A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Collection
2.2.1. Satellite Imagery
2.2.2. Ground Control Data
2.3. Methods
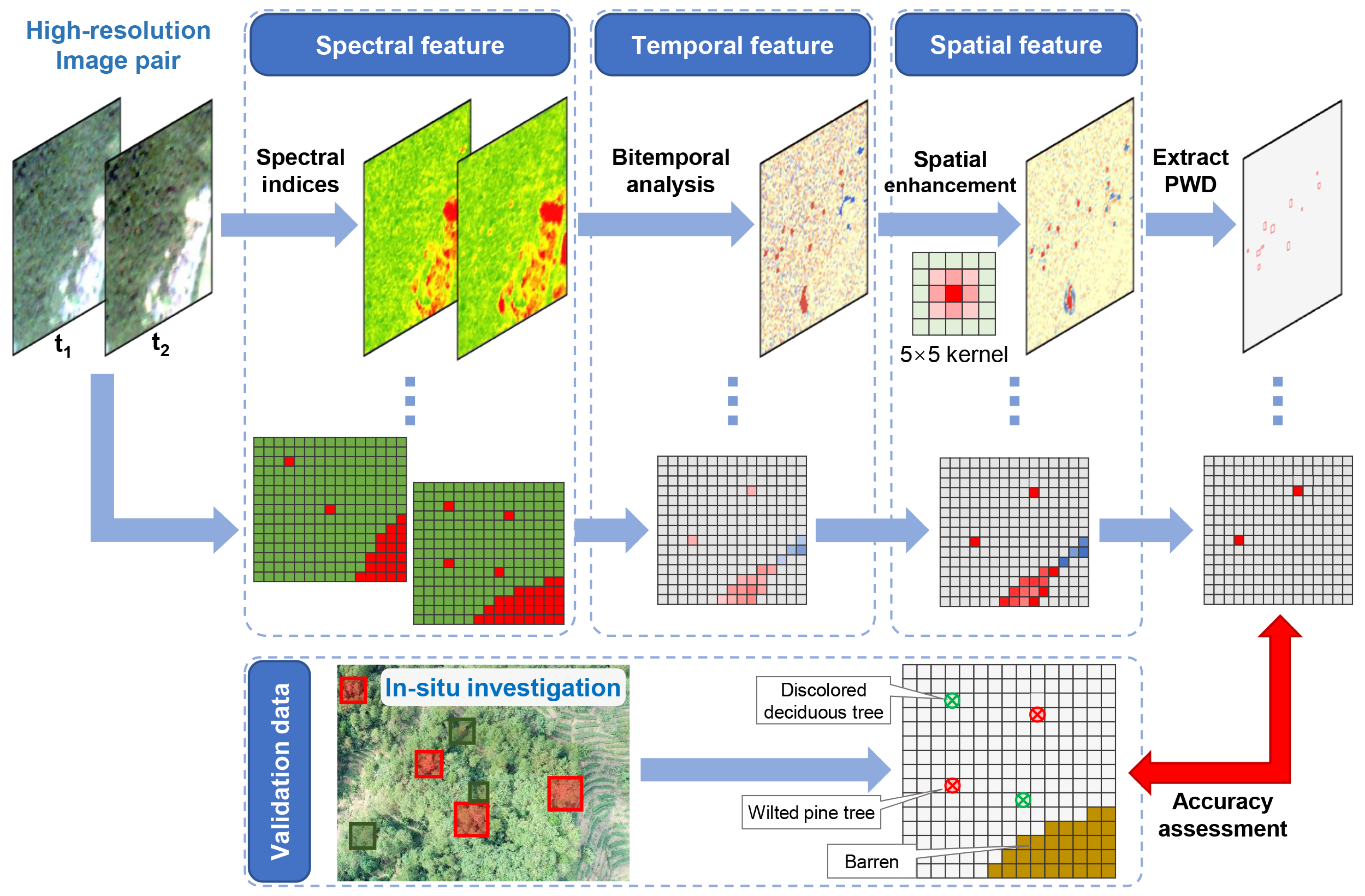
2.3.1. General Workflow
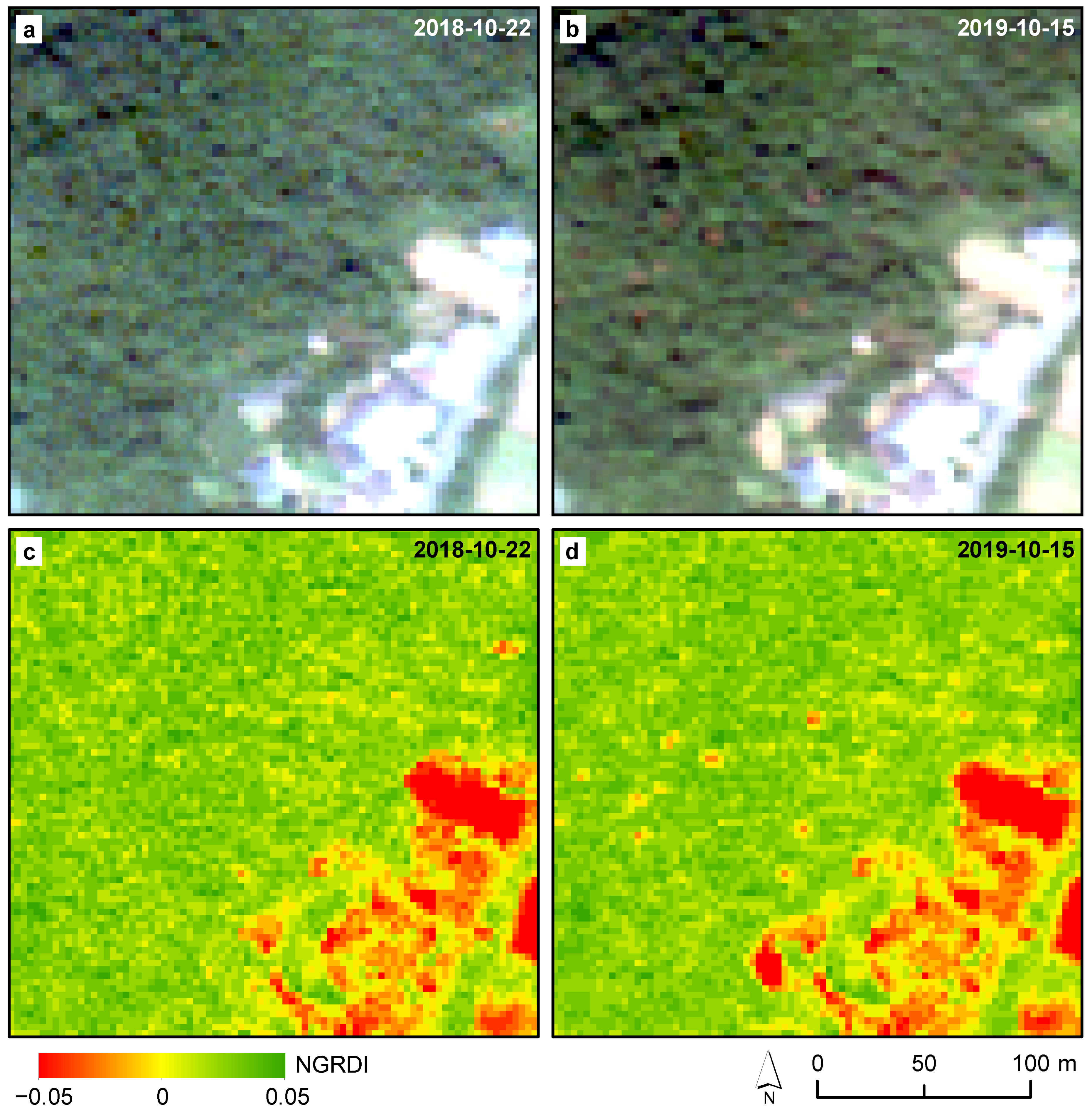
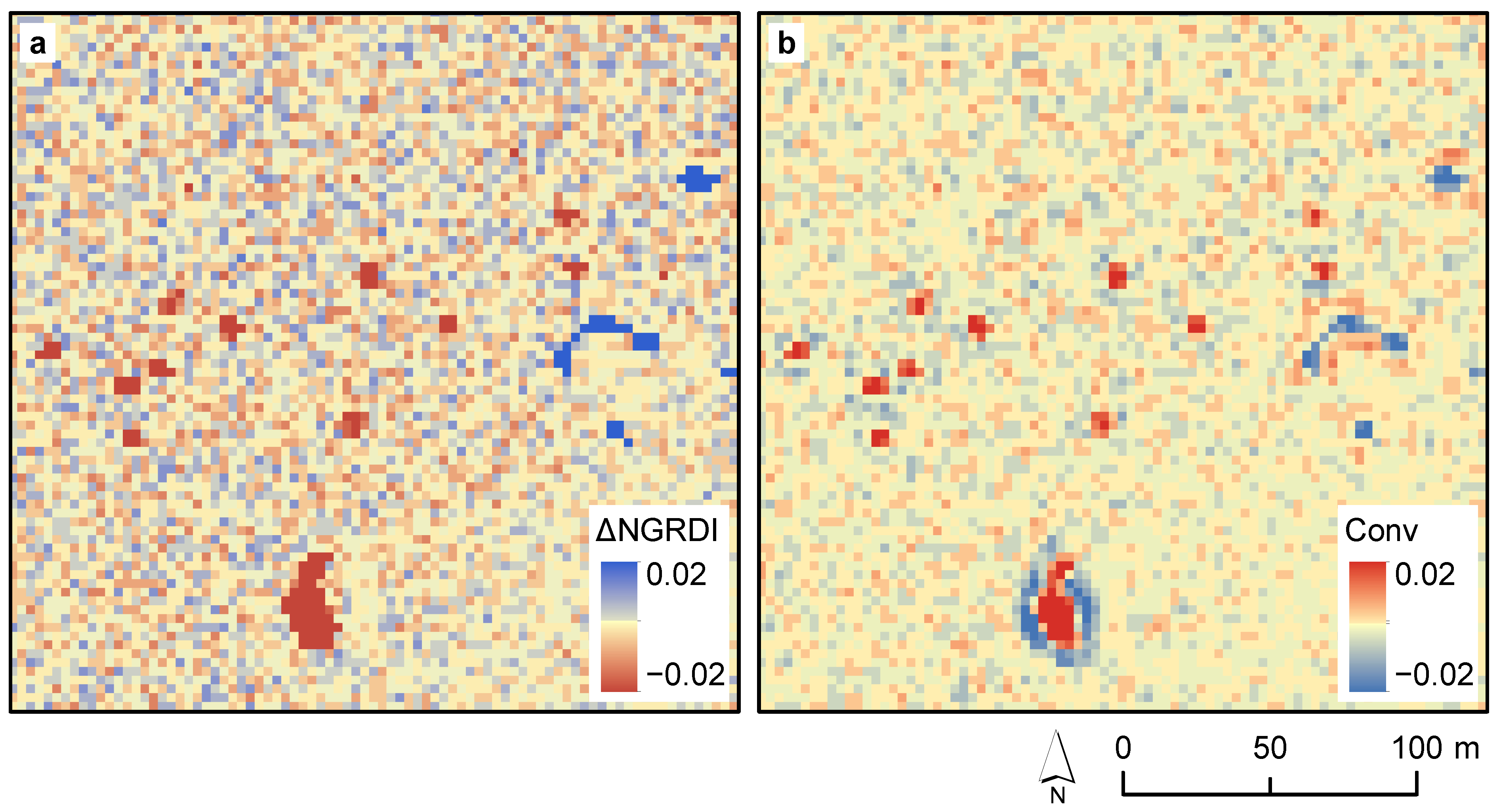
2.3.2. Bi-Temporal Change Detection
2.3.3. Spatial Enhancement
2.3.4. Extraction of PWD
2.3.5. Accuracy Assessment
3. Results
3.1. Spatial and Temporal Patterns of PWD-Induced Wilting
3.2. Comparison of Spatiotemporal Change Detection and Single-Date Classification
4. Discussion
4.1. General Framework of Tree-Scale PWD Monitoring
4.2. Advantages of Spatiotemporal Change Detection Method
4.3. Error Source of the Spatiotemporal Change Detection Method
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| 0.01 | 0.011 | 0.012 | 0.013 | 0.014 | 0.015 | 0.016 | 0.017 | 0.018 | 0.019 | 0.02 | ||
| N | ||||||||||||
| 1 | 30.8% | 29.5% | 28.8% | 27.4% | 25.9% | 24.8% | 24.0% | 22.7% | 21.5% | 19.4% | 15.6% | |
| 2 | 39.5% | 38.1% | 36.2% | 35.0% | 33.5% | 31.6% | 29.6% | 26.5% | 23.5% | 23.3% | 21.9% | |
| 4 | 47.4% | 45.9% | 43.9% | 42.0% | 41.2% | 40.1% | 37.2% | 36.3% | 32.7% | 30.5% | 26.4% | |
| 6 | 60.0% | 58.7% | 56.0% | 53.4% | 51.0% | 48.5% | 48.5% | 45.3% | 40.5% | 40.5% | 34.8% | |
| 9 | 69.4% | 66.8% | 64.5% | 64.3% | 63.7% | 61.2% | 60.0% | 56.8% | 52.4% | 51.4% | 46.3% | |
| 12 | 80.0% | 78.9% | 76.9% | 76.5% | 73.6% | 71.3% | 69.7% | 69.3% | 65.4% | 57.7% | 57.4% | |
| 16 | 89.2% | 88.6% | 88.0% | 86.4% | 85.9% | 84.7% | 80.7% | 77.6% | 69.2% | 68.6% | 55.2% | |
| 20 | 91.7% | 90.4% | 88.6% | 86.2% | 85.4% | 82.7% | 78.0% | 73.2% | 67.9% | 65.5% | 62.9% | |
| 25 | 94.8% | 93.2% | 92.4% | 90.8% | 88.6% | 85.1% | 80.5% | 75.8% | 71.5% | 66.1% | 53.5% | |
| 30 | 96.0% | 95.7% | 94.6% | 92.4% | 90.6% | 89.2% | 86.2% | 81.2% | 79.6% | 76.2% | 74.2% | |
