Abstract
Precise stand species classification and volume estimation are key research topics for automated forest inventory. This study aims to explore the feasibility of light detection and ranging (lidar) height, intensity, and ratio parameters for discriminating dominant species (Pinus densiflora, Larix kaempferi, and Quercus spp.) and estimating volume at plot scale. To achieve these objectives, multiple linear discriminant and regression analyses were utilized after a separate selection of explanatory variables from extracted 38 lidar height, intensity, and ratio parameters. A kappa accuracy of 0.75 was achieved in discriminating the plot-dominant species from three different species by adopting a combination of nine selected explanatory variables. Further investigation found that dispersion and mean of lidar intensity within a plot are key classifiers of identifying three species. Species-specific optimal plot volume models for Pinus densiflora, Larix kaempferi, and Quercus spp. were evaluated by coefficients of determination of 0.71, 0.74, and 0.56, respectively. Compared to species classification, height-related lidar variables play a key role in modeling forest plot volume. Several explanatory variables for each modeling practice were correlated to canopy vertical and horizontal structures and were enough to represent species-specific characteristics in both approaches for species classification and plot volume estimation. Additionally, observed different variable combinations for two important applications imply that future studies should use proper variable combinations for each purpose.
1. Introduction
Precise and quantitative information of forests is essential for forest management and planning [1,2,3]. Forest stand volume is one of the most important structural variables characterizing economic and environmental value of the forest stand. As it can estimate stand biomass and carbon content, accurate stand volume estimation is critical for understanding forest carbon dynamics. Field surveying can provide accurate and extensive forest inventories nationwide; however, it is not only labor-intensive and time-consuming, but also difficult to seamlessly cover large forested area. Together with increasing needs of high quality and large-scale forest information, in this context, remote sensing has become a more powerful tool in forest management [4,5,6,7].
Laser surveying techniques known as light detection and ranging (lidar) have been widely used to characterize a large-scale three-dimensional forest structure and its dynamics [1,7,8,9]. Studies have proven the potential of lidar for estimating forest stand volume [10,11,12]. Approaches for estimating stand volume using lidar data can be categorized into (a) individual tree detection-based approaches [1,9,13] and (b) height distributional approaches at the stand or plot level [13,14]. The individual tree detection based approaches for estimating forest stand volume, tree height, and diameter at breast height (DBH) are required for calculating volume using an allometric function [1]. However, direct estimation of DBH is problematic due to stem concealment by obstructive upper canopy structures. To obtain more accurate DBH estimates, several studies have suggested ways to apply the statistical relationship between measured DBH and crown width derived from lidar data [13]. Although stand volume could be estimated by using height and DBH derived from lidar data, modeling procedures for DBH estimates and the necessity of accurately isolating individual trees tend to increase the uncertainty of stand volume estimates.
To avoid the complications and limitations of approaches based on individual tree detection, stand volume has been estimated directly using height distribution parameters of large- and small-footprint lidar systems at the stand or plot level [10,15,16]. This approach assumes that stand volume is closely related to actual vertical and horizontal forest structures and that lidar height distributions can represent the forest structures [15]. In other words, the forest structure from tree top to ground can be explained by structurally arranged large- and small-footprint lidar data [17]. In practice, large-footprint full-waveform lidar can provide height distribution data depending on the time-varying intensity of the returned energy within the laser pulse; however, this system is not appropriate for demonstrating finer scale forest attributes, including volume and biomass [18]. Small-footprint discrete lidar can produce a height distribution of forest vertical structures by accumulating all returns per sampling unit. This distribution then can be employed to estimate fine-scale and stand-level forest volume [15,19].
Tree species information is also critical for correctly valuing forests in terms of economic, ecological, and technical perspectives. Inaccurate species identification can result in prominent bias in the estimates of stem volume and biomass as allometric dependencies are species-specific [1,20]. The approach using height distribution data for volume estimation has been used to discriminate tree species at the individual tree and plot or stand level [21,22,23]. These species discrimination analyses have been successfully used to differentiate forest species between coniferous and broadleaf forests [24]. Both height and intensity data from lidar clearly show the potential to classify individual or stand dominant species by characterizing spectral, vertical, and horizontal profiles of canopy structure.
