Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Objects of the Study
2.2. Field Measurements
2.3. Airborne Hyperspectral HySpex Data
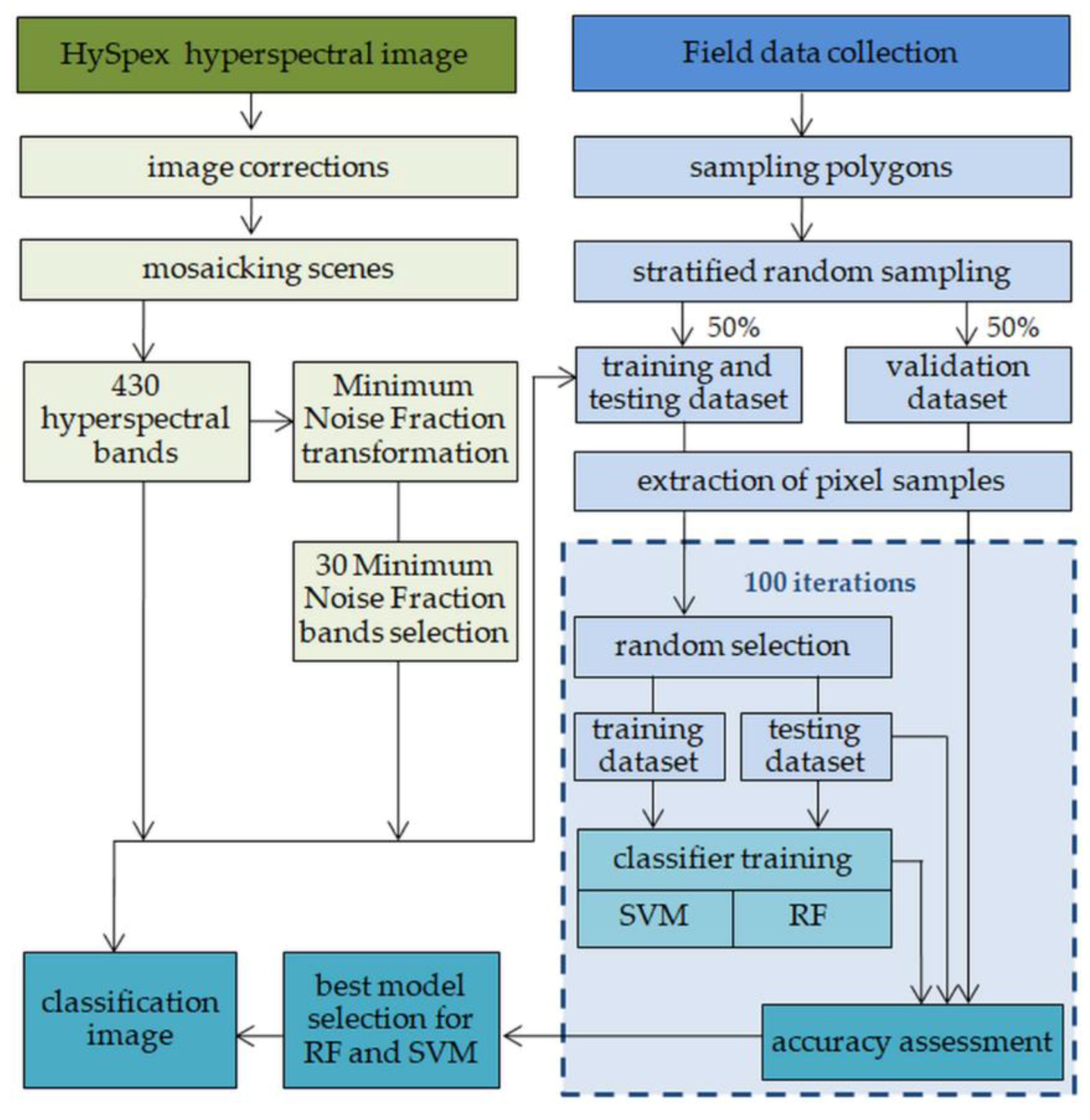
2.4. Classification Process and Accuracy Assessment
- Sub-sample the training test data set in order to create a training data set with a set number of samples per class;
- Train SVM and RF classifiers;
- Assess accuracy using test and validation data sets; and
- Save trained classifier models and accuracy measures for further analysis.
- Overall accuracy—the ratio of the total number of correctly classified pixels to the total number of reference pixels [45];
- Cohen’s Kappa—the similarity of the analyzed classification compared to the random classification (a Kappa value of 0 means full similarity while 1 means no similarity) [46];
- Producer’s Accuracy (PA)—the ratio of correctly classified pixels of a given class to all pixels in the validation data set for this class [45];