| 36 | 96.5% | 95.8% | 95.1% | 94.5% | 94.4% | 91.8% | 91.5% | 87.1% | 84.6% | 75.8% | 67.1% | |
| 0.01 | 0.011 | 0.012 | 0.013 | 0.014 | 0.015 | 0.016 | 0.017 | 0.018 | 0.019 | 0.02 | ||
| N | ||||||||||||
| 1 | 56.9% | 62.4% | 66.7% | 71.3% | 76.2% | 80.7% | 82.1% | 83.6% | 87.8% | 92.1% | 93.2% | |
| 2 | 58.0% | 66.8% | 71.6% | 77.3% | 79.3% | 81.6% | 83.5% | 84.2% | 87.3% | 91.6% | 92.8% | |
| 4 | 58.3% | 69.3% | 73.2% | 78.6% | 80.2% | 82.5% | 82.9% | 85.5% | 86.9% | 89.8% | 92.6% | |
| 6 | 58.8% | 65.9% | 68.2% | 70.3% | 73.3% | 82.1% | 83.1% | 84.9% | 85.5% | 88.2% | 91.5% | |
| 9 | 58.0% | 62.4% | 63.9% | 69.0% | 72.1% | 81.9% | 82.5% | 84.4% | 85.2% | 87.9% | 90.1% | |
| 12 | 55.7% | 65.6% | 69.8% | 70.2% | 76.8% | 81.5% | 82.2% | 83.8% | 85.0% | 86.5% | 87.7% | |
| 16 | 54.4% | 61.9% | 71.9% | 76.1% | 78.3% | 81.2% | 82.0% | 83.2% | 84.5% | 86.6% | 89.1% | |
| 20 | 45.7% | 47.0% | 53.3% | 57.8% | 60.3% | 72.6% | 75.1% | 78.3% | 79.2% | 80.9% | 82.3% | |
| 25 | 39.2% | 44.7% | 45.0% | 46.8% | 48.5% | 48.9% | 50.0% | 50.4% | 51.7% | 52.1% | 62.9% | |
| 30 | 31.1% | 37.0% | 39.1% | 39.3% | 40.3% | 41.8% | 42.8% | 43.5% | 44.2% | 44.3% | 44.7% | |
| 36 | 17.7% | 20.3% | 22.9% | 26.8% | 28.3% | 29.9% | 31.7% | 33.5% | 34.8% | 35.2% | 35.1% | |
References
- Kuroda, K.; Yamada, T.; Mineo, K.; Tamura, H. Effects of cavitation on the development of pine wilt disease caused by Bursaphelenchus xylophilus. Jpn. J. Phytopathol. 1988. [Google Scholar] [CrossRef]
- Vollenweider, P.; Günthardt-Goerg, M.S. Diagnosis of abiotic and biotic stress factors using the visible symptoms in foliage. Environ. Pollut. 2005. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B. Pine wilt disease in China. In Pine Wilt Disease; Springer: Tokyo, Japan, 2008; pp. 18–25. [Google Scholar] [CrossRef]
