A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements
Abstract
1. Introduction
2. Materials and Methods
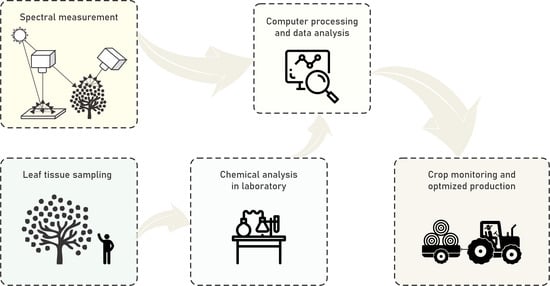
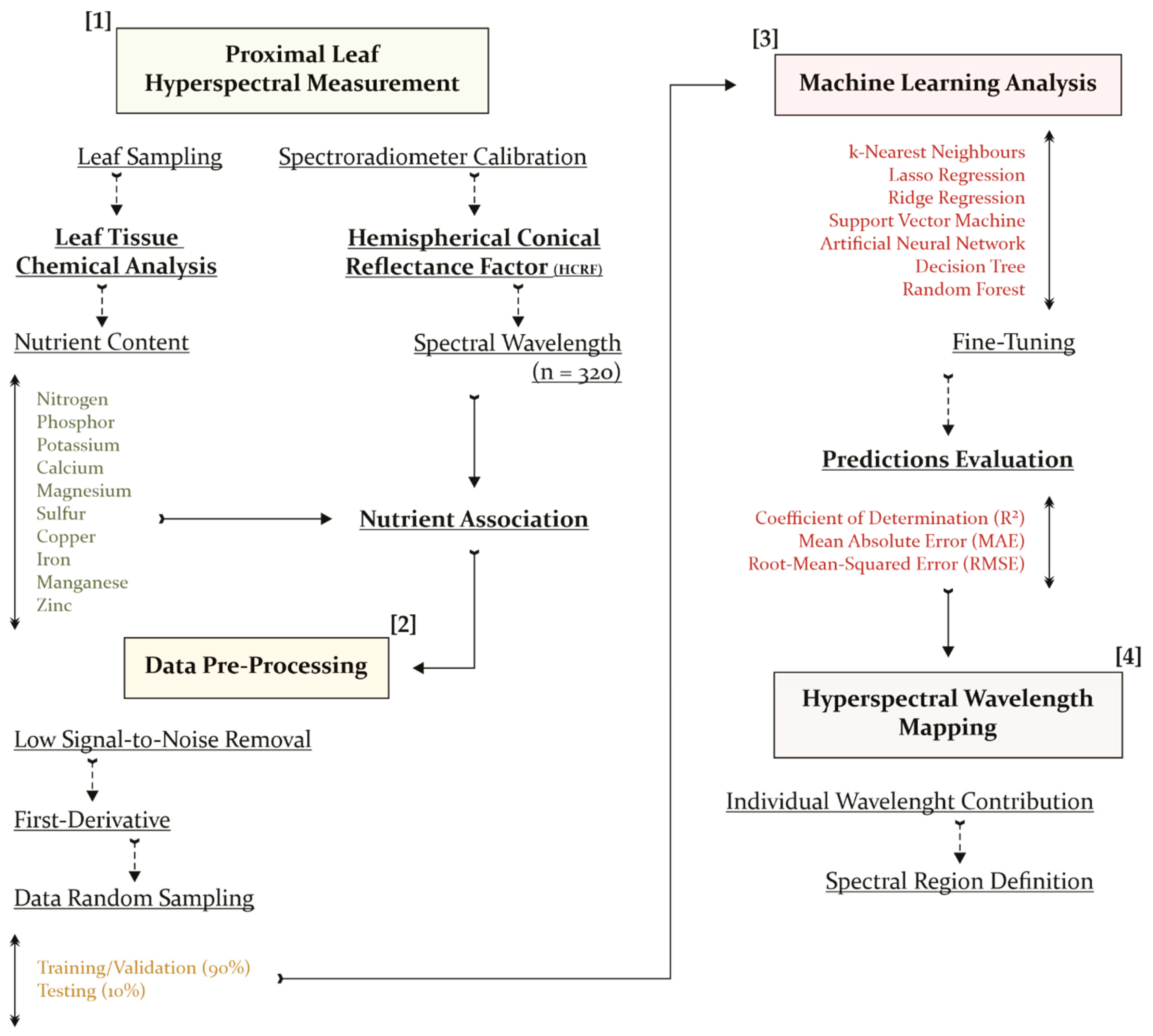
2.1. Study Area and Data Acquisition
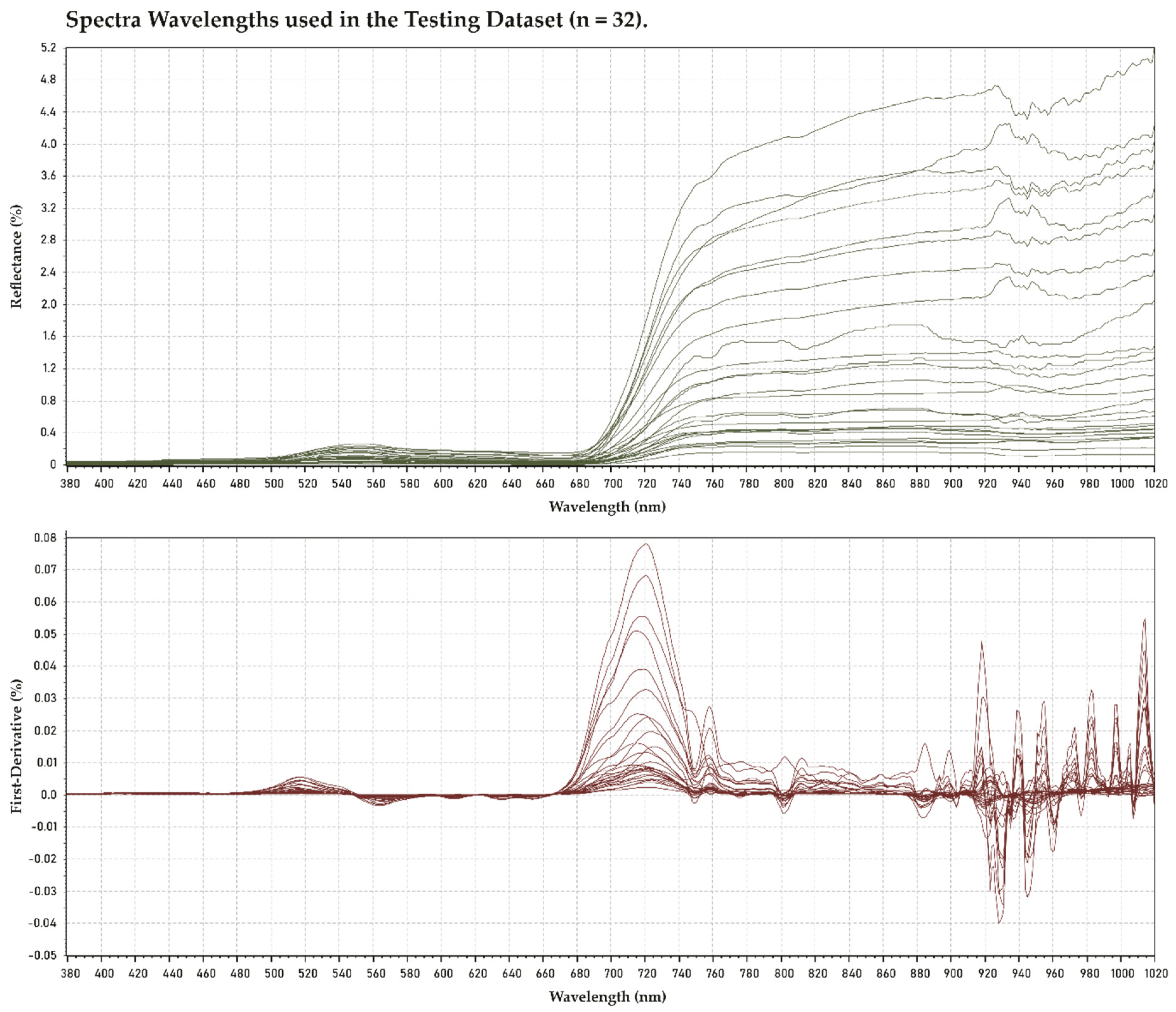
2.2. Hyperspectral Measurement Processing
2.3. Machine Learning Analysis and Hyperspectral Mapping
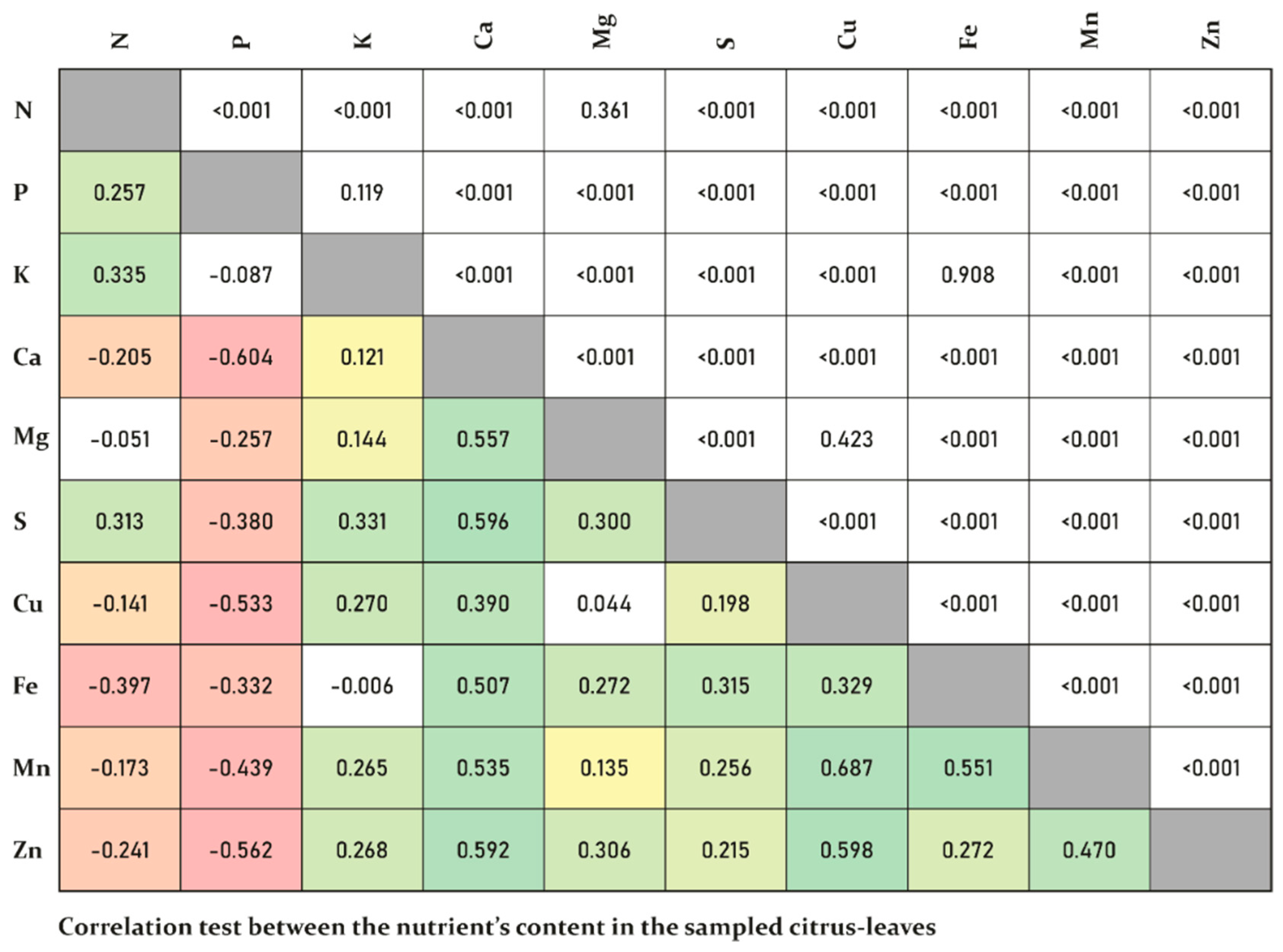
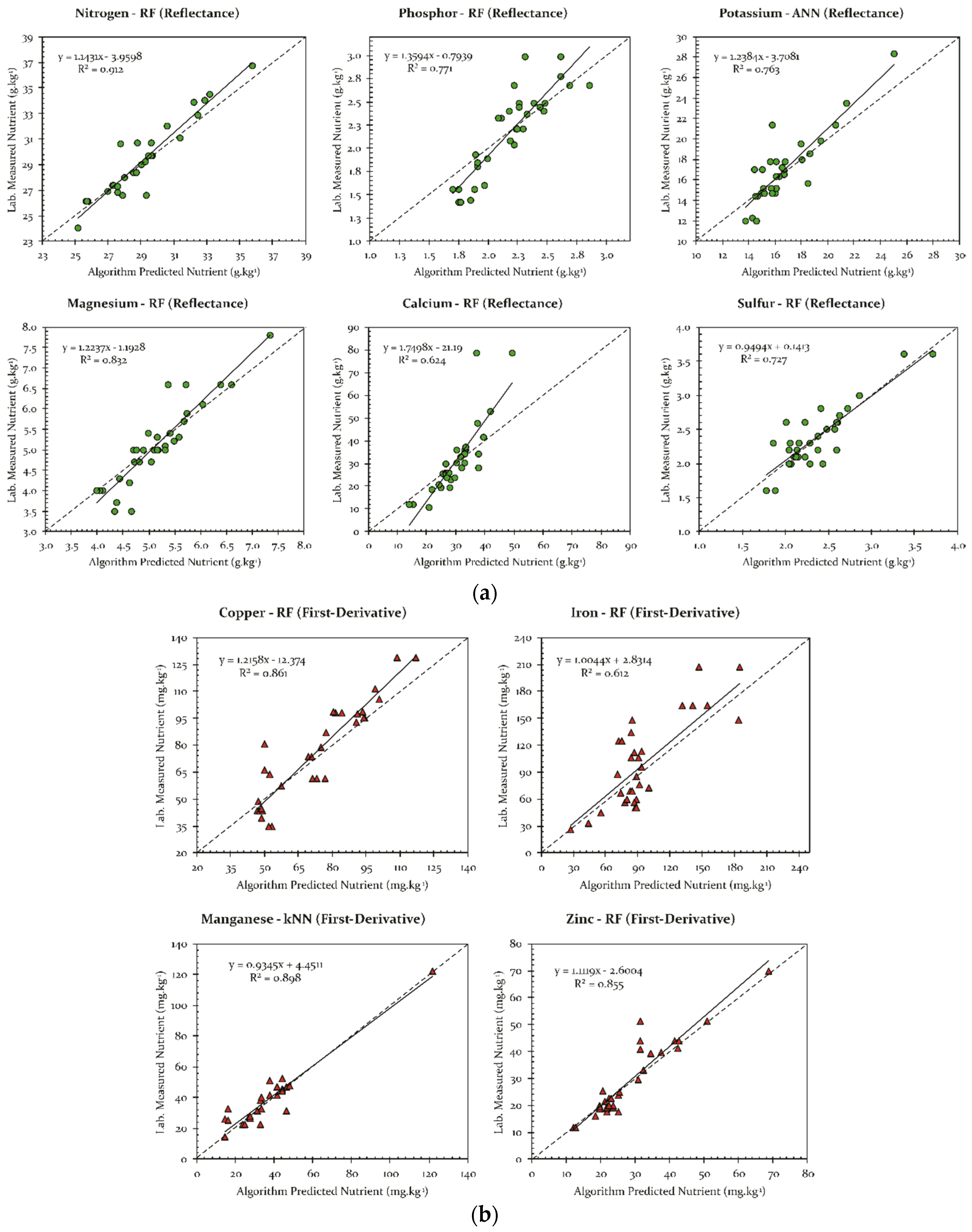
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, F.; Wang, L.; Liu, J.; Wang, Y.; Chang, Q. Evaluation of leaf N concentration in winter wheat based on discrete wavelet transform analysis. Remote Sens. 2019, 11, 1331. [Google Scholar] [CrossRef]
- Li, Z.; Jin, X.; Yang, G.; Drummond, J.; Yang, H.; Clark, B.; Li, Z.; Zhao, C. Remote sensing of leaf and canopy nitrogen status in winter wheat (Triticum aestivum L.) based on N-PROSAIL model. Remote Sens. 2018, 10, 1463. [Google Scholar] [CrossRef]
- Osco, L.P.; Marques Ramos, A.P.; Saito Moriya, É.A.; de Souza, M.; Marcato Junior, J.; Matsubara, E.T.; Imai, N.N.; Creste, J.E. Improvement of leaf nitrogen content inference in Valencia-orange trees applying spectral analysis algorithms in UAV mounted-sensor images. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101907. [Google Scholar] [CrossRef]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X. A comparative assessment of different modeling algorithms for estimating leaf nitrogen content in winter wheat using multispectral images from an unmanned aerial vehicle. Remote Sens. 2018, 10, 2026. [Google Scholar] [CrossRef]
- Loggenberg, K.; Strever, A.; Greyling, B.; Poona, N. Modelling water stress in a Shiraz vineyard using hyperspectral imaging and machine learning. Remote Sens. 2018, 10, 202. [Google Scholar] [CrossRef]
- Gerhards, M.; Schlerf, M.; Rascher, U.; Udelhoven, T.; Juszczak, R.; Alberti, G.; Miglietta, F.; Inoue, Y. Analysis of airborne optical and thermal imagery for detection of water stress symptoms. Remote Sens. 2018, 10, 1139. [Google Scholar] [CrossRef]
- Johnson, K.; Sankaran, S.; Ehsani, R. Identification of Water Stress in Citrus Leaves Using Sensing Technologies. Agronomy 2013, 3, 747–756. [Google Scholar] [CrossRef]