- User’s Accuracy (UA)—the ratio of pixels correctly classified in a given class to all pixels classified in this category [45]; and
3. Results
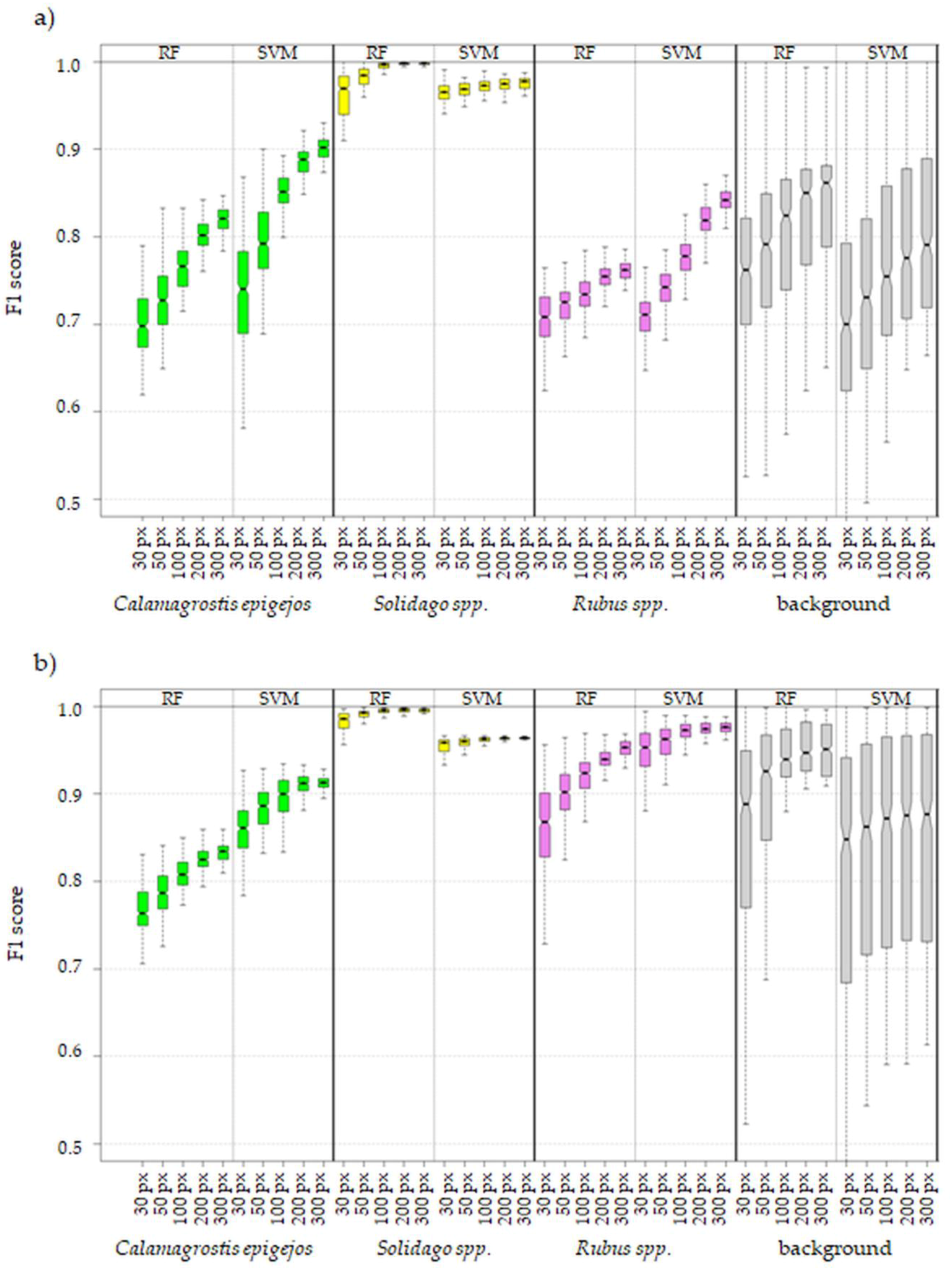
3.1. Statistical Analysis of Investigated Classification Scenarios
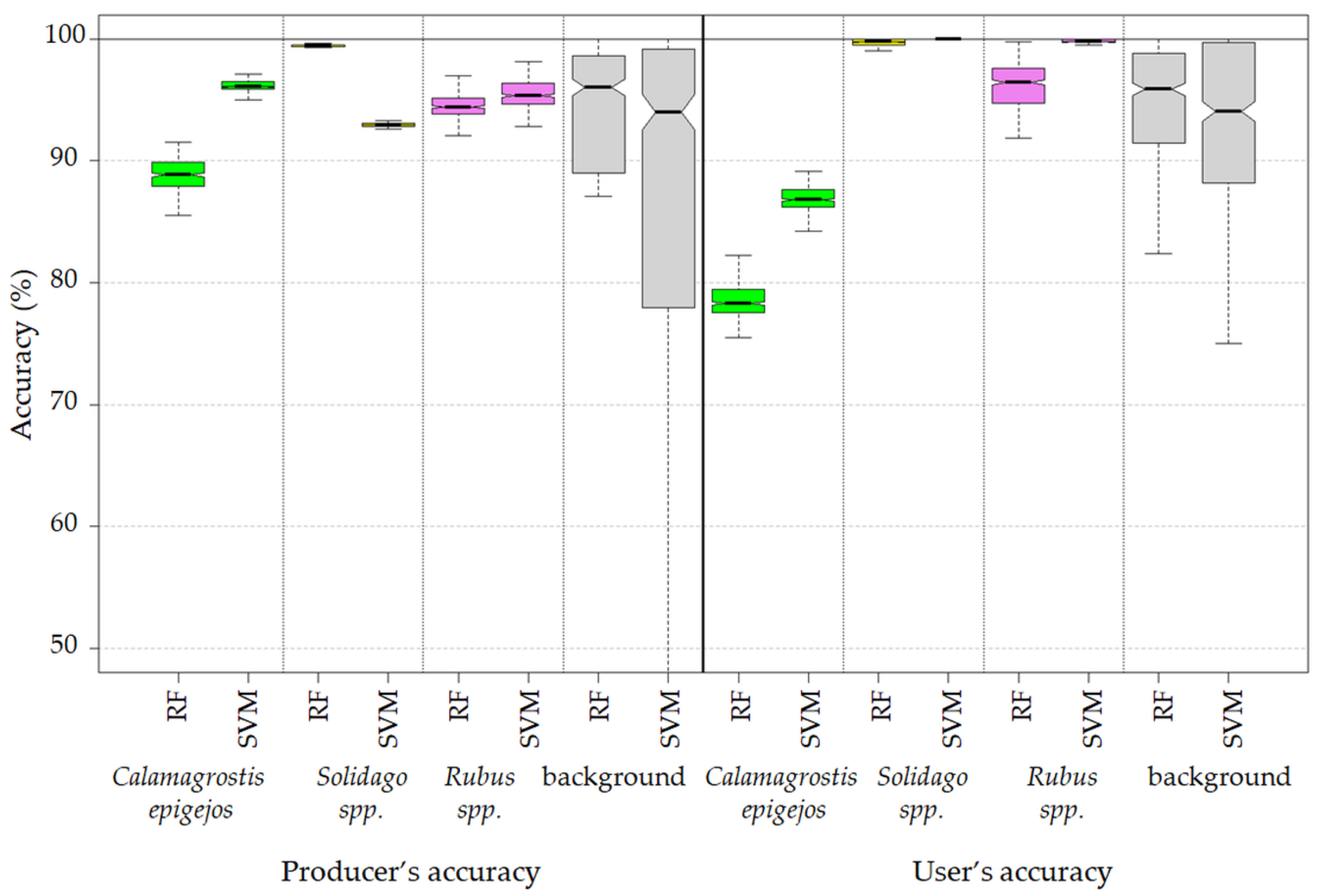
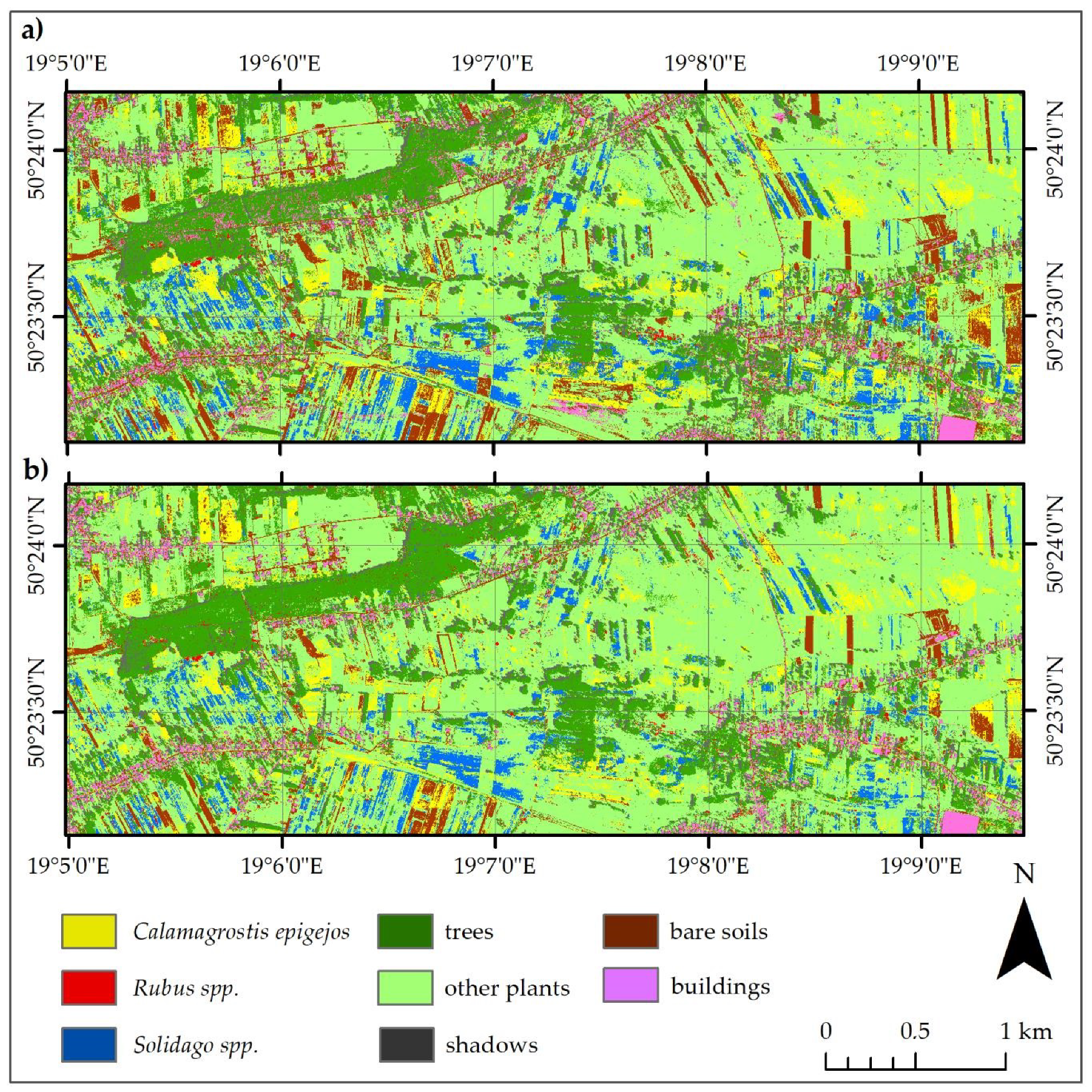
3.2. Best Model Plant Species Identification Accuracy
4. Discussion
5. Conclusions
- The accuracy assessment method presented in the paper allows us to confirm that all analyzed species can be identified in heterogenous habitats (achieved classification results F1 oscillated around 0.90). The species created a unique set of spectral properties, which are recognizable by the SVM and RF classifiers, and separating training and validation sets at the level of the reference polygons, and not at the level of individual pixels, is justified. This allows one to avoid overestimating the accuracy of the results due to spatial correlation of pixels from the same reference polygon. We have shown a clear need to divide classes into training and validation at the polygon level in order to minimize spatial correlation between samples and in order to achieve unbiased and accurate classification metrics. A spatially separate and unchanging validation set can be used to improve the quality of the obtained accuracy scores and compare the results more objectively. Unfortunately, this type of approach makes it more difficult to use iterative methods of assessing accuracy or significantly reduces the number of observations in a data set that can be used for classifier training. What is more, the principles of a constant and unchanging validation set are not optimal, which may negatively affect the quality of the resulting post-classification images. A set of 30 MNF bands allows for more accurate identification of the analyzed invasive and expansive plant species than that of the 430 original spectral bands of the HySpex image.