Reviewing the literature for related research reveals an opportunity to develop a sequential analytic framework that identify stand-dominant species and estimate stand volume. This study thus aims to achieve these two main objectives using height, intensity, and ratio parameters derived from airborne lidar data. Both objectives focused on the relationship between forest attributes and various lidar distributional characteristics at the plot scale, thus this study can provide insight of how different sets of lidar height, intensity, and ratio variables play a role in species classification and plot volume estimation. The detailed first objective is to identify explanatory parameters and their combination for classifying plot-dominant species between homogeneously distributed Korean red pine (Pinus densiflora), Japanese Larch (Larix kaempferi), and oaks (Quercus spp.) with determining statistical criteria. The second objective is to examine the appropriate parameters for estimating species-specific plot volume based on classification results under critical statistical criteria and select an optimal volume model for each plot-dominant species. Following results and possible limitations are compared and discussed.
2. Materials
2.1. Study Area
The study sites are located in Obin-ri (Coordinates: upper left 127°27′04.08′′E, 37°31′15.80″N and lower right 127°29′22.89″E, 37°30′15.45″N) and at the Korea University Yangpyeong Experimental Forest (upper left 127°40′45.77′′E, 37°30′56.22″N and lower right 127°43′19.11″E, 37°29′36.76″N), Yangpyeong-gun, central South Korea (Figure 1). The forests of the study region range from 21 m to 220 m above sea level. The main tree species in these sites are Pinus densiflora, Larix kaempferi, and Quercus spp. For this study, a total of 90 plots (30 plots for each species) were surveyed and investigated. Each 30 plots for each species were split into 20 training and 10 testing sites. The training sites were used for developing stand dominant species classification and volume estimation model, then the testing sites were used for verifying the developed models.
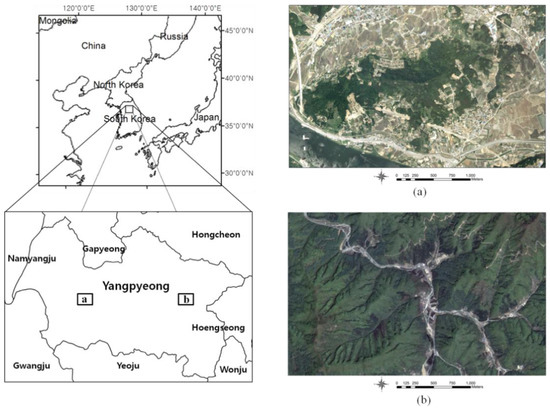
Figure 1.
Geographical location of study area with digital aerial photographs. (a) Obin-ri; (b) Korea University Yangpyeong Experimental Forest.
2.2. Lidar Data Acquisition
A small-footprint lidar system (ALTM 3070 developed by Optech, Inc.) was used to acquire the lidar data. The flight took place on 4 May 2009, and LiDAR data acquisition was performed at an altitude of 1400 m and with a point density of 5–8 points per square meter. Each point return provides the x, y, z position and intensity information derived from a near-infrared (1064 nm) laser pulse. The radiometric resolution, scan frequency, and scan width of the recorded lidar data were 12 bits, 70 Hz, and ±20°, respectively. This study only used lidar data within a ±10° scanning angle to reduce the scan angle effect on tree height and volume estimation [25]. Prior to extracting parameters from the lidar data, every lidar return was classified by the automatic procedure of the TerraScam Program [26]. The lidar returns were first classified into two groups, ground returns and above-ground returns. Ground returns were determined to be reflected from the ground within the plots, and above-ground returns included all returns other than ground returns. Then, every return of the lidar data recorded as height above sea level was converted to local height measure using a digital terrain model (DTM) generated from ground returns.
2.3. Ground Data Acquisition
A field survey was performed on 25–27 September 2010 across two study regions, i.e., the Obin-ri and the Korea University Yangpyeong Experimental Forest. Training and testing plots are 20 and 10 square plots (0.04 ha or 400 m2, 20 m sides) of each plot-dominant species, respectively [27]. For the survey, species, DBH, height, and age of every individual trees > 5 cm DBH were measured and tree density within each plot was recorded (Table S1). The dominant tree species, which accounts for more than 75% of the total tree species within the plot, was determined by counting every individual tree species [28]. The volume of each individual tree was calculated using species-specific allometric equations from the field-measured DBH and tree height; thereafter, plot volume was summed from every individual trees within each plot [28]. The species-specific allometric equations for volume estimation were developed by Korea Forest Research Institute [29]. The survey followed a standard procedure guided by the 5th National Forest Inventory [28].