- Osco, L.P.; dos Santos de Arruda, M.; Marcato Junior, J.; da Silva, N.B.; Ramos, A.P.M.; Moryia, É.A.S.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus-trees in UAV multispectral imagery. ISPRS J. Photogramm. Remote Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in rgb imagery using semi-supervised deep learning neural networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Zhang, K.; Ge, X.; Shen, P.; Li, W.; Liu, X.; Cao, Q.; Zhu, Y.; Cao, W.; Tian, Y. Predicting rice grain yield based on dynamic changes in vegetation indexes during early to mid-growth stages. Remote Sens. 2019, 11, 387. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Lipping, T. Crop yield prediction with deep convolutional neural networks. Comput. Electron. Agric. 2019, 163, 104859. [Google Scholar] [CrossRef]
- Hunt, M.L.; Blackburn, G.A.; Carrasco, L.; Redhead, J.W.; Rowland, C.S. High resolution wheat yield mapping using Sentinel-2. Remote Sens. Environ. 2019, 233, 111410. [Google Scholar] [CrossRef]
- Cui, B.; Zhao, Q.; Huang, W.; Song, X.; Ye, H.; Zhou, X. A new integrated vegetation index for the estimation of winter wheat leaf chlorophyll content. Remote Sens. 2019, 11, 974. [Google Scholar] [CrossRef]
- Guo, T.; Tan, C.; Li, Q.; Cui, G.; Li, H. Estimating leaf chlorophyll content in tobacco based on various canopy hyperspectral parameters. J. Ambient Intell. Humaniz. Comput. 2019, 10, 3239–3247. [Google Scholar] [CrossRef]
- Peng, Z.; Guan, L.; Liao, Y.; Lian, S. Estimating total leaf chlorophyll content of gannan navel orange leaves using hyperspectral data based on partial least squares regression. IEEE Access 2019, 7, 155540–155551. [Google Scholar] [CrossRef]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-based remote sensing technique to detect citrus canker disease utilizing hyperspectral imaging and machine learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef]
- Yao, Z.; Lei, Y.; He, D. Early visual detection of wheat stripe rust using visible/near-infrared hyperspectral imaging. Sensors (Switzerland) 2019, 19, 952. [Google Scholar] [CrossRef]
- Pham, T.D.; Yokoya, N.; Bui, D.T.; Yoshino, K.; Friess, D.A. Remote sensing approaches for monitoring mangrove species, structure, and biomass: Opportunities and challenges. Remote Sens. 2019, 11, 230. [Google Scholar] [CrossRef]
- Brinkhoff, J.; Dunn, B.W.; Robson, A.J.; Dunn, T.S.; Dehaan, R.L. Modeling mid-season rice nitrogen uptake using multispectral satellite data. Remote Sens. 2019, 11, 1837. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, H.; Xu, Z.; Hu, J.; Shi, X.; Hua, S.; Yue, J.; Yang, G. Estimating maize-leaf coverage in field conditions by applying a machine learning algorithm to UAV remote sensing images. Appl. Sci. 2019, 9, 2389. [Google Scholar] [CrossRef]