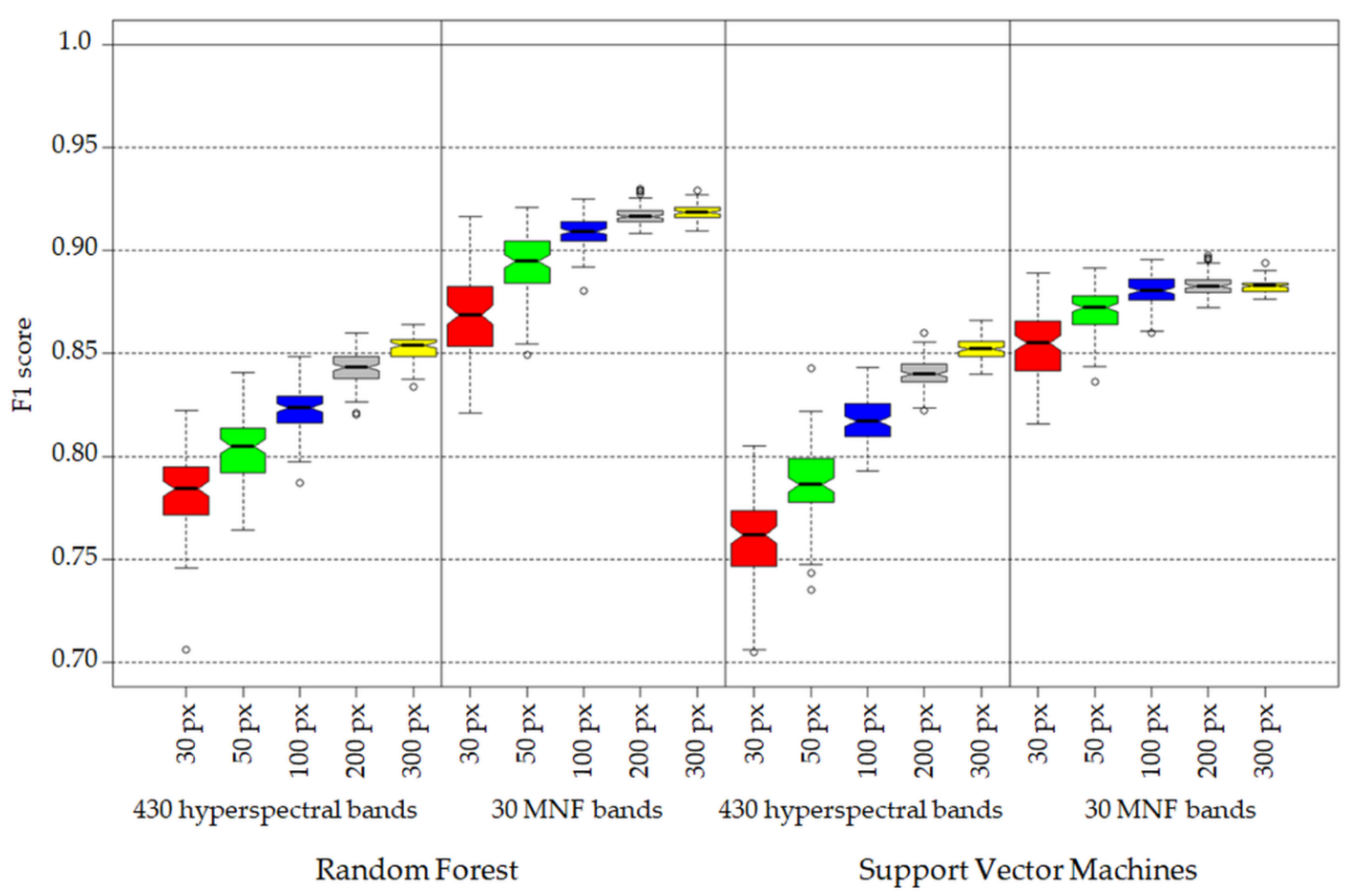
- Increase of number of pixels per class in training data set has a greater effect on achieved accuracy measures in the case of 430 spectral bands data set (difference in medians between 30 and 300 pixel data sets around 8 percentage points (p.p.) in the case of RF and 9 p.p. in the case of SVM algorithm) then in the case of 30 MNF bands data set (median difference between 30 and 300 pixel data set: 5 p. p. for RF and 3 p.p for SVM, Figure 7).
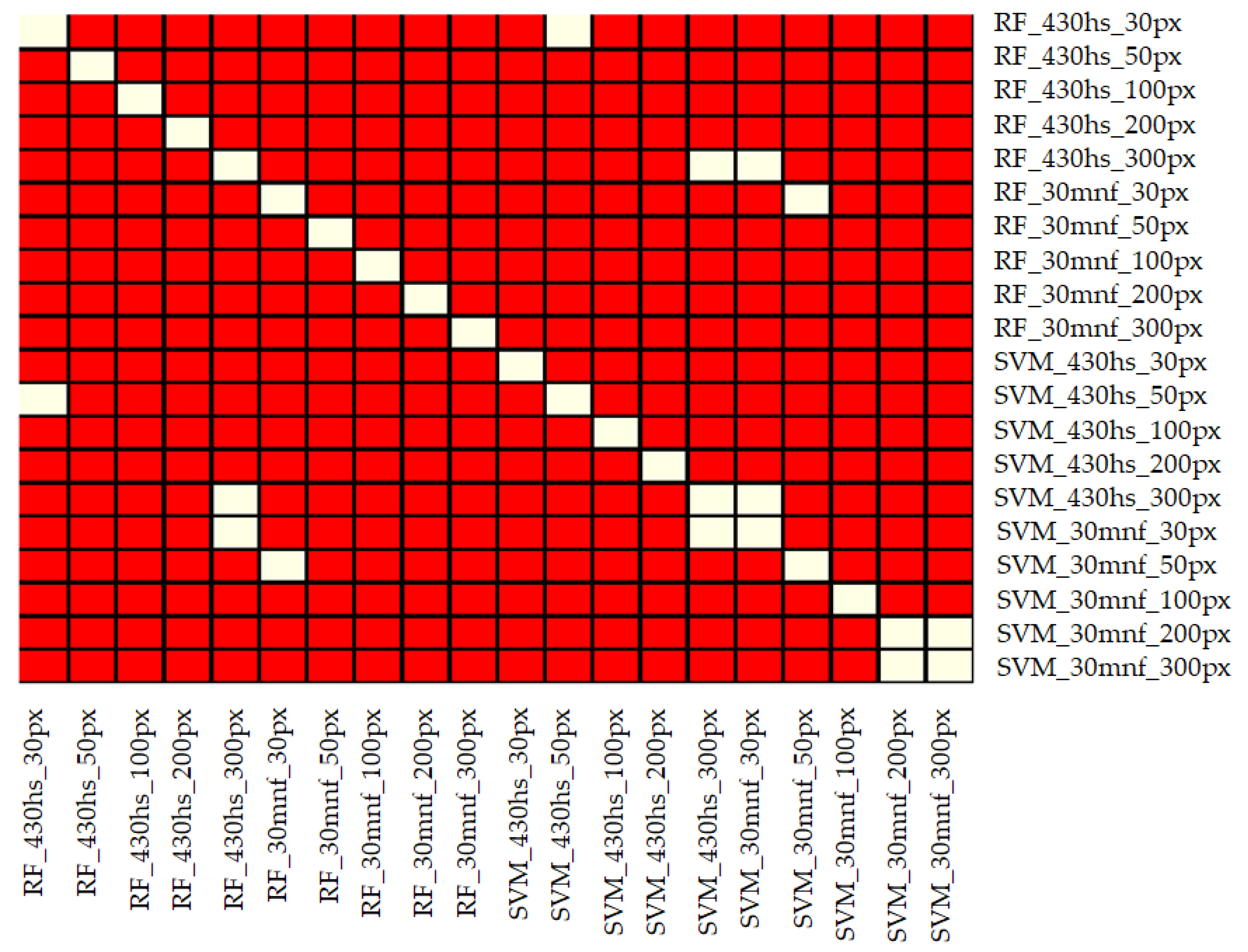
- Three hundred pixels per class is the preferred number of samples in the training set for classification of the analyzed plant species with the help of the SVM and RF methods. Fewer pixels result in a significant decrease in classification accuracy and less stable results. In our case, we managed to find the optimal number of pixels in training data sets per class only in the case of the SVM classifier applied to MNF data. Figure 6 shows that there was no significant statistical difference between tests performed on MNF bands with 200 and 300 pixel samples per class. Hence, 200 pixels per class in the training data set for 30 MNF bands and the SVM classifier is optimal.