The position of the center of each sample and test plot was acquired using the GPS Pathfinder Pro XR, which is manufactured by Trimble. This position information was used to spatially join and geo-match field surveying data with lidar data. The acquired GPS data were processed by a differential correction method using supplementary information received from a continuous GPS station near the study area. The corrected positions of the geo-referenced plots were obtained with position errors of less than 1 m. This process aimed to correct some errors that could be related to differences between the satellites and receivers, the atmosphere, satellite orbits, and reflective surfaces near the receiver when surveying with a single GPS receiver [30].
3. Methods
This study utilized multiple linear discriminant and regression analysis for plot-level species classification and volume estimation, respectively. Each process used the LiDAR height, intensity, ratio parameters, and field measurements of the plot-dominant species and volume as independent and dependent variables, respectively (Figure 2). Multiple linear discriminant analysis was adopted to classify plot-dominant species using the independent dataset, and the results were then verified by a cross-validation procedure with separate testing data. Thereafter, multiple linear regression analysis was performed on the LiDAR dataset, with the results of each plot-dominant species then evaluated using a corrected Akaike’s Information Criterion (AICc) [31] for selecting the optimal equation candidates. The candidate models showing the best performance were then verified and selected as an optimal volume model using testing datasets.
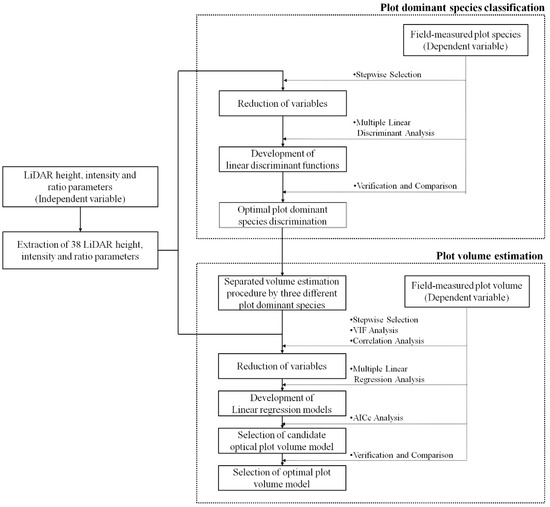
Figure 2.
Flowchart of plot dominant species classification and plot volume estimation.
3.1. Extraction of Lidar Height, Intensity, and Ratio Parameters
I extracted lidar height, intensity, and ratio parameters for constructing independent variables from two separate LiDAR return types, i.e., canopy and total returns. Canopy LiDAR returns are defined as returns that are higher than 1.0 m normalized height from DTM, while total returns include every return falling in the target plot [8,16]. According to Chen et al. [13], the height threshold for discriminating canopy returns from total returns might vary due to various forest stand conditions. This study set the threshold at 1.0 m as it effectively differentiates canopy crowns (i.e., lower than minimum crown base heights) and ground (Table S1). Then, 38 variables for the modeling process were extracted based on a dataset classified into canopy and total returns.
The lidar variables include percentile, mean, maximum, minimum, median, mode, standard deviation, coefficient of variation, kurtosis, skewness, and range of height and intensity data as those are closely related to volume and species information [32] (Table 1). These height and intensity variables were extracted from only canopy returns. The intensity variables can be significant descriptors of tree specifications, however, the intensity values might be affected by variations in laser path length and target reflectivity [33]. Thus, for intensity variables, I calibrated and normalized it by following an approach suggested by Kwak et al. [34].

Table 1.
Definition of lidar height, intensity, and ratio metrics.
To consider return transmission from the canopy to ground, this study added not only the number of returns from the canopy and total, but also the canopy return ratio (CRR, Equation (1)) and canopy intensity ratio (CIR, Equation (2)) (Table 1).
3.2. Plot-Dominant Species Classification
3.2.1. Selection of Explanatory Variables
Multiple linear discriminant analysis is a multivariate technique for separating distinct sets of observations and allocating new observations into predefined classes, i.e., plot-dominant species in this study [35]. This analysis is fundamentally based on minimizing the expected misclassification cost. Fisher’s linear discriminant analysis, a widely used discriminant analysis function [35], was used to classify three plot-dominant species (Pinus densiflora, Larix kaempferi, and Quercus spp.) in this study.