- Delloye, C.; Weiss, M.; Defourny, P. Retrieval of the canopy chlorophyll content from Sentinel-2 spectral bands to estimate nitrogen uptake in intensive winter wheat cropping systems. Remote Sens. Environ. 2018, 216, 245–261. [Google Scholar] [CrossRef]
- Vanbrabant, Y.; Tits, L.; Delalieux, S.; Pauly, K.; Verjans, W.; Somers, B. Multitemporal chlorophyll mapping in pome fruit orchards from remotely piloted aircraft systems. Remote Sens. 2019, 11, 1468. [Google Scholar] [CrossRef]
- Osco, L.P.; Ramos, A.P.M.; Moriya, E.A.S.; Bavaresco, L.G.; Lima, B.C.; Estrabis, N.; Pereira, D.R.; Creste, J.E.; Marcato Junior, J.; Gonçalves, W.N.; et al. Modeling hyperspectral response of water-stress induced lettuce plants using artificial neural networks. Remote Sens. 2019, 11, 2797. [Google Scholar] [CrossRef]
- de Oliveira, D.M.; Fontes, L.M.; Pasquini, C. Comparing laser induced breakdown spectroscopy, near infrared spectroscopy, and their integration for simultaneous multi-elemental determination of micro- and macronutrients in vegetable samples. Anal. Chim. Acta 2019, 1062, 28–36. [Google Scholar] [CrossRef]
- Chen, J.; Li, F.; Wang, R.; Fan, Y.; Raza, M.A.; Liu, Q.; Wang, Z.; Cheng, Y.; Wu, X.; Yang, F.; et al. Estimation of nitrogen and carbon content from soybean leaf reflectance spectra using wavelet analysis under shade stress. Comput. Electron. Agric. 2019, 156, 482–489. [Google Scholar] [CrossRef]
- Cuq, S.; Lemetter, V.; Kleiber, D.; Levasseur-Garcia, C. Assessing macro-element content in vine leaves and grape berries of vitis vinifera by using near-infrared spectroscopy and chemometrics. Int. J. Environ. Anal. Chem. 2019. [Google Scholar] [CrossRef]
- Santoso, H.; Tani, H.; Wang, X.; Segah, H. Predicting oil palm leaf nutrient contents in kalimantan, indonesia by measuring reflectance with a spectroradiometer. Int. J. Remote Sens. 2019, 40, 7581–7602. [Google Scholar] [CrossRef]
- Osco, L.P.; Ramos, A.P.M.; Pereira, D.R.; Moriya, E.A.S.; Imai, N.N.; Matsubara, E.T. Predicting canopy nitrogen content in citrus-trees using random forest algorithm associated to spectral vegetation indices from UAV-Imagery. Remote Sens. 2019, 11, 2925. [Google Scholar] [CrossRef]
- Allen, V.; Barker, D.J.P. Handbook of Plant Nutrition; Taylor et Francis, 2015. Available online: https://www.bokus.com/bok/9781439881989/handbook-of-plant-nutrition/ (accessed on 4 February 2020).