- Both the Support Vector Machine method and the Random Forest method allowed us to obtain very accurate images of the distribution of analyzed species in the research area. However, the SVM classifier worked better for the classification of blackberry and wood small-reed (i.e., for classes that are not uniform and do not differ spectrally from their surroundings). On the other hand, the Random Forest algorithm allows one to obtain a higher accuracy for homogeneous classes that stand out spectrally (i.e., goldenrod and background classes). Still, the SVM image was found to be more reliable, despite its relatively lower accuracy for the background classes. Most classification errors occurred between background classes rather than individual species.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mooney, H.A.; Cleland, E.E. The evolutionary impact of invasive species. Proc. Natl. Acad. Sci. USA 2001, 98, 5446–5451. [Google Scholar] [CrossRef] [PubMed]
- Tokarska-Guzik, B.; Dajdok, Z.; Zając, M.; Zając, A.; Urbisz, A.; Danielewicz, W.; Hołdyński, C. Rośliny Obcego Pochodzenia w Polsce ze Szczególnym Uwzględnieniem Gatunków Inwazyjnych; Generalna Dyrekcja Ochrony Środowiska: Warszawa, Poland, 2012; ISBN 978-83-62940-34-9. [Google Scholar]
- Hulme, P.E.; Pyšek, P.; Nentwig, W.; Vilà, M. Will Threat of Biological Invasions Unite the European Union? Science 2009, 324, 40–41. [Google Scholar] [CrossRef] [PubMed]
- Holub, P.; Tůma, I.; Fiala, K. The effect of nitrogen addition on biomass production and competition in three expansive tall grasses. Environ. Pollut. 2012, 170, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Pruchniewicz, D.; Żołnierz, L. The influence of environmental factors and management methods on the vegetation of mesic grasslands in a central European mountain range. Flora-Morphol. Distrib. Funct. Ecol. Plants 2014, 209, 687–692. [Google Scholar] [CrossRef]
- Babai, D.; Molnár, Z. Small-scale traditional management of highly species-rich grasslands in the Carpathians. Agric. Ecosyst. Environ. 2014, 182, 123–130. [Google Scholar] [CrossRef]
- Sedláková, I.; Fiala, K. Ecological problems of degradation of alluvial meadows due to expanding Calamagrostis epigejos. Ekol. Bratisl. 2001, 20, 226–233. [Google Scholar]
- Huang, C.-Y.; Asner, G. Applications of Remote Sensing to Alien Invasive Plant Studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef]
- Sabat-Tomala, A.; Jarocińska, A.M.; Zagajewski, B.; Magnuszewski, A.S.; Sławik, Ł.M.; Ochtyra, A.; Raczko, E.; Lechnio, J.R. Application of HySpex hyperspectral images for verification of a two-dimensional hydrodynamic model. Eur. J. Remote Sens. 2018, 51, 637–649. [Google Scholar] [CrossRef]
- Curatola Fernández, G.; Obermeier, W.; Gerique, A.; López Sandoval, M.; Lehnert, L.; Thies, B.; Bendix, J. Land Cover Change in the Andes of Southern Ecuador—Patterns and Drivers. Remote Sens. 2015, 7, 2509–2542. [Google Scholar] [CrossRef]
- Rapinel, S.; Clément, B.; Magnanon, S.; Sellin, V.; Hubert-Moy, L. Identification and mapping of natural vegetation on a coastal site using a Worldview-2 satellite image. J. Environ. Manag. 2014, 144, 236–246. [Google Scholar] [CrossRef]
- Hestir, E.L.; Khanna, S.; Andrew, M.E.; Santos, M.J.; Viers, J.H.; Greenberg, J.A.; Rajapakse, S.S.; Ustin, S.L. Identification of invasive vegetation using hyperspectral remote sensing in the California Delta ecosystem. Remote Sens. Environ. 2008, 112, 4034–4047. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Despain, D.G.; Clark, R.N.; Livo, K.E. Mapping vegetation in Yellowstone National Park using spectral feature analysis of AVIRIS data. Remote Sens. Environ. 2003, 84, 437–456. [Google Scholar] [CrossRef]
- Okin, G.S.; Roberts, D.A.; Murray, B.; Okin, W.J. Practical limits on hyperspectral vegetation discrimination in arid and semiarid environments. Remote Sens. Environ. 2001, 77, 212–225. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (randomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Pengra, B.W.; Johnston, C.A.; Loveland, T.R. Mapping an invasive plant, Phragmites australis, in coastal wetlands using the EO-1 Hyperion hyperspectral sensor. Remote Sens. Environ. 2007, 108, 74–81. [Google Scholar] [CrossRef]