Thirty-eight independent variables were first extracted to discriminate plot-dominant species. However, the use of all of the candidate variables to separate plot-dominant species would be inefficient due to the need for intensive, time-consuming collection and management of all of the data. In addition, the use of too many variables is referred to as overfitting, a condition in which the results are dependent on sampling errors [35]. Therefore, a reduced discriminant analysis, with essential explanatory variables, need to be performed using a stepwise selection method based on the Wilks’ λ criteria approach (0.05 significance level used in this study). The Wilks’ λ, also known as the ratio of generalized variance, is a test statistic used in multivariate analysis of variance to test whether differences exist between the means of specified groups, i.e., plot-dominant-species groups in this study [35]. Further, reduced parameters from the stepwise technique were also evaluated for their discriminant ability by using both box-whisker plots and Tukey’s honestly significant difference (HSD) test. From the 38 lidar height, intensity, and ratio parameters, several parameters were selected as explanatory variables for plot-dominant species discrimination by stepwise selection. The discriminating power of each variable was then evaluated.
3.2.2. Development and Assessment of Linear Discriminant Analysis
Fisher’s linear discriminant analysis is a linear dimensionality reduction technique using canonical discriminant functions [36]. The procedure constructs a discriminant function by maximizing the ratio between the groups’ and within the groups’ variances [35]. Fisher’s method yields a linear function that divides the variable space into two dimensions by developing a canonical discriminant function. This canonical discriminant function is commonly written as Equation (3):
where D is the discriminant score, bi is the discriminant coefficient of the ith independent variable, and Xi is the ith independent variable. Canonical discriminant functions can be used to calculate the discriminant score of each plot for determining the centroid of scores related with species group separation. The centroid of each species group was calculated by averaging discriminant scores derived from these functions. These canonical functions might be evaluated by explanation degree for discriminant score variations referred from canonical correlation coefficients [35]. In addition, standardized canonical discriminant function coefficients are used to evaluate which variable has higher discriminating power in each developed canonical discriminant function [35].
A cross-validation approach (leave-one-out method) was used to assess the accuracy of the discriminant analysis [37]. The classification result was evaluated based on an originally grouped—and cross-validated—accuracy assessment process by hit ratio explaining the correctly corresponding discrimination performance. Additionally, Cohen’s kappa coefficients were calculated to measure the agreement between the classifications of the best performance combination case. The Kappa class value was used to rate the agreement as poor (0.40), fair (0.40–0.55), good (0.55–0.70), very good (0.70–0.85), and excellent (>0.85) [38]. Among manifold variable combinations, the variable combination showing the best performance was applied to determine plot-dominant species and this information was sequentially used to develop the species-specific volume model. In addition to such evaluation procedures, this study verified the variable combinations showing the best performance in the training plots by applying these to the 30 separate testing plots.
3.3. Plot Volume Estimation
3.3.1. Selection of Explanatory Variables
To estimate the plot volume dominated by different species, multiple linear regression modeling was separately performed. The 38 independent variables from the lidar height, intensity, and ratio metrics were first utilized for regression modeling of the plot volume. The use of the fully developed model using all candidate variables would be inefficient due to the same reason as in the discriminant analysis; thus, this study reduced the model using stepwise selection methods at the 0.05 significance level [13]. However, such selected variables under stepwise selection might have linear dependency relationships, i.e., a problem referred to as multicollinearity. The multicollinearity can disturb the estimation of a least square estimator in the regression procedure, so that the estimated value may be unreliable due to increased variation. O’Brien [39] suggested a variance inflation factor (VIF) that can evaluate the multicollinearity between selected independent variables. Moreover, Kutner et al. [40] mentioned that a VIF below 10 is suitable for selecting independent variables, but values above 10 indicate a multicollinearity problem. Therefore, I only selected independent variables that have VIF lower than 10. Then, highly correlated independent variables were further eliminated (Pearson’s correlation coefficients over 0.5) [18].
3.3.2. Development and Assessment of Linear Volume Models
In this study, multiple linear regression analysis was performed using combinations of selected variables for each species with 10 or fewer regressed functions selected, as shown in Equation (4):
where y is the plot volume measured in the field survey; x1, x2, x3, …, xn are the selected variables derived from lidar height, intensity, and ratio parameters; α, β1, β2, β3, …, βn are the estimated regression parameters; and ε is its residuals.