- Malmir, M.; Tahmasbian, I.; Xu, Z.; Farrar, M.B.; Bai, S.H. Prediction of macronutrients in plant leaves using chemometric analysis and wavelength selection. J. Soils Sediments 2019. [Google Scholar] [CrossRef]
- Ye, X.; Abe, S.; Zhang, S. Estimation and mapping of nitrogen content in apple trees at leaf and canopy levels using hyperspectral imaging. Precis. Agric. 2019. [Google Scholar] [CrossRef]
- Ling, B.; Goodin, D.G.; Raynor, E.J.; Joern, A. Hyperspectral analysis of leaf pigments and nutritional elements in tallgrass prairie vegetation. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yan, C.; Lu, S.; Wang, P.; Qiu, G.Y.; Li, R. Estimation of chlorophyll content in intertidal mangrove leaves with different thicknesses using hyperspectral data. Ecol. Indic. 2019, 106, 105511. [Google Scholar] [CrossRef]
- Shi, J.; Li, W.; Zhai, X.; Guo, Z.; Holmes, M.; Elrasheid Tahir, H.; Zou, X. Nondestructive diagnostics of magnesium deficiency based on distribution features of chlorophyll concentrations map on cucumber leaf. J. Plant Nutr. 2019, 42, 2773–2783. [Google Scholar] [CrossRef]
- Román, J.R.; Rodríguez-Caballero, E.; Rodríguez-Lozano, B.; Roncero-Ramos, B.; Chamizo, S.; Águila-Carricondo, P.; Cantón, Y. Spectral response analysis: An indirect and non-destructive methodology for the chlorophyll quantification of biocrusts. Remote Sens. 2019, 11, 1350. [Google Scholar] [CrossRef]
- Bruning, B.; Liu, H.; Brien, C.; Berger, B.; Lewis, M.; Garnett, T. The Development of hyperspectral distribution maps to predict the content and distribution of nitrogen and water in wheat (Triticum aestivum). Front. Plant Sci. 2019. [Google Scholar] [CrossRef]
- Lu, J.; Yang, T.; Su, X.; Qi, H.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.; Tian, Y. Monitoring leaf potassium content using hyperspectral vegetation indices in rice leaves. Precis. Agric. 2019. [Google Scholar] [CrossRef]
- Jull, H.; Künnemeyer, R.; Schaare, P. Nutrient quantification in fresh and dried mixtures of ryegrass and clover leaves using laser-induced breakdown spectroscopy. Precis. Agric. 2018, 19, 823–839. [Google Scholar] [CrossRef]
- Galvez-Sola, L.; García-Sánchez, F.; Pérez-Pérez, J.G.; Gimeno, V.; Navarro, J.M.; Moral, R.; Martínez-Nicolás, J.J.; Nieves, M. Rapid estimation of nutritional elements on citrus leaves by near infrared reflectance spectroscopy. Front. Plant Sci. 2015, 6, 1–8. [Google Scholar] [CrossRef]
- Yang, J.; Du, L.; Gong, W.; Shi, S.; Sun, J.; Chen, B. Analyzing the performance of the first-derivative fluorescence spectrum for estimating leaf nitrogen concentration. Opt. Express 2019, 27, 3978. [Google Scholar] [CrossRef]
- Yang, J.; Cheng, Y.; Du, L.; Gong, W.; Shi, S.; Sun, J.; Chen, B. Selection of the optimal bands of first-derivative fluorescence characteristics for leaf nitrogen concentration estimation. Appl. Opt. 2019, 58, 5720–5727. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, J.; Zou, M.; Liu, X.; Du, X.; Wang, Q.; Liu, Y.; Liu, Y.; Li, J. Prediction of cadmium concentration in brown rice before harvest by hyperspectral remote sensing. Environ. Sci. Pollut. Res. 2019, 26, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.M. Machine Learning, 1st ed.; McGraw-Hill, Inc.: New York, NY, USA, 1997. [Google Scholar]
- Ball, J.E.; Anderson, D.T.; Chan, C.S. Comprehensive survey of deep learning in remote sensing: Theories, tools, and challenges for the community. J. Appl. Remote Sens. 2017, 11, 1. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. ISPRS Journal of Photogrammetry and Remote Sensing Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Gao, J.; Meng, B.; Liang, T.; Feng, Q.; Ge, J.; Yin, J. ISPRS Journal of Photogrammetry and Remote Sensing Modeling alpine grassland forage phosphorus based on hyperspectral remote sensing and a multi-factor machine learning algorithm in the east of Tibetan. ISPRS J. Photogramm. Remote Sens. 2019, 147, 104–117. [Google Scholar] [CrossRef]
- Feng, P.; Wang, B.; Liu, D.L.; Yu, Q. Machine learning-based integration of remotely-sensed drought factors can improve the estimation of agricultural drought in South-Eastern Australia. Agric. Syst. 2019, 173, 303–316. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef]
- Singhal, G.; Bansod, B.; Mathew, L.; Goswami, J.; Choudhury, B.U.; Raju, P.L.N. Chlorophyll estimation using multi-spectral unmanned aerial system based on machine learning techniques. Remote Sens. Appl. Soc. Environ. 2019, 15, 100235. [Google Scholar] [CrossRef]
- Chanda, S.; Hazarika, A.K.; Choudhury, N.; Islam, S.A.; Manna, R.; Sabhapondit, S.; Tudu, B.; Bandyopadhyay, R. Support vector machine regression on selected wavelength regions for quantitative analysis of caffeine in tea leaves by near infrared spectroscopy. J. Chemom. 2019, 33, 10. [Google Scholar] [CrossRef]
- Shah, S.H.; Angel, Y.; Houborg, R.; Ali, S.; McCabe, M.F. A random forest machine learning approach for the retrieval of leaf chlorophyll content in wheat. Remote Sens. 2019, 11, 920. [Google Scholar] [CrossRef]
- Fu, P.; Meacham-Hensold, K.; Guan, K.; Bernacchi, C.J. Hyperspectral leaf reflectance as proxy for photosynthetic capacities: An ensemble approach based on multiple machine learning algorithms. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Obreza, T.A.; Morgan, K.T. (Eds.) Nutrition of Florida Citrus Trees, 2nd ed.; IFAS Extension; University of Florida: Gainesville, FL, USA, 2008. [Google Scholar]
- Köppen, W.; Volken, E.; Brönnimann, S. The thermal zones of the Earth according to the duration of hot, moderate and cold periods and to the impact of heat on the organic world. Meteorol. Zeitschrift 2011, 20, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Nitrogen Determination by Kjeldahl Method PanReac AppliChem ITW Reagents. Available online: https://www.itwreagents.com/uploads/20180114/A173_EN.pdf (accessed on 27 February 2019).
- Malavolta, E.; Vitti, G.C.; Oliveira, S.A. Evaluation of Nutritional Status of Plants: Principles and Perspectives, 2nd ed.; POTAFOS: Piracicaba, SP, Brazil, 1997; 319p. [Google Scholar]
- Anderson, K.; Rossini, M.; Labrador, J.P.; Balzarolo, M.; Arthur, A.; Fava, F.; Julitta, T.; Vescovo, L. Inter-comparison of hemispherical conical reflectance factors (HCRF) measured with four fiber-based spectrometers. Opt. Express. 2013, 21, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.; Philpot, W. Derivative Analysis of Hyperspectral Data. Remote Sens. Environ. 1998, 66, 41–51. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- RapidMiner. RapidMiner Python Package. Available online: https://github.com/rapidminer/pythonrapidminer (accessed on 5 December 2019).
- XGBoost. eXtreme Gradient Boosting. Available online: https://github.com/dmlc/xgboost (accessed on 5 December 2019).
