call to the machine learning model. |
| ensemble_size | 50 (default) | The number of models added to the ensemble built by Ensemble selection from libraries of models. |
| ensemble_nbest | 50 (default) | The number of best models for building an ensemble model. |
| resampling_strategy | CV; folds = 3 | (CV = cross-validation); to handle overfitting |
| seed | 47 | Used to seed SMAC. |
| training/testing split | (0.6; 0.4) | Data partitioning way |
| Indices | Equations |
|---|---|
| Recall | TP/(TP + FN) |
| Precision | TP/(TP + FP) |
| Specificity | TN/(TN + FP) |
| Accuracy | TP/(TP + TN + FP + FN) |
| F1-score | 2 * Precision * Recall/(Precision + Recall) |
| False Positive Rate (FPR) | 1 − Specificity = FP/(FP + TN) |
| True Positive Rate (TPR) | Sensitivity = TP/(TP + FN) |
| ML Algorithms | ||||||
|---|---|---|---|---|---|---|
| Field | AMPs | AutoML (1200 s Run) | AutoML (60 s Run) | RF | SVM | ANN |
| F1 (RC + G) | CM | 0.79 | 0.76 ** | 0.79 | 0.83 | 0.86 * |
| MA | 0.59 | 0.62 * | 0.62 * | 0.62 * | 0.55 ** | |
| STM | 0.57 * | 0.31 | 0.48 | 0.38 ** | 0.48 | |
| F2 (WS) | CM | 0.79 | 0.79 | 0.79 | 0.83 * | 0.72 ** |
| MA | 0.55 * | 0.52 | 0.48 | 0.52 | 0.45 ** | |
| STM | 0.52 * | 0.45 ** | 0.48 | 0.45 ** | 0.52 * | |
| F3 (P + O) | CM | 0.55 * | 0.41 ** | 0.55 * | 0.48 | 0.55 * |
| MA | 0.66 | 0.72 | 0.76 * | 0.62 ** | 0.76 * | |
| STM | 0.66 | 0.69 * | 0.69 * | 0.57 ** | 0.59 | |
| F4 (SB + RC) | CM | 0.57 | 0.59 * | 0.56 | 0.59 * | 0.48 ** |
| MA | 0.85 * | 0.78 | 0.67 | 0.78 | 0.63 ** | |
| STM | 0.56 | 0.59 | 0.59 | 0.52 ** | 0.63 * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.-Y.; Burnside, N.G.; de Lima, R.S.; Peciña, M.V.; Sepp, K.; Cabral Pinheiro, V.H.; de Lima, B.R.C.A.; Yang, M.-D.; Vain, A.; Sepp, K. An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches. Remote Sens. 2021, 13, 3190. https://doi.org/10.3390/rs13163190
Li K-Y, Burnside NG, de Lima RS, Peciña MV, Sepp K, Cabral Pinheiro VH, de Lima BRCA, Yang M-D, Vain A, Sepp K. An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches. Remote Sensing. 2021; 13(16):3190. https://doi.org/10.3390/rs13163190
Chicago/Turabian StyleLi, Kai-Yun, Niall G. Burnside, Raul Sampaio de Lima, Miguel Villoslada Peciña, Karli Sepp, Victor Henrique Cabral Pinheiro, Bruno Rucy Carneiro Alves de Lima, Ming-Der Yang, Ants Vain, and Kalev Sepp. 2021. "An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches" Remote Sensing 13, no. 16: 3190. https://doi.org/10.3390/rs13163190
APA StyleLi, K.-Y., Burnside, N. G., de Lima, R. S., Peciña, M. V., Sepp, K., Cabral Pinheiro, V. H., de Lima, B. R. C. A., Yang, M.-D., Vain, A., & Sepp, K. (2021). An Automated Machine Learning Framework in Unmanned Aircraft Systems: New Insights into Agricultural Management Practices Recognition Approaches. Remote Sensing, 13(16), 3190. https://doi.org/10.3390/rs13163190