- Sun, H.; Zhou, Y.; Li, X.; Zhang, Y.; Wang, Y. The occurrence of major forestry pests nationwide in 2020 and the forecast of their occurrence in 2021. For. Pest Dis. 2021, 45–48. (In Chinese) [Google Scholar] [CrossRef]
- Cheng, G.; Lü, Q.; Feng, Y.; Li, Y.; Wang, Y.; Zhang, X. Temporal and spatial dynamic pattern of pine wilt disease distribution in China predicted under climate change scenario. Sci. Silvae Sin. 2015, 51, 119–126. [Google Scholar]
- He, S.; Wen, J.; Luo, Y.; Zong, S.; Zhao, Y.; Han, J. The predicted geographical distribution of Bursaphelenchus xylophilus in China under climate warming. Chin. J. Appl. Entomol. 2012, 49, 236–243. [Google Scholar]
- Li, Y.; Zhang, X. Analysis on the trend of invasion and expansion of Bursaphelenchus xylophilus. For. Pest Dis. 2018, 037, 1–4. [Google Scholar] [CrossRef]
- Zhao, B.G.; Tao, J.; Ju, Y.W.; Wang, P.K.; Ye, J.L. The role of wood-inhabiting bacteria in pine wilt disease. J. Nematol. 2011, 43, 129. [Google Scholar] [PubMed]
- Huang, M.; Gong, J.; Zhang, J. Study on Pine Wilt Disease Remote Sensing Monitoring and Diffusion Simulation; China Environmental Science Press: Beijing, China, 2012. [Google Scholar]
- Senf, C.; Seidl, R.; Hostert, P. Remote sensing of forest insect disturbances: Current state and future directions. Int. J. Appl. Earth Obs. Geoinf. 2017, 60, 49–60. [Google Scholar] [CrossRef]
- Beck, P.S.A.; Zarco-Tejada, P.; Strobl, P.; San Miguel, J. The feasibility of detecting trees affected by the Pine Wood Nematode using remote sensing. In European Commission Joint Research Centre Institute for Environment and Sustainability; Publications Office of the European Union: Luxembourg, 2015. [Google Scholar]
- Kim, S.R.; Lee, W.K.; Lim, C.H.; Kim, M.; Kafatos, M.C.; Lee, S.H.; Lee, S.S. Hyperspectral analysis of pine wilt disease to determine an optimal detection index. Forests 2018, 9, 115. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Z.; Zheng, L.; Han, C.; Wang, X.; Xu, J.; Wang, X. Research progress on the early monitoring of pine wilt disease using hyperspectral techniques. Sensors 2020, 20, 3729. [Google Scholar] [CrossRef]