- Amancio, D.R.; Comin, C.H.; Casanova, D.; Travieso, G.; Bruno, O.M.; Rodrigues, F.A.; Da Fontoura Costa, L. A systematic comparison of supervised classifiers. PLoS ONE 2014, 9, 1–14. [Google Scholar] [CrossRef]
- Pôças, I.; Tosin, R.; Gonçalves, I.; Cunha, M. Toward a generalized predictive model of grapevine water status in Douro region from hyperspectral data. Agric. For. Meteorol. 2020, 280, 107793. [Google Scholar] [CrossRef]
- Zha, H.; Miao, Y.; Wang, T.; Li, Y.; Zhang, J.; Sun, W. Sensing-Based Rice Nitrogen Nutrition Index Prediction with Machine Learning. Remote Sens. 2020, 12, 215. [Google Scholar] [CrossRef]
- Hennessy, A.; Clarke, K.; Lewis, M. Hyperspectral Classification of Plants: A Review of Waveband Selection Generalisability. Remote Sens. 2020, 12, 113. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Meeker, M.; La Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef] [PubMed]
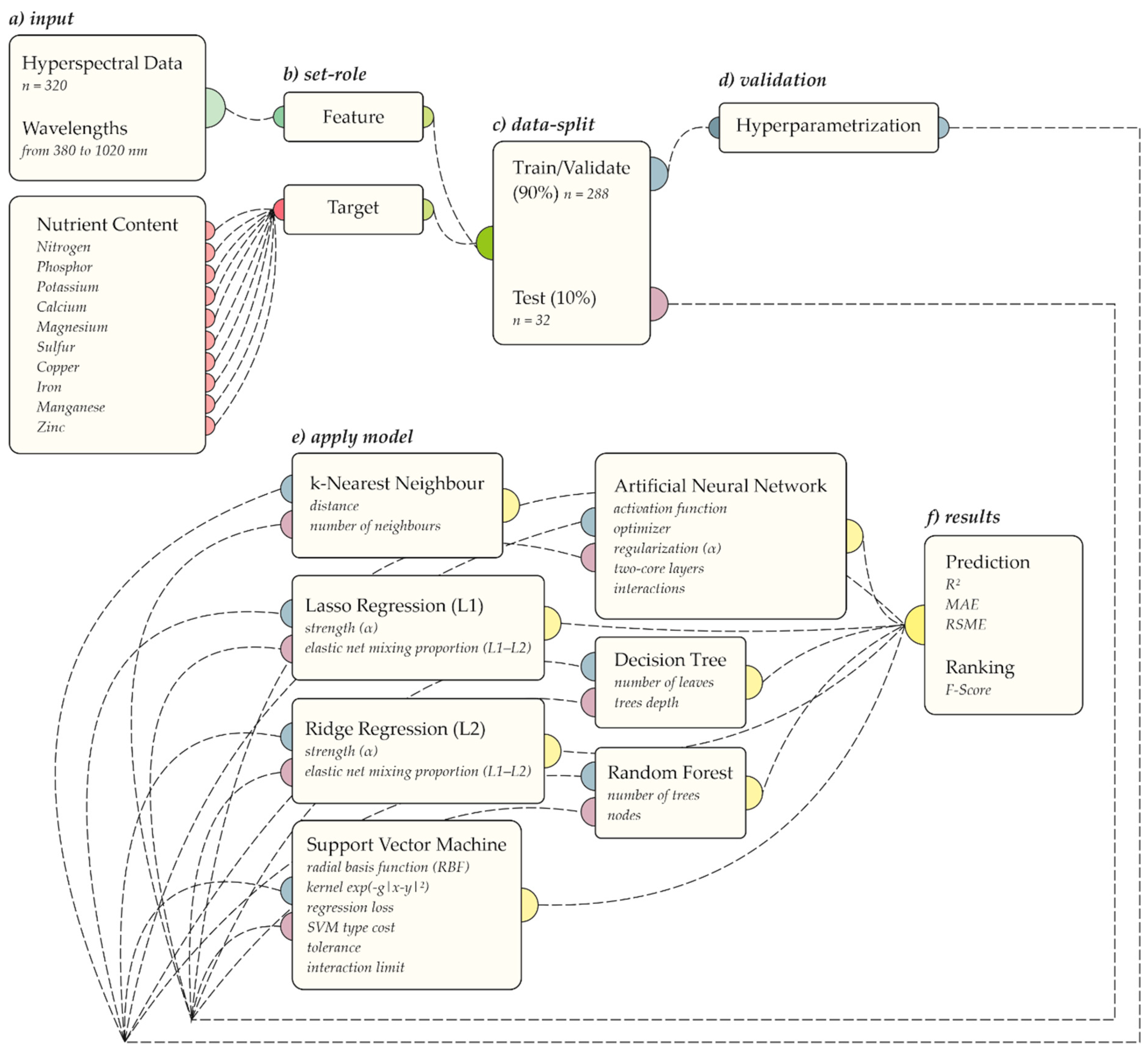
| Algorithm | Hyperparameter | Criteria |
|---|---|---|
| kNN | Distance Number of Neighbors | Euclidian k-Neighbors = 5 |
| Lasso Regression (L1) | Strength (α) Elastic Net Mixing Proportion (L1–L2) | 1.0 0.57:0.43 |
| Ridge Regression (L2) | Strength (α) Elastic Net Mixing Proportion (L1–L2) | 0.1 0.57:0.43 |
| SVM | Radial Basis Function (RBF) Kernel exp(-g|x-y|2) | g = automatic Regression Loss = 1.00 SVM Type Cost = 1 Tolerance = 0.001 Interaction (limit) = Unlimited |
| ANN | Activation: Logistic Function Adam Optimizer Regularization (α) = 0.0001 | Neurons (First Hidden Layer) = 400 Neurons (Second Hidden Layer) = 200 Interactions = 10,000 |
| DT | Number of Leaves Trees Depth | Leaves (minimal) = 2 Tree-depth (maximum) = 100 |
| RF | Number of Trees Nodes | Trees = 100 Nodes (maximum) = 5 |
| Method | N | P | K | Ca | Mg | S | Cu | Fe | Mn | Zn |
|---|---|---|---|---|---|---|---|---|---|---|
| kNN | ||||||||||
| MAE (Ref.)—Cross-Validation | 0.682 | 0.122 | 1.070 | 6.125 | 0.201 | 0.179 | 6.793 | 17.022 | 4.890 | 5.058 |
| MAE (1st Der.)—Cross-Validation | 0.884 | 0.174 | 1.235 | 6.358 | 0.357 | 0.154 | 6.589 | 16.789 | 3.012 | 4.246 |
| MAE (Ref.)—Leave-One-Out | 0.700 | 0.158 | 1.107 | 6.549 | 0.224 | 0.144 | 6.723 | 17.686 | 4.705 | 5.157 |
| MAE (1st Der.)—Leave-One-Out | 0.912 | 0.178 | 1.301 | 6.897 | 0.377 | 0.163 | 6.453 | 16.949 | 3.192 | 4.349 |
| Lasso Regression | ||||||||||
| MAE (Ref.)—Cross-Validation | 1.986 | 0.083 | 1.322 | 7.010 | 0.523 | 0.118 | 17.552 | 31.849 | 8.004 | 7.128 |
| MAE (1st Der.)—Cross-Validation | 1.056 | 0.079 | 1.756 | 6.849 | 0.446 | 0.287 | 17.389 | 30.595 | 7.341 | 9.058 |
| MAE (Ref.)—Leave-One-Out | 1.898 | 0.095 | 1.352 | 7.101 | 0.538 | 0.107 | 17.515 | 32.256 | 8.058 | 7.185 |
| MAE (1st Der.)—Leave-One-Out | 1.285 | 0.087 | 1.975 | 6.942 | 0.455 | 0.284 | 17.395 | 30.112 | 7.578 | 9.088 |
| Ridge Regression | ||||||||||
| MAE (Ref.)—Cross-Validation | 2.058 | 0.245 | 1.022 | 9.388 | 0.589 | 0.284 | 17.383 | 27.218 | 9.085 | 6.143 |
| MAE (1st Der.)—Cross-Validation | 1.359 | 0.234 | 1.766 | 6.238 | 0.687 | 0.235 | 17.287 | 33.185 | 7.898 | 8.897 |
| MAE (Ref.)—Leave-One-Out | 2.045 | 0.277 | 1.079 | 9.183 | 0.587 | 0.225 | 17.858 | 27.351 | 9.056 | 6.041 |