- Lass, L.W.; Thill, D.C.; Shafii, B.; Prather, T.S. Detecting Spotted Knapweed (Centaurea maculosa) with Hyperspectral Remote Sensing. Weed Technol. 2002, 16, 426–432. [Google Scholar] [CrossRef]
- Skowronek, S.; Ewald, M.; Isermann, M.; Van De Kerchove, R.; Lenoir, J.; Aerts, R.; Warrie, J.; Hattab, T.; Honnay, O.; Schmidtlein, S.; et al. Mapping an invasive bryophyte species using hyperspectral remote sensing data. Biol. Invasions 2017, 19, 239–254. [Google Scholar] [CrossRef]
- Ishii, J.; Washitani, I. Early detection of the invasive alien plant Solidago altissima in moist tall grassland using hyperspectral imagery. Int. J. Remote Sens. 2013, 34, 5926–5936. [Google Scholar] [CrossRef]
- Atkinson, J.T.; Ismail, R.; Robertson, M. Mapping Bugweed (Solanum mauritianum) Infestations in Pinus patula Plantations Using Hyperspectral Imagery and Support Vector Machines. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 17–28. [Google Scholar] [CrossRef]
- Sluiter, R.; Pebesma, E.J. Comparing techniques for vegetation classification using multi- and hyperspectral images and ancillary environmental data. Int. J. Remote Sens. 2010, 31, 6143–6161. [Google Scholar] [CrossRef]
- Vapnik, V.; Lerner, A. Pattern recognition using generalized portrait method. Autom. Remote Control 1963, 24, 774–780. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Calpe-Maravilla, J.; Martin-Guerrero, J.D.; Soria-Olivas, E.; Alonso-Chorda, L.; Moreno, J. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1530–1542. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Jarocińska, A.; Bzdęga, K.; Tokarska-Guzik, B. Classification of Expansive Grassland Species in Different Growth Stages Based on Hyperspectral and LiDAR Data. Remote Sens. 2018, 10, 2019. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of herbaceous vegetation using airborne hyperspectral imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef]
- McPheeters, K.D.; Skirvin, R.M.; Hall, H.K. Brambles (Rubus spp.). In Crops II.; Bajaj, Y.P.S., Ed.; Springer: Heidelberg, Germany, 1988; pp. 104–123. ISBN 3642735223. [Google Scholar]
- Grime, J.P.; Hodgson, J.G.; Hunt, R. Comparative Plant Ecology; Springer: Dordrecht, The Netherlands, 1988; Volume 51, ISBN 978-0-412-74170-8. [Google Scholar]
- Balandier, P.; Marquier, A.; Casella, E.; Kiewitt, A.; Coll, L.; Wehrlen, L.; Harmer, R. Architecture, cover and light interception by bramble (Rubus fruticosus): a common understorey weed in temperate forests. Forestry 2013, 86, 39–46. [Google Scholar] [CrossRef]
- Wolanin, M.M.; Wolanin, M.N.; Oklejewicz, K. Occurrence of brambles (Rubus L.) in young forest plantations on the Kolbuszowa Plateau. For. Res. Pap. 2017, 78, 179–186. [Google Scholar] [CrossRef][Green Version]
- Dehaan, R.; Louis, J.; Wilson, A.; Hall, A.; Rumbachs, R. Discrimination of blackberry (Rubus fruticosus sp. agg.) using hyperspectral imagery in Kosciuszko National Park, NSW, Australia. ISPRS J. Photogramm. Remote Sens. 2007, 62, 13–24. [Google Scholar] [CrossRef]
- Pruchniewicz, D.; Żołnierz, L. The influence of Calamagrostis epigejos expansion on the species composition and soil properties of mountain mesic meadows. Acta Soc. Bot. Pol. 2016, 86, 1–11. [Google Scholar] [CrossRef]
- Aiken, S.G.; Dore, W.G.; Lefkovitch, L.P.; Armstrong, K.C. Calamagrostis epigejos (Poaceae) in North America, especially Ontario. Can. J. Bot. 1989, 67, 3205–3218. [Google Scholar] [CrossRef]
- Rebele, F.; Lehmann, C. Biological flora of central europe: Calamagrostis epigejos (L.) Roth. Flora 2001, 196, 325–344. [Google Scholar] [CrossRef]
- Rebele, F. Competition and coexistence of rhizomatous perennial plants along a nutrient gradient. Plant Ecol. 2000, 147, 77–94. [Google Scholar] [CrossRef]
- Kabuce, N.; Priede, A. NOBANIS-Invasive Alien Species Fact Sheet Solidago Canadensis. 2019. Available online: www.nobanis.org (accessed on 16 November 2019).