- Xu, H.C.; Luo, Y.Q.; Zhang, T.T.; Shi, Y.J. Changes of reflectance spectra of pine needles in different stage after being infected by pine wood nematode. Spectrosc. Spectr. Anal. 2011, 31, 1352–1356. [Google Scholar] [CrossRef]
- Raffa, K.F.; Aukema, B.H.; Bentz, B.J.; Carroll, A.L.; Hicke, J.A.; Turner, M.G.; Romme, W.H. Cross-scale drivers of natural disturbances prone to anthropogenic amplification: The dynamics of bark beetle eruptions. BioScience 2008, 58, 501–517. [Google Scholar] [CrossRef]
- Kim, J.B.; Park, J.H.; Jo, M.H. A spectral characteristic analysis of damage pine wilt disease area in ikonos image. In Proceedings of the 23rd Asian Conference on Remote Sensing, Kathmandu, Nepal, 25–29 November 2002. [Google Scholar]
- White, J.C.; Wulder, M.A.; Brooks, D.; Reich, R.; Wheate, R.D. Detection of red attack stage mountain pine beetle infestation with high spatial resolution satellite imagery. Remote Sens. Environ. 2005, 96, 340–351. [Google Scholar] [CrossRef]
- Coops, N.C.; Johnson, M.; Wulder, M.A.; White, J.C. Assessment of QuickBird high spatial resolution imagery to detect red attack damage due to mountain pine beetle infestation. Remote Sens. Environ. 2006, 103, 67–80. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Coops, N.C.; Butson, C.R. Multi-temporal analysis of high spatial resolution imagery for disturbance monitoring. Remote Sens. Environ. 2008, 112, 2729–2740. [Google Scholar] [CrossRef]
- Takenaka, Y.; Katoh, M.; Deng, S.; Cheung, K. Detecting forests damaged by pine wilt disease at the individual tree level using airborne laser data and worldview-2/3 images over two seasons. In International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences; Copernicus Publications: Göttingen, Germany, 2017; Volume XLII-3/W3, pp. 181–184. [Google Scholar] [CrossRef]