| MAE (1st Der.)—Leave-One-Out | 1.350 | 0.202 | 1.768 | 6.156 | 0.678 | 0.202 | 17.084 | 33.584 | 7.984 | 8.789 |
| SVM | ||||||||||
| MAE (Ref.)—Cross-Validation | 0.789 | 0.134 | 1.012 | 5.888 | 0.456 | 0.179 | 18.894 | 20.170 | 3.374 | 7.071 |
| MAE (1st Der.)—Cross-Validation | 1.058 | 0.158 | 1.415 | 5.979 | 0.568 | 0.199 | 10.152 | 23.158 | 6.385 | 5.759 |
| MAE (Ref.)—Leave-One-Out | 0.798 | 0.159 | 1.028 | 5.978 | 0.456 | 0.178 | 18.265 | 20.588 | 3.318 | 7.052 |
| MAE (1st Der.)—Leave-One-Out | 1.158 | 0.178 | 1.456 | 5.899 | 0.589 | 0.158 | 10.128 | 23.480 | 6.318 | 5.878 |
| ANN | ||||||||||
| MAE (Ref.)—Cross-Validation | 0.789 | 0.157 | 1.025 | 7.126 | 0.285 | 0.134 | 7.895 | 29.389 | 5.058 | 6.388 |
| MAE (1st Der.)—Cross-Validation | 1.058 | 0.193 | 1.453 | 6.087 | 0.456 | 0.146 | 9.185 | 19.241 | 4.358 | 5.268 |
| MAE (Ref.)—Leave-One-Out | 0.744 | 0.155 | 1.064 | 7.235 | 0.259 | 0.138 | 7.563 | 29.289 | 5.568 | 6.456 |
| MAE (1st Der.)—Leave-One-Out | 1.057 | 0.189 | 1.487 | 6.023 | 0.482 | 0.105 | 9.458 | 19.568 | 4.238 | 5.215 |
| DT | ||||||||||
| MAE (Ref.)—Cross-Validation | 0.689 | 0.158 | 1.056 | 6.054 | 0.315 | 0.113 | 7.874 | 19.498 | 4.286 | 5.158 |
| MAE (1st Der.)—Cross-Validation | 1.055 | 0.123 | 1.126 | 6.586 | 0.467 | 0.205 | 6.894 | 31.218 | 4.878 | 4.238 |
| MAE (Ref.)—Leave-One-Out | 0.641 | 0.102 | 1.085 | 6.088 | 0.305 | 0.106 | 7.415 | 19.352 | 4.984 | 5.512 |
| MAE (1st Der.)—Leave-One-Out | 1.028 | 0.112 | 1.285 | 6.547 | 0.489 | 0.202 | 6.897 | 31.189 | 4.489 | 4.354 |
| RF | ||||||||||
| MAE (Ref.)—Cross-Validation | 0.689 | 0.078 | 1.112 | 3.025 | 0.210 | 0.087 | 7.289 | 18.548 | 4.898 | 3.789 |
| MAE (1st Der.)—Cross-Validation | 0.638 | 0.101 | 1.207 | 6.238 | 0.389 | 0.179 | 6.046 | 16.189 | 3.874 | 1.789 |
| MAE (Ref.)—Leave-One-Out | 0.677 | 0.089 | 1.103 | 3.045 | 0.207 | 0.088 | 7.358 | 18.895 | 4.984 | 3.898 |
| MAE (1st Der.)—Leave-One-Out | 0.622 | 0.107 | 1.201 | 6.125 | 0.379 | 0.189 | 6.215 | 16.189 | 3.789 | 1.875 |
| Summary | Macronutrient (g/kg) | Micronutrient (mg/kg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | P | K | Ca | Mg | S | Cu | Fe | Mn | Zn | |
| Mean | 29.55 | 2.13 | 17.07 | 30.72 | 5.36 | 2.36 | 72.20 | 86.95 | 36.69 | 27.77 |
| Std. Dev. | 2.95 | 0.43 | 3.34 | 13.18 | 1.39 | 0.38 | 26.09 | 39.50 | 19.11 | 13.09 |
| Median | 29.45 | 2.17 | 16.70 | 28.85 | 5.25 | 2.35 | 69.90 | 78.35 | 33.10 | 22.80 |
| Min. | 24.00 | 1.21 | 11.80 | 10.70 | 2.70 | 1.60 | 25.50 | 26.20 | 14.30 | 10.90 |
| Max. | 36.70 | 2.98 | 28.30 | 78.60 | 9.90 | 3.60 | 128.90 | 207.30 | 122.10 | 69.80 |
| Coeff. Var. | 9.98 | 20.39 | 19.60 | 42.90 | 25.97 | 16.37 | 36.14 | 45.44 | 52.105 | 47.16 |
| Method | N | P | K | Ca | Mg | S | Cu | Fe | Mn | Zn |
|---|---|---|---|---|---|---|---|---|---|---|
| kNN | ||||||||||
| R2 | 0.852 | 0.623 | 0.621 | 0.179 | 0.797 | 0.119 | 0.834 | 0.437 | 0.592 | 0.431 |
| MAE | 0.704 | 0.163 | 1.087 | 6.765 | 0.285 | 0.204 | 7.083 | 18.142 | 5.105 | 6.005 |
| RMSE | 1.245 | 0.278 | 2.041 | 13.905 | 0.445 | 0.416 | 11.362 | 36.248 | 11.707 | 10.157 |
| Lasso Regression | ||||||||||
| R2 | 0.394 | 0.452 | 0.315 | 0.157 | 0.413 | 0.660 | 0.215 | 0.180 | 0.189 | 0.128 |