- Guzikowa, M.; Maycock, P.F. The invasion and expansion of three North American species of goldenrod (Solidago canadensis L. sensu lato. S. gigantea Ait. and S. graminifolia (L) Salisb in Poland. Acta Soc. Bot. Pol. 2014, 55, 367–384. [Google Scholar] [CrossRef]
- Weber, E. Current and Potential Ranges of Three Exotic Goldenrods (Solidago) in Europe. Conserv. Biol. 2001, 15, 122–128. [Google Scholar] [CrossRef]
- Werner, P.A.; Gross, R.S.; Bradbury, I.K. The biology of Canadian Weeds. 45. Solidago canadensis L. Can. J. Plant Sci. 1980, 60, 1393–1409. [Google Scholar] [CrossRef]
- Shui-Liang, G.; Fang, F. Physiological Adaptation of the Invasive Plant Solidago canadensis to Environments. Chinese J. Plant Ecol. 2003, 27, 47–52. [Google Scholar] [CrossRef]
- Yang, R.-Y.; Tang, J.-J.; Yang, Y.-S.; Chen, X. Invasive and non-invasive plants differ in response to soil heavy metal lead contamination. Bot. Stud. 2007, 48, 453–458. [Google Scholar]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Raczko, E.; Ochtyra, A.; Jarocińska, A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sens. 2018, 10, 570. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Lillesand, T.; Kiefer, R.; Chipman, J. Remote Sensing and Image Interpretation, 7th ed.; Wiley: Hoboken, NJ, USA, 2015; p. 736. ISBN 978-1-118-34328-9. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Sasaki, Y. The truth of the F-measure. Teach. Tutor Mater. 2007, 1, 1–5. [Google Scholar]
- Van Rijsbergen, C.J. Information Retrieval, 2nd ed.; Butterworth-Heinemann: Newton, MA, USA, 1979; p. 208. ISBN 978-0-408-70929-3. [Google Scholar]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocińska, A.; Sławik, Ł. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Forster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of Feature Reduction Algorithms for Classifying Tree Species with Hyperspectral Data on Three Central European Test Sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images. Remote Sens. 2018, 10, 1111. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A Transformation for Ordering Multispectral Data in Terms of Image Quality with Implications for Noise Removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef]
- Mather, P.M.; Koch, M. Computer Processing of Remotely-Sensed Images; John Wiley & Sons, Ltd: Chichester, UK, 2011; ISBN 9780470666517. [Google Scholar]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, 110–122. [Google Scholar] [CrossRef]
- Shen, G.; Sakai, K.; Hoshino, Y. High Spatial Resolution Hyperspectral Mapping for Forest Ecosystem at Tree Species Level. Agric. Inf. Res. 2010, 19, 71–78. [Google Scholar] [CrossRef]
- Kopeć, D.; Zakrzewska, A.; Halladin-Dąbrowska, A.; Wylazłowska, J.; Kania, A.; Niedzielko, J. Using Airborne Hyperspectral Imaging Spectroscopy to Accurately Monitor Invasive and Expansive Herb Plants: Limitations and Requirements of the Method. Sensors 2019, 19, 2871. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Förster, M. Grassland habitat mapping by intra-annual time series analysis -Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Wojtuń, B.; Rogass, C.; Mielke, C.; Lavender, S. Subalpine and alpine vegetation classification based on hyperspectral APEX and simulated EnMAP images. Int. J. Remote Sens. 2017, 38. [Google Scholar] [CrossRef]
- Burai, P.; Laposi, R.; Enyedi, P.; Schmotzer, A.; Bognar, V.K. Mapping invasive vegetation using AISA Eagle airborne hyperspectral imagery in the Mid-Ipoly-Valley. In Proceedings of the 2011 3rd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lisbon, Portugal, 6–9 June 2011; pp. 1–4. [Google Scholar]
- Chance, C.M.; Coops, N.C.; Plowright, A.A.; Tooke, T.R.; Christen, A.; Aven, N. Invasive Shrub Mapping in an Urban Environment from Hyperspectral and LiDAR-Derived Attributes. Front. Plant Sci. 2016, 7, 1–19. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O. Evaluating the potential of freely available multispectral remotely sensed imagery in mapping American bramble (Rubus cuneifolius). S. Afr. Geogr. J. 2018, 100, 291–307. [Google Scholar] [CrossRef]
- Zagajewski, B. Assesment of Neural Networks and Imaging Spectroscopy for Vegetation Classification of the High Tatras; Olędzki, J., Ed.; Klub Teledetekcji Środowiska Polskiego Towarzystwa Geograficznego: Warsaw, Poland, 2010; Volume 43, ISBN 0071-8076. [Google Scholar]


| Parameters | HySpex VNIR-1800 | HySpex SWIR-384 |
|---|---|---|
| Spectral range | 416–995 nm | 954–2510 nm |
| Number of spectral bands | 182 (163 1) | 288 |
| Spectral sampling | 3.26 nm | 5.45 nm |
| Spatial resolution | 0.5 m | 1 m |
| Spatial pixels 2 | 1800 | 384 |
| Field of View (FOV) across track | 17–34◦ | 16–32◦ |
| Instantaneous Field of View (IFOV) | 0.01–0.04◦ | 0.04–0.08◦ |
| Data Used | Algorithm (Parameters) | Number of Pixels | Mean for 100 Iterations | |||||
|---|---|---|---|---|---|---|---|---|
| Training Set per Class/All | Testing Set | Testing Set | Validation Set (4835 pixels) | |||||
| F1 for All Classes | F1 for all Classes | F1 for 3 Plant Species | F1 for Background Classes | Kappa | ||||
| 430 hyperspectral bands | RF (mtry =140; ntree = 500) | 30/240 | 4612 | 0.847 | 0.784 | 0.789 | 0.856 | 0.759 |