This study utilized Akaike’s information criterion (AIC) to select an optimal plot volume model for each dominant species. To remedy potential bias introduced by the size of sample, AIC was substituted into the corrected AIC (AICc) in this study (Equation (5)),
where k is the number of parameters in the model and n the number of observations. and stand for the plot volume and its estimate from the model. The smaller AICc is, the more closely the model approaches reality. When comparing different regression models, the estimated AICc values are generally normalized by subtracting the minimum AICc values, according to Equation (6):
where AICci is the AICc value of the ith model and AICcmin is the minimum AICci value. The results of this transformation allow the following criteria. Regression models with Δi ≥ 2 were rejected and models with Δi ≤ 2 were selected because only models with Δi ≤ 2 are accepted as providing substantial support. With these AIC criteria, other supplementary statistics including R2, adjusted R2, RMSE, and SSE of the models were also considered and compared when selecting the optimal plot volume model.
4. Results
4.1. Plot-Dominant Species
4.1.1. Explanatory Variables for Plot-Dominant Species Classification
Explanatory variables of the LiDAR height, intensity, and ratio parameters for plot-dominant species classification were chosen through the stepwise selection method based on Wilks’ λ criteria at a significance level of 0.05. The selected metrics were the 80th and 90th percentiles and the standard deviation of height (HEI,80, HEI,90, and HEI,std), the mean, mode, standard deviation, coefficient of variation, and skewness of intensity (INT,mean, INT,mode, INT,std, INT,cv, and INT,skew), and the canopy return ratio (CRR). In order to determine the statistical significance of the differences in the three plot-dominant species, the Wilks’ λ statistic, F-value, and Tukey’s HSD test were examined (Table 2).

Table 2.
Results of stepwise selection and Tukey’s HSD test for plot dominant species discrimination (both under 0.05 significant level).
The Wilks’ λ and F-value are generalized tests for determining the probability level of equality of population centroids, assuming equality of dispersion. The variables including lower λ statistics and higher F statistics at a high level of significance indicated high discrimination ability in plot-dominant-species classification. According to these statistics, HEI,80, HEI,90, and INT,std were the most effective discrimination parameters among the nine selected explanatory variables. The results of multiple species comparisons by Tukey’s HSD test showed which parameter was the distinctively meaningful factor for differentiating between two particular species. Tukey’s HSD tests revealed significant differences between Larix kaempferi and other species for three variables (HEI,80, HEI,90, and INT,mode). In the case of Pinus densiflora and Quercus spp., four (HEI,80, HEI,90, INT,mode, and INT,cv) and two (HEI,80 and HEI,90) variables showed differences with other species under critical statistical criteria (p < 0.001). Similar patterns can be found in the box–whisker plots (Figure S1).
4.1.2. Evaluation of Plot-Dominant-Species Discrimination
A set of explanatory variables was used to differentiate three plot-dominant species using the linear discriminant analysis. The 9 selected lidar height, intensity, and ratio parameters generated 511 combinations () of independent variables for discriminant analysis. Every possible combination classified plot-dominant species of the 60 sampled plots and was evaluated by original grouped and cross-validated accuracy assessments. The best performance in species discrimination was obtained from a combination of all explanatory variables, including HEI,80, HEI,90, HEI,std, INT,mean, INT,mode, INT,std, INT,cv, INT,skew, and CRR. Original grouped and cross-validated accuracy of this model were 95.0 and 93.3%, respectively. On the other hand, using only one variable, INT,mean, produced the lowest classification accuracies of 46.7% (original grouped accuracy) and 45.0% (cross-validated accuracy). Among the 511 possible combinations, Table 3 shows 10 variable combinations in order of cross-validated results. The cross-validated results of the 10 combinations range from 88.3 to 93.3% and most combinations have more than six variables showing that a general tendency between number of variables and model accuracy (Figure S2).

Table 3.
Accuracy assessment results of multiple linear discriminant analysis by various combinations (upper ranked 10 combinations).
The all of the combinations generated canonical discriminant functions to calculate discriminant scores and classify plot-dominant species. In particular, the highest accuracy was achieved by the combination with all variables (93.3%). This combination had first and second canonical discriminant functions that could explain 86.3 and 67.4% of the discriminant score variance, respectively, refers from canonical correlation coefficient. According to the result of the x2 test, both functions had a p-value that was lower than 0.05, which meant that this function significantly discriminated Pinus densiflora, Larix kaempferi, