2.3. Laser Scanner Data
ALS data were acquired under leaf-on conditions using a fixed-wing aircraft. The acquisition took place on 18 June 2017 using a LMS-Q1560 laser scanner system (Riegel, Horn, Austria) and was part of the governmental effort to construct a new detailed terrain model for Norway. The study area was located within a 1169 km
2 ALS block for which ground control points were established across the entire block for calibration of the height of the laser measurements. The contracted minimum point density for the block expressed as number of first echoes per 10 m × 10 m cell tessellating the block was 5 points m
−2. The data satisfied this criterion within our study area. In fact, in certain parts of the 12 ha study area the point density was >25 points m
−2 due to side overlap between adjacent, parallel strips and a single flight line perpendicular to the main direction of the scanned block. This dataset was used by the data vendor (TerraTec, Oslo, Norway) to produce the official national terrain model by classifying the points as ground and non-ground echoes using the progressive triangular irregular network (TIN) densification algorithm [
30] in the TerraScan software [
31].
The official national terrain model was used as terrain reference surface for the study. However, a harmonization of the point density was considered important because order statistics were used in the analysis of the tree and vegetation height. Order statistics, such as maximum height, are monotone increasing functions of number of points for a given target area [
32]. In order to keep the point density stable across the study area, we discarded all data from the perpendicular flight line and from the overlap zone between adjacent, parallel strips. The resulting mean point density across the 12 ha area was reduced to 6.5 points m
−2 for “first” and “single” echoes.
Normalized height values were computed for all “first” and “single” echoes relative to the official TIN by linear interpolation. Only “first” and “single” echoes with normalized height values were used in the subsequent analysis. All classified ground and non-ground points with negative normalized height values were assigned the value zero. All classified ground points were assumed to lie on the official terrain surface and where therefore assigned the value zero.
2.4. Unmanned Aerial Systems Image Data
UAS image data were acquired under leaf-on conditions using a eBee fixed-wing drone (senseFly Ltd, Cheseaux-Lausanne, Switzerland) weighing approximately 0.41 kg without payload [
33]. The acquisition took place on 21 June 2017, three days after the ALS acquisition, using a Canon IXUS127 HS (Canon Inc., Tokyo, Japan) red, green, and blue camera producing three separate 16.1 megapixel images in the red (660 nm), green (520 nm), and blue (450 nm) wavelengths. The drone was equipped with an inertial measurement unit and an on-board Global Navigation Satellite System (GNSS) to control the flight parameters and provide rough positioning during flight operations [
33]. The eBee flight plan was managed through senseFly’s eMotion 2 software, ver. 2 [
33], installed on a laptop computer. The longitudinal and lateral image overlaps were set to 90% and 80% respectively, although only a longitudinal overlap of 70% was achieved during the survey. The ground pixel resolution was set to 3.9 cm.
Prior to the image acquisition, the position of ten ground control points (GCPs) were determined and measured using the same RTK-based procedure as the one used to record positions of the GRPs. The GCP targets consisted of a set of 1 × 1 cross-shaped 4 cm × 46 cm timber planks painted orange to insure good contrast with the background vegetation.
The UAS images were processed in Agisoft PhotoScan Professional software, ver. 1.4.3 (Agisoft LLC, St.Petersburg, Russia), to produce a 3D point cloud [
34]. The processing steps followed in the PhotoScan software together with the parameters used are described in
Table 3.
After initial testing, an adaptive camera model fitting was used to perform the alignment. This function automatically selects the camera parameters to be included in the adjustment based on their reliability. The position of the GCPs were imported in the software to improve the estimates of the camera position and orientation. The GCP positions were manually refined and the camera alignment was optimized based on the GCPs to allow a more accurate model reconstruction. The average RMSEs associated with the estimated camera and GCP locations compared to the PhotoScan-estimated values were 0.92 m and 0.06 m, respectively. A dense point cloud was constructed using a medium quality parameter to reduce excessive processing time and a mild depth filtering parameter to remove outliers and reduce noise while allowing height variation between the 3D points. The point density of the resulting dense point cloud was around 50 points m−2.