| 50/400 | 4452 | 0.876 | 0.803 | 0.811 | 0.871 | 0.785 | ||
| 100/800 | 4052 | 0.905 | 0.823 | 0.831 | 0.866 | 0.808 | ||
| 200/1600 | 3252 | 0.931 | 0.843 | 0.851 | 0.869 | 0.832 | ||
| 300/2400 | 2452 | 0.935 | 0.853 | 0.859 | 0.869 | 0.842 | ||
| SVM (kernel = radial; cost = 1000; gamma = 0.1) | 30/240 | 4612 | 0.843 | 0.760 | 0.803 | 0.807 | 0.737 | |
| 50/400 | 4452 | 0.888 | 0.787 | 0.833 | 0.853 | 0.771 | ||
| 100/800 | 4052 | 0.935 | 0.817 | 0.867 | 0.820 | 0.808 | ||
| 200/1600 | 3252 | 0.964 | 0.840 | 0.893 | 0.827 | 0.833 | ||
| 300/2400 | 2452 | 0.972 | 0.852 | 0.907 | 0.826 | 0.847 | ||
| 30 MNF bands | RF (mtry = 13; tree = 500) | 30/240 | 4612 | 0.952 | 0.869 | 0.868 | 0.965 | 0.853 |
| 50/400 | 4452 | 0.974 | 0.893 | 0.890 | 0.952 | 0.888 | ||
| 100/800 | 4052 | 0.988 | 0.910 | 0.908 | 0.942 | 0.906 | ||
| 200/1600 | 3252 | 0.994 | 0.917 | 0.920 | 0.940 | 0.917 | ||
| 300/2400 | 2452 | 0.995 | 0.918 | 0.926 | 0.932 | 0.92 | ||
| SVM (kernel = radial; cost = 1000; gamma = 0.1) | 30/240 | 4612 | 0.961 | 0.854 | 0.918 | 0.860 | 0.850 | |
| 50/400 | 4452 | 0.980 | 0.871 | 0.933 | 0.850 | 0.874 | ||
| 100/800 | 4052 | 0.993 | 0.881 | 0.943 | 0.847 | 0.885 | ||
| 200/1600 | 3252 | 0.998 | 0.883 | 0.949 | 0.846 | 0.889 | ||
| 300/2400 | 2452 | 0.999 | 0.883 | 0.951 | 0.838 | 0.891 | ||
| Class | C. epigejos | Rubus spp. | Solidago spp. | Shadows | Trees | Other Plants | Soils | Buildings | Total | UA (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| C. epigejos | 454 | 0 | 0 | 0 | 0 | 21 | 66 | 0 | 541 | 83.92 |
| Rubus spp. | 0 | 406 | 0 | 0 | 0 | 0 | 0 | 0 | 406 | 100.00 |
| Solidago spp. | 0 | 0 | 781 | 0 | 0 | 0 | 0 | 0 | 781 | 100.00 |
| Shadows | 0 | 0 | 0 | 415 | 0 | 0 | 0 | 5 | 420 | 98.81 |
| Trees | 0 | 6 | 0 | 0 | 344 | 2 | 0 | 0 | 352 | 97.73 |
| Other plants | 18 | 14 | 0 | 0 | 11 | 1375 | 44 | 0 | 1462 | 94.05 |
| Soils | 3 | 3 | 56 | 0 | 12 | 0 | 309 | 87 | 470 | 65.74 |
| Buildings | 9 | 2 | 3 | 3 | 60 | 0 | 0 | 326 | 403 | 80.89 |
| Total | 484 | 431 | 840 | 418 | 427 | 1398 | 419 | 418 | 4835 | |
| PA (%) | 93.80 | 94.20 | 92.98 | 99.28 | 80.56 | 98.35 | 73.75 | 77.99 |
| Class | C. epigejos | Rubus spp. | Solidago spp. | Shadows | Trees | Other Plants | Soils | Buildings | Total | UA (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| C. epigejos | 437 | 0 | 0 | 0 | 0 | 33 | 84 | 0 | 554 | 78.88 |
| Rubus spp. | 0 | 394 | 0 | 0 | 2 | 0 | 23 | 0 | 419 | 94.03 |
| Solidago spp. | 0 | 0 | 833 | 0 | 0 | 0 | 0 | 0 | 833 | 100.00 |
| Shadows | 0 | 0 | 0 | 418 | 0 | 0 | 0 | 10 | 428 | 97.66 |
| Trees | 0 | 2 | 1 | 0 | 414 | 5 | 0 | 0 | 422 | 98.10 |
| Other plants | 23 | 35 | 6 | 0 | 11 | 1360 | 31 | 27 | 1493 | 91.09 |
| Soils | 24 | 0 | 0 | 0 | 0 | 0 | 278 | 5 | 307 | 90.55 |
| Buildings | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 376 | 379 | 99.21 |
| Total | 484 | 431 | 840 | 418 | 427 | 1398 | 419 | 418 | 4835 | |
| PA (%) | 90.29 | 91.42 | 99.17 | 100.00 | 96.96 | 97.28 | 66.35 | 89.95 |
| Plant Species | Sensor | Raster Data | Algorithm | UA (%) | PA (%) | F1 (%) | Reference |
|---|---|---|---|---|---|---|---|
| Calamagrostis epigejos | HySpex | 430 HS | RF | 77 | 87 | 82 | Present paper |
| SVM | 89 | 92 | 90 | ||||
| HySpex | 30 MNF | RF | 79 | 90 | 83 | ||
| SVM | 84 | 94 | 91 | ||||
| Calamagrostis epigejos | HySpex | 30 MNF + 42 discrete LiDAR data | RF | 88 | 63 | 73 | [26] |
| Calamagrostis villosa | APEX | 30 MNF | SVM | 51 | 68 | [60] | |
| Solidagospp. | HySpex | 430 HS | RF | 99 | 99 | 99 | Present paper |
| SVM | 97 | 98 | 97 | ||||
| HySpex | 30 MNF | RF | 100 | 99 | 99 | ||
| SVM | 100 | 93 | 96 | ||||
| Solidago gigantea | HySpex | 30 MNF | RF | 73 | [58] | ||
| Solidagospp. | AISA Eagle | 15 MNF | Maximum Likelihood | 71 | 100 | [61] | |
| 129 HS | Spectral Angle Mapper | 61 | 69 | ||||
| Solidago altissima | AISA Eagle | 3 MNF | Generalized Linear Models | 94 | 80 | [19] | |
| Rubusspp. | HySpex | 430 HS | RF | 70 | 83 | 76 | Present paper |
| SVM | 79 | 90 | 84 | ||||
| 30 MNF | RF | 94 | 92 | 95 | |||
| SVM | 100 | 94 | 97 | ||||
| Rubus fruticosus sp. agg. | HyMap | 20 HS | MTMF | 81 | 92 | [32] | |
| MF | 61 | 53 | |||||
| SAM | 71 | 58 | |||||
| 128 HS | MTMF | 90 | 77 | ||||
| MF | 49 | 35 | |||||
| SAM | 75 | 45 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabat-Tomala, A.; Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data. Remote Sens. 2020, 12, 516. https://doi.org/10.3390/rs12030516
Sabat-Tomala A, Raczko E, Zagajewski B. Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data. Remote Sensing. 2020; 12(3):516. https://doi.org/10.3390/rs12030516
Chicago/Turabian StyleSabat-Tomala, Anita, Edwin Raczko, and Bogdan Zagajewski. 2020. "Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data" Remote Sensing 12, no. 3: 516. https://doi.org/10.3390/rs12030516
APA StyleSabat-Tomala, A., Raczko, E., & Zagajewski, B. (2020). Comparison of Support Vector Machine and Random Forest Algorithms for Invasive and Expansive Species Classification Using Airborne Hyperspectral Data. Remote Sensing, 12(3), 516. https://doi.org/10.3390/rs12030516