- Dennison, P.E.; Brunelle, A.R.; Carter, V.A. Assessing canopy mortality during a mountain pine beetle outbreak using GeoEye-1 high spatial resolution satellite data. Remote Sens. Environ. 2010. [Google Scholar] [CrossRef]
- Hicke, J.A.; Logan, J. Mapping whitebark pine mortality caused by a mountain pine beetle outbreak with high spatial resolution satellite imagery. Int. J. Remote Sens. 2009, 30, 4427–4441. [Google Scholar] [CrossRef]
- Poona, N.K.; Ismail, R. Discriminating the occurrence of pitch canker infection in Pinus radiata forests using high spatial resolution QuickBird data and artificial neural networks. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Munich, Germany, 22–27 July 2012. [Google Scholar] [CrossRef]
- Sharma, R. Detection of Mountain Pine Beetle Infestations Using Landsat TM Tasseled Cap Transformations. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 2001. [Google Scholar] [CrossRef]
- Skakun, R.S.; Wulder, M.A.; Franklin, S.E. Sensitivity of the thematic mapper enhanced wetness difference index to detect mountain pine beetle red-attack damage. Remote Sens. Environ. 2003, 86, 433–443. [Google Scholar] [CrossRef]
- Gower, S.T.; Kucharik, C.J.; Norman, J.M. Direct and indirect estimation of leaf area index, fAPAR, and net primary production of terrestrial ecosystems. Remote Sens. Environ. 1999, 70, 29–51. [Google Scholar] [CrossRef]
- Reed, D.E.; Ewers, B.E.; Pendall, E.; Frank, J.; Kelly, R. Bark beetle-induced tree mortality alters stand energy budgets due to water budget changes. Theor. Appl. Climatol. 2018, 131, 153–165. [Google Scholar] [CrossRef]
- Tao, H.; Li, C.; Cheng, C.; Jiang, L.; Hu, H. Progress in Remote Sensing Monitoring for Pine Wilt Disease Induced Tree Mortality: A Review. For. Res. 2020, 33, 172–183. [Google Scholar]