At this point we would like to clarify that the DAP methodology applied in this study is what is commonly known in the literature as structure-from-motion (SfM). Using an iterative least-squares solution, camera position and orientation, and scene geometry are simultaneously reconstructed by identification of matching features, or tie points, in multiple images. The output from SfM is fixed into a relative, not absolute, coordinate system [
36]. The GCPs were used to transform the data to the absolute coordinate system adopted in this study. For the sake of simplicity, we refer to this methodology as DAP.
Normalized height values were computed for the DAP points relative to the official TIN by linear interpolation. Because the absolute height values of the DAP data were determined according to the elevation of the ten GCPs, we compared the elevation of the GCPs to the elevation of the official TIN. There was a mean difference in elevation of 0.055 m with a standard deviation of the differences of 0.028 m. The mean difference was subtracted from the normalized heights of all the DAP points. All DAP points with negative normalized height values after this subtraction were assigned the value zero.
2.6. Analysis—Objective #1
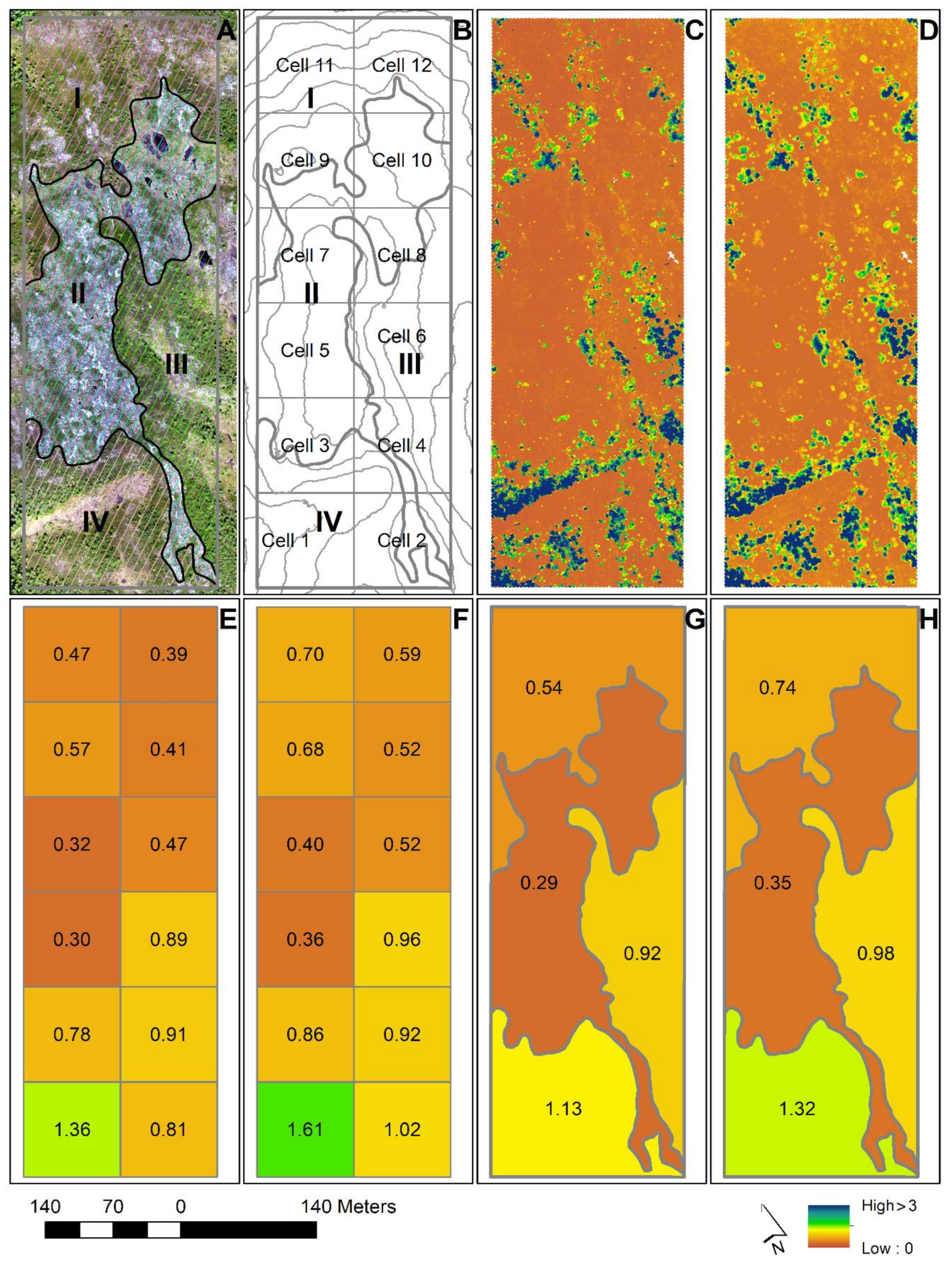
Under objective #1, we assessed and compared the performance of 3D remotely sensed data from ALS and from DAP for prediction of tree height of small pioneer trees and evaluated how tree size and tree species affected the predictive ability of the two types of 3D data. The main steps of the analysis are shown in
Figure 3 for the sake of clarity and overview.