| MAE | 2.145 | 0.193 | 1.542 | 7.304 | 0.627 | 0.137 | 19.881 | 34.140 | 8.470 | 7.852 |
| RMSE | 2.526 | 0.335 | 2.745 | 14.091 | 0.757 | 0.258 | 24.744 | 43.751 | 16.513 | 12.573 |
| Ridge Regression | ||||||||||
| R2 | 0.351 | 0.153 | 0.354 | 0.169 | 0.139 | 0.056 | 0.232 | 0.222 | 0.190 | 0.137 |
| MAE | 2.468 | 0.284 | 1.347 | 9.923 | 0.698 | 0.298 | 19.321 | 28.456 | 9.456 | 6.541 |
| RMSE | 2.912 | 0.417 | 2.597 | 13.989 | 0.916 | 0.431 | 24.485 | 42.158 | 16.502 | 12.982 |
| SVM | ||||||||||
| R2 | 0.638 | 0.404 | 0.530 | 0.336 | 0.458 | 0.400 | 0.277 | 0.308 | 0.742 | 0.447 |
| MAE | 0.902 | 0.247 | 1.546 | 6.551 | 0.505 | 0.233 | 19.692 | 21.421 | 3.666 | 7.891 |
| RMSE | 1.952 | 0.349 | 2.275 | 12.501 | 0.752 | 0.344 | 23.754 | 40.201 | 9.309 | 9.741 |
| ANN | ||||||||||
| R2 | 0.860 | 0.656 | 0.762 | 0.481 | 0.733 | 0.438 | 0.841 | 0.340 | 0.698 | 0.595 |
| MAE | 0.840 | 0.177 | 1.265 | 7.637 | 0.359 | 0.174 | 8.377 | 30.259 | 5.880 | 6.949 |
| RMSE | 1.211 | 0.265 | 1.619 | 11.052 | 0.510 | 0.332 | 11.120 | 39.251 | 10.078 | 8.567 |
| DT | ||||||||||
| R2 | 0.743 | 0.661 | 0.613 | 0.576 | 0.759 | 0.452 | 0.731 | 0.453 | 0.730 | 0.640 |
| MAE | 0.787 | 0.178 | 1.434 | 6.375 | 0.345 | 0.166 | 8.835 | 20.016 | 5.090 | 5.681 |
| RMSE | 1.644 | 0.263 | 2.064 | 12.123 | 0.484 | 0.328 | 14.472 | 35.726 | 9.525 | 8.0811 |
| RF | ||||||||||
| R2 | 0.912 | 0.771 | 0.699 | 0.624 | 0.832 | 0.727 | 0.754 | 0.527 | 0.854 | 0.741 |
| MAE | 0.706 | 0.093 | 1.146 | 3.525 | 0.234 | 0.100 | 7.828 | 19.375 | 5.093 | 4.246 |
| RMSE | 1.059 | 0.216 | 1.818 | 9.404 | 0.405 | 0.231 | 13.850 | 33.233 | 7.007 | 6.846 |
| Method | N | P | K | Ca | Mg | S | Cu | Fe | Mn | Zn |
|---|---|---|---|---|---|---|---|---|---|---|
| kNN | ||||||||||
| R2 | 0.669 | 0.453 | 0.329 | 0.311 | 0.512 | 0.172 | 0.752 | 0.512 | 0.898 | 0.587 |
| MAE | 0.944 | 0.180 | 1.341 | 6.994 | 0.398 | 0.184 | 7.456 | 17.846 | 3.594 | 4.948 |
| RMSE | 1.867 | 0.335 | 2.717 | 12.039 | 0.689 | 0.404 | 13.903 | 33.740 | 5.859 | 8.655 |
| Lasso Regression | ||||||||||
| R2 | 0.257 | 0.401 | 0.161 | 0.287 | 0.401 | 0.158 | 0.292 | 0.190 | 0.234 | 0.130 |
| MAE | 1.489 | 0.197 | 1.986 | 7.048 | 0.486 | 0.304 | 19.513 | 33.146 | 7.934 | 9.588 |
| RMSE | 2.804 | 0.354 | 3.040 | 12.258 | 0.789 | 0.407 | 23.503 | 43.487 | 16.055 | 12.561 |
| Ridge Regression | ||||||||||
| R2 | 0.210 | 0.310 | 0.157 | 0.302 | 0.129 | 0.222 | 0.273 | 0.183 | 0.300 | 0.158 |
| MAE | 1.650 | 0.265 | 1.997 | 6.978 | 0.789 | 0.248 | 19.212 | 34.240 | 8.242 | 9.047 |
| RMSE | 2.987 | 0.380 | 3.101 | 12.268 | 0.987 | 0.358 | 23.819 | 44.289 | 15.348 | 11.978 |
| SVM | ||||||||||
| R2 | 0.373 | 0.323 | 0.330 | 0.531 | 0.270 | 0.459 | 0.679 | 0.423 | 0.649 | 0.513 |
| MAE | 1.357 | 0.250 | 1.714 | 6.745 | 0.678 | 0.240 | 11.870 | 24.653 | 6.676 | 6.159 |
| RMSE | 2.262 | 0.373 | 2.716 | 10.511 | 0.844 | 0.326 | 15.829 | 36.705 | 10.855 | 9.397 |
| ANN | ||||||||||
| R2 | 0.721 | 0.554 | 0.445 | 0.566 | 0.564 | 0.582 | 0.800 | 0.444 | 0.838 | 0.731 |
| MAE | 1.287 | 0.219 | 1.680 | 6.495 | 0.559 | 0.197 | 10.030 | 20.466 | 4.617 | 5.605 |
| RMSE | 1.712 | 0.302 | 2.471 | 10.113 | 0.652 | 0.287 | 12.493 | 36.028 | 7.372 | 6.983 |
| DT | ||||||||||