- Hart, S.J.; Veblen, T.T. Detection of spruce beetle-induced tree mortality using high- and medium-resolution remotely sensed imagery. Remote Sens. Environ. 2015, 168, 134–145. [Google Scholar] [CrossRef]
- Wulder, M.A.; Dymond, C.C.; White, J.C.; Leckie, D.G.; Carroll, A.L. Surveying mountain pine beetle damage of forests: A review of remote sensing opportunities. For. Ecol. Manag. 2006, 221, 1–41. [Google Scholar] [CrossRef]
- Planet Labs Inc. Planet Imagery and Archive. 2020. Available online: https://www.planet.com/products/planet-imagery/ (accessed on 17 January 2021).
- Planet Team. Planet Application Program Interface: In Space for Life on EARTH; Planet Team: San Francisco, CA, USA, 2017. [Google Scholar]
- Canty, M.J.; Nielsen, A.A. Automatic radiometric normalization of multitemporal satellite imagery with the iteratively re-weighted MAD transformation. Remote Sens. Environ. 2008, 112, 1025–1036. [Google Scholar] [CrossRef]
- Guo, Q.; Kelly, M.; Gong, P.; Liu, D. An object-based classification approach in mapping tree mortality using high spatial resolution imagery. GISci. Remote Sens. 2007, 44, 24–47. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, H.K. Detection of the pine trees damaged by pine wilt disease using high spatial remote sensing data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2006, 36. [Google Scholar] [CrossRef]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.; McMurtrey, J.E.; Walthall, C.L. Evaluation of digital photography from model aircraft for remote sensing of crop biomass and nitrogen status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Afdhalia, F.; Supriatna, S.; Shidiq, I.P.A.; Manessa, M.D.M.; Ristya, Y. Detection of rice varieties based on spectral value data using UAV-based images. In Proceedings of the Sixth International Symposium on LAPAN-IPB Satellite, International Society for Optics and Photonics, Bogor, Indonesia, 24 December 2019; Volume 11372, p. 1137222. [Google Scholar]