A first step of the assessment was to analyse to what extent the different 3D data were sensitive to the small trees, i.e., if positive height values of the point clouds could be expected for a tree. Previous research (e.g., [
9]) suggests that this will depend on factors such as tree height, size of the tree crown, tree species, the point density of the remotely sensed data which in the current study clearly differed between ALS and the DAP 3D data (
Section 2.3 and
Section 2.4), and degree of laser pulse penetration into the tree crowns for ALS as opposed to a likely depiction of the outer surface of a tree crown with DAP data.
A logistic regression analysis with binary response supported this assessment. Among the 532 field-measured trees (
Table 1), two trees had a substantially higher maximum height in the 3D remotely sensed datasets (1.51–2.86 m) than field-measured tree height. These two trees (trees #48 and #2100) had likely overhanging branches from taller, neighbouring trees and they were discarded from all subsequent analysis. They were both spruce trees. Fifteen trees with maximum heights in the remotely sensed datasets 0.20–0.97 cm greater than the corresponding field-measured tree heights were retained because we were unable to identify a specific reason for this pattern. We were thus rather conservative in the treatment of potential outliers. The logistic regression analysis was based on the remaining 530 trees (
Table 4).
For each of the two remotely sensed datasets (ALS, DAP) every tree was classified as POSITIVE if the maximum height for the tree polygon (
hALSmax or
hDAPmax) had a positive value. If the maximum value was zero or the tree polygon did not contain any points for a given 3D remotely sensed dataset, the tree was classified as ZERO. The analysis was carried out in two steps. First, a general logistic regression model reflecting all effects mentioned above was fitted. This model of the probability of POSITIVE was formulated as follows:
where
is the probability of maximum height of a tree polygon with a value greater than zero using observations from datasets (DATA) ALS and DAP. DATA
DAP is a dummy variable for DAP (DATA
DAP = 1 if DAP). Further, SP
pine is a dummy variable for pine (SP
pine = 1 if pine), SP
birch is a dummy variable for birch (SP
birch = 1 if birch),
h (m) is the tree height measured in field, and
A (m
2) is the elliptic tree crown area according to the field recordings of crown diameters. The betas (
,
,
,
,
,
) are parameters to be estimated. Maximum-likelihood computation for fitting of the logistic model in Equation (1) was performed with the LOGISTIC procedure of the SAS package [
37].
It should be noted that the reference in the model is the ALS dataset and the tree species spruce. Thus, the estimated parameters for the DATA and SP variables express differences relative to this reference (differences in intercept of the model). The effects of, for example, DAP relative to ALS will be expressed directly by the parameter estimate of the former variable. Finally, a Wald chi-square test was performed to test the null hypothesis that the parameter estimates for the two dummy variables for tree species were equal.
One of the results of the first step of the logistic regression analysis was that the effects of tree species on probability of detected trees differed significantly in the statistical sense between some of the species (p < 0.001, p = 0.037, and p = 0.080, respectively), see Table 7. On the other hand, the effect of dataset was not significant in the statistical sense (p = 0.733; Table 7). Further, both tree height and crown area were statistically significant (p < 0.001, Table 7).
Although some effects in the basic model in Equation (1) were significant and others not, some of the effects are likely confounded which may lead to incorrect interpretations. For example, the point density of the DAP point cloud was around 50 points m
−2 whereas the corresponding density in the ALS data was 6.5 points m
−2. It is therefore reasonable that the area of a tree crown polygon is more critical for a crown polygon having a positive height value in the ALS data than in the DAP data. Likewise, tree species may affect the probability of positive height values differently in the two 3D remotely sensed datasets since laser pulses tend to penetrate the tree crowns before an echo is triggered while DAP may better capture the surface of a crown. Crowns of different species have different densities of biological matter (foliage and branches) and different shapes which may influence the point clouds for the two 3D remote sensing techniques differently. A more complex model was therefore formulated. In the model in Equation (1), it was assumed that the effect of dataset was similar for each individual tree species, i.e., that the different datasets only affected the intercept of the model. In addition to the basic effects accommodated by Equation (1), we allowed the effects of tree species to vary between the two 3D remotely sensed datasets. This was accommodated by introducing separate regression coefficients for tree species for the different datasets. Further, in the former model, it was assumed that the effect of dataset was constant across the entire range of tree heights and tree crown areas. In the second step of the analysis, we allowed the effects of dataset to vary according to the magnitude of the tree height and the tree crown area as well. This was accommodated by introducing separate regression coefficients for tree height and crown area for each individual dataset in the model:
Similar to the model in Equation (1), the ALS dataset and the species spruce represent the reference in the model in Equation (2). Thus, the estimated parameter for the DATA variable (β1) will express the overall difference in intercept relative to ALS, while parameters for the two SP variables (β2 and β3) will express the overall difference in intercept relative to spruce. The height and crown area parameter estimates (β6 and β8) will express the general effects of these two variables. The estimated parameters for the respective products of the DATA variable and the two species variables (β4 and β5), and the DATA variable and height and crown area (β7 and β9), will express differences in parameters for pine, birch, h, and A for DAP relative to ALS. Finally, a Wald chi-square test was performed to test the null hypothesis that the parameter estimates for pine and birch (β2 and β3) were equal. Likewise, Wald chi-square tests were performed to test the null hypotheses that the parameter estimates of the products of the DATA variable and the two SP variables (β4 and β5) were equal.
The second part of the analysis under objective #1 entailed modeling of tree height of the pioneer trees and evaluation of how tree size and tree species affect the predictive ability of the two types of the 3D remotely sensed data. Following a similar strategy as in the logistic regression analysis, we formulated a model with tree height observed in field as dependent variable and maximum height for each tree crown polygon in each of the 3D remotely sensed datasets and the factors to be evaluated as independent variables:
where
hmax = hALSmax when the dataset was ALS and
hmax = hDAPmax when the dataset was DAP. The other variables in the model were defined as above. The analysis was based on 389 of the trees for which
hmax ≥ 0 in both datasets (
Table 5). The least squares method for fitting the model was applied by using the REG procedure of the SAS statistical software package [
37].
F-tests were performed to test the null hypotheses that (1) the parameter estimates for the two SP variables (
β2 and
β3) were equal and that (2) the parameter estimates of the products of the two SP variables and
hmax (
β6 and
β7) were equal.
Finally, leave-one-out cross validation was adopted to assess the predictive ability of the two 3D datasets. However, the model in Equation (3) assumed the error variance to be the same for both 3D datasets. Separate models would be required if the error variances could be assumed to be different ([
38], p. 173). The cross validation was therefore performed for separate models constructed according to:
where
hmax = hALSmax when the model was constructed by using the ALS data.
hmax = hDAPmax when the model was constructed by using the DAP data.
In the cross validation of the two respective models, the prediction accuracy was assessed separately for different classes according to tree height and different tree species. The assessment was based on the differences between predicted and observed tree height for individual trees according to the statistics (1) mean difference, (2) standard deviation of the differences (Stdev), and root mean square error (RMSE). These statistics were also calculated across all trees for each individual 3D dataset and the null hypothesis of homogeneity of prediction variances among the two 3D datasets was tested by Levene’s
F-test [
39] in the GLM procedure of the SAS package [
37].