| R2 | 0.703 | 0.633 | 0.491 | 0.491 | 0.479 | 0.474 | 0.786 | 0.509 | 0.728 | 0.584 |
| MAE | 1.298 | 0.136 | 1.223 | 6.850 | 0.597 | 0.232 | 7.013 | 32.559 | 4.976 | 4.832 |
| RMSE | 2.767 | 0.274 | 2.368 | 13.015 | 0.832 | 0.314 | 25.209 | 33.861 | 15.036 | 11.391 |
| RF | ||||||||||
| R2 | 0.866 | 0.765 | 0.548 | 0.501 | 0.507 | 0.453 | 0.861 | 0.612 | 0.879 | 0.855 |
| MAE | 0.738 | 0.119 | 1.225 | 6.668 | 0.424 | 0.209 | 6.509 | 17.280 | 4.050 | 2.075 |
| RMSE | 1.185 | 0.219 | 2.231 | 10.839 | 0.693 | 0.328 | 10.389 | 31.640 | 6.377 | 5.121 |
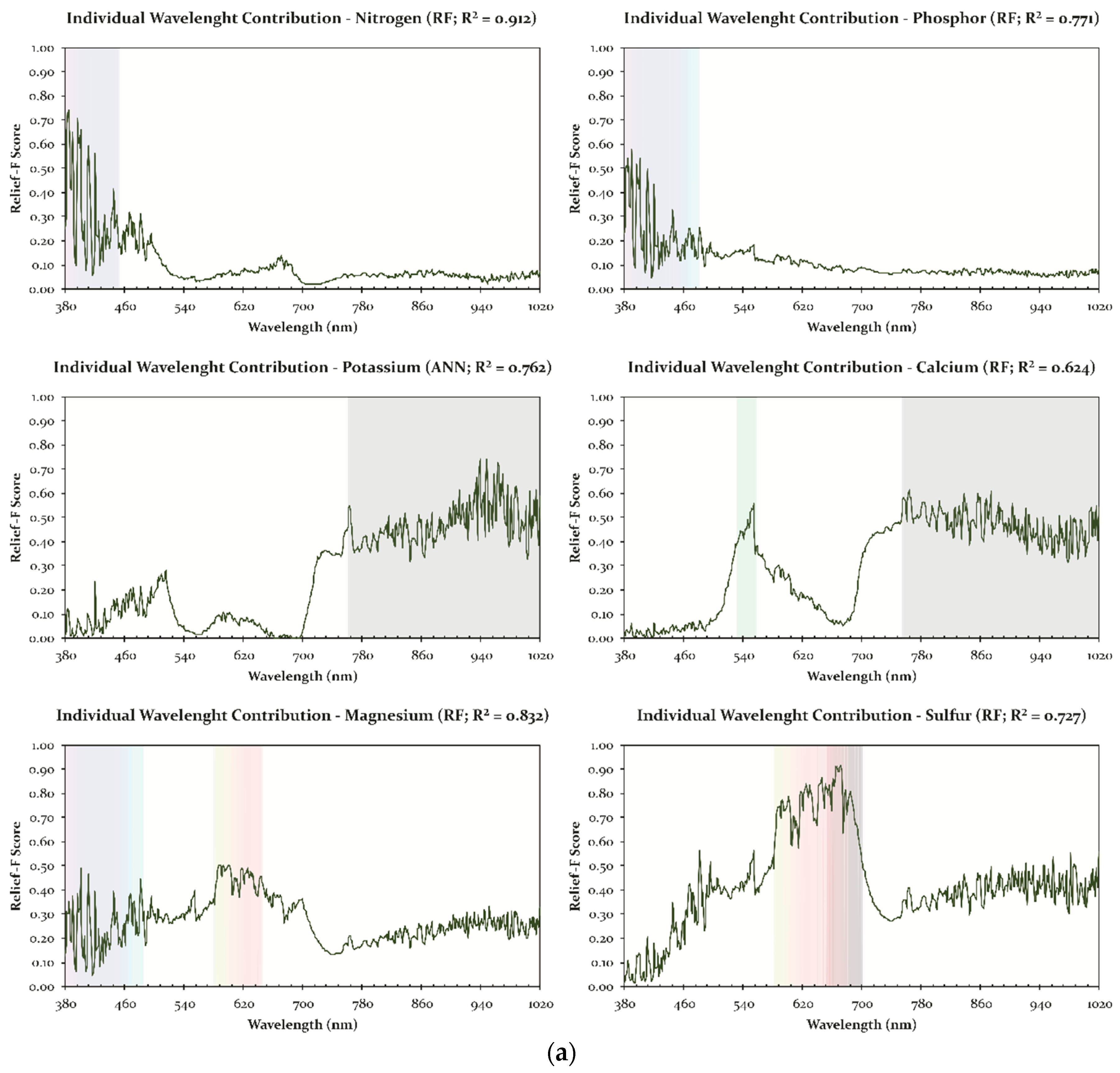
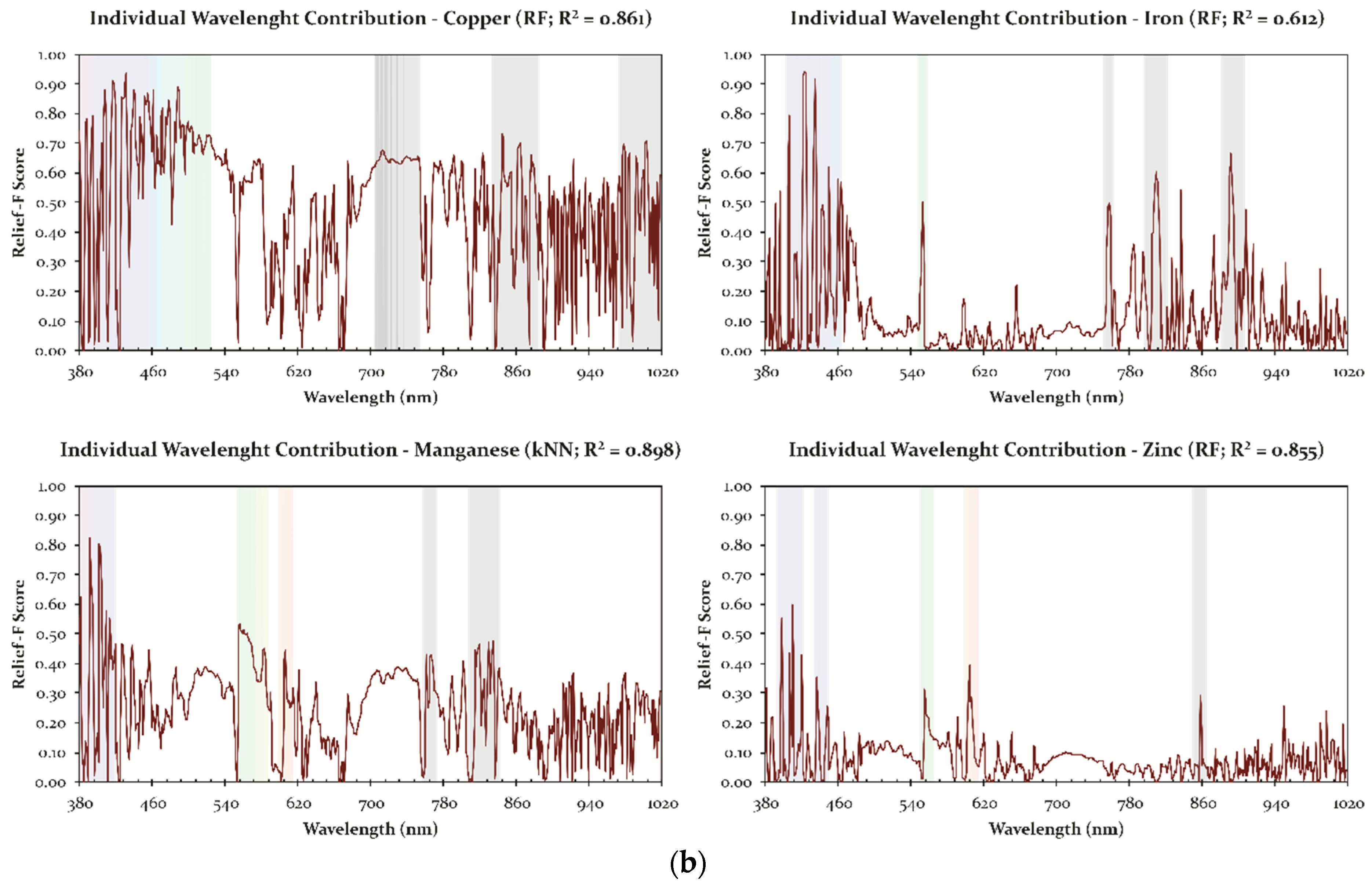
| Nutrient | Class | Method | R2 | MAE | Spectral Data | Contributive Wavelengths/Spectral Regions (nm) * |
|---|---|---|---|---|---|---|
| N | Macro | RF | 0.912 | 0.706 | Reflectance | 384–412; 421; 423; 432; 433; 435; 440–455; 464–472; 480–487 |
| P | Macro | RF | 0.771 | 0.093 | Reflectance | 385–411; 438–456; 472–477; 502; 521; 527; 544–555; |
| K | Macro | ANN | 0.763 | 1.265 | Reflectance | 762–764; 816; 838; 857; 903 908; 915–925; 934–957; 973–1020 |
| Ca | Macro | RF | 0.624 | 3.525 | Reflectance | 545–551; 749–787; 843–888; 901–1020 |
| Mg | Macro | RF | 0.832 | 0.234 | Reflectance | 390; 411–412; 445; 496; 554; 586–630; 643; 656–669 |
| S | Macro | RF | 0.727 | 0.100 | Reflectance | 579; 590; 595; 609–612; 618; 624–632; 645–680; 684–689; 700 |
| Cu | Micro | RF | 0.861 | 6.509 | First-Derivative | 388; 394; 416–419; 430–432; 440; 452–456; 461; 475; 512; 523; 823; 863–865; 951; 977–979; |
| Fe | Micro | RF | 0.612 | 17.280 | First-Derivative | 391–396; 405; 421–424; 433–436; 474–477; 552; 758; 810; 837; 890–892; 910; 926 |
| Mn | Micro | kNN | 0.898 | 3.594 | First-Derivative | 381; 392–410; 414; 438; 555–568; 582; 819; 607; 761–767; 823–835; 841 |
| Zn | Micro | RF | 0.855 | 2.075 | First-Derivative | 381; 398; 407–411; 420; 449; 555–559; 604–607; 858 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osco, L.P.; Ramos, A.P.M.; Faita Pinheiro, M.M.; Moriya, É.A.S.; Imai, N.N.; Estrabis, N.; Ianczyk, F.; Araújo, F.F.d.; Liesenberg, V.; Jorge, L.A.d.C.; et al. A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sens. 2020, 12, 906. https://doi.org/10.3390/rs12060906
Osco LP, Ramos APM, Faita Pinheiro MM, Moriya ÉAS, Imai NN, Estrabis N, Ianczyk F, Araújo FFd, Liesenberg V, Jorge LAdC, et al. A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sensing. 2020; 12(6):906. https://doi.org/10.3390/rs12060906
Chicago/Turabian StyleOsco, Lucas Prado, Ana Paula Marques Ramos, Mayara Maezano Faita Pinheiro, Érika Akemi Saito Moriya, Nilton Nobuhiro Imai, Nayara Estrabis, Felipe Ianczyk, Fábio Fernando de Araújo, Veraldo Liesenberg, Lúcio André de Castro Jorge, and et al. 2020. "A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements" Remote Sensing 12, no. 6: 906. https://doi.org/10.3390/rs12060906
APA StyleOsco, L. P., Ramos, A. P. M., Faita Pinheiro, M. M., Moriya, É. A. S., Imai, N. N., Estrabis, N., Ianczyk, F., Araújo, F. F. d., Liesenberg, V., Jorge, L. A. d. C., Li, J., Ma, L., Gonçalves, W. N., Marcato Junior, J., & Eduardo Creste, J. (2020). A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sensing, 12(6), 906. https://doi.org/10.3390/rs12060906