- Bustamante, B.D.R.; Bustamante, S.G.H. Method of Anomalies Detection in Persea Americana Leaves with Thermal and NGRDI Imagery. In Proceedings of the 5th Brazilian Technology Symposium; Springer: Cham, Switzerland, 2021; pp. 287–296. [Google Scholar]
- Elazab, A.; Ordóñez, R.A.; Savin, R.; Slafer, G.A.; Araus, J.L. Detecting interactive effects of N fertilization and heat stress on maize productivity by remote sensing techniques. Eur. J. Agron. 2016, 73, 11–24. [Google Scholar] [CrossRef]
- Jannoura, R.; Brinkmann, K.; Uteau, D.; Bruns, C.; Joergensen, R.G. Monitoring of crop biomass using true colour aerial photographs taken from a remote controlled hexacopter. Biosyst. Eng. 2015, 129, 341–351. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E.; Eddins, S. Digital Image Processing with Matlab; Prentice-Hall: Hoboken, NJ, USA, 2004. [Google Scholar]
- Healey, S.P.; Cohen, W.B.; Yang, Z.; Krankina, O.N. Comparison of Tasseled Cap-based Landsat data structures for use in forest disturbance detection. Remote Sens. Environ. 2005, 97, 301–310. [Google Scholar] [CrossRef]






| Site | Area (ha) | Forest Type | Number of Wilted Pine Trees in Different Canopy Width | ||
|---|---|---|---|---|---|
| >3 m | ≤3 m | Total | |||
| A | 5.1 | Mixed | 12 | 3 | 15 |
| B | 9.8 | Mixed | 6 | 1 | 7 |
| C | 14.3 | Mixed | 90 | 14 | 104 |
| D | 12.6 | Mixed | 28 | 2 | 30 |
| E | 8.5 | Pure | 67 | 5 | 72 |
| Total | 50.3 | 203 | 25 | 228 | |
| Category | Feature | Abbrev | Type | Criterion |
|---|---|---|---|---|
| Temporal | NGRDI observation in the first image (22 October 2018) | Pixel-wise | >0 | |
| NGRDI observation in the second image (15 October 2018) | Pixel-wise | <0 | ||
| Spatial | Output of the spatial convolution on the | Pixel-wise | ≥ | |
| Pixel count in a candidate bounding box (BB) | Object-wise | ≤N |
| Site | Total Wilted Trees for Validation | True Positive (Tree Count) | Omission Errors (Tree Count) | Commission Errors (BB Count) | Producer’s Accuracy | User’s Accuracy |
|---|---|---|---|---|---|---|
| A | 12 | 11 | 1 | 2 | 91.7% | 84.6% |
| B | 6 | 5 | 1 | 2 | 83.3% | 71.4% |
| C | 90 | 79 | 11 | 15 | 87.8% | 83.0% |
| D | 28 | 22 | 6 | 4 | 78.6% | 84.0% |
| E | 67 | 55 | 12 | 14 | 82.1% | 78.1% |
| Total | 203 | 172 | 31 | 37 | 84.7% | 81.2% |
| Site | Total Wilted Trees for Validation | True Positive (Tree Count) | Omission Errors (Tree Count) | Commission Errors (BB Count) | Producer’s Accuracy | User’s Accuracy |
|---|---|---|---|---|---|---|
| A | 12 | 12 | 0 | 8 | 100.0% | 60.0% |
| B | 6 | 5 | 1 | 4 | 83.3% | 55.6% |
| C | 36 * | 30 | 6 | 16 | 83.3% | 63.6% |
| D | 11 * | 9 | 2 | 6 | 81.8% | 60.0% |
| E | 67 | 53 | 14 | 16 | 79.1% | 76.1% |
| Total | 132 | 109 | 23 | 50 | 82.6% | 67.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Ye, H.; Lu, W.; Huang, W.; Wu, B.; Hao, Z.; Sun, H. A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 2083. https://doi.org/10.3390/rs13112083
Zhang B, Ye H, Lu W, Huang W, Wu B, Hao Z, Sun H. A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sensing. 2021; 13(11):2083. https://doi.org/10.3390/rs13112083
Chicago/Turabian StyleZhang, Biyao, Huichun Ye, Wei Lu, Wenjiang Huang, Bo Wu, Zhuoqing Hao, and Hong Sun. 2021. "A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery" Remote Sensing 13, no. 11: 2083. https://doi.org/10.3390/rs13112083
APA StyleZhang, B., Ye, H., Lu, W., Huang, W., Wu, B., Hao, Z., & Sun, H. (2021). A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sensing, 13(11), 2083. https://doi.org/10.3390/rs13112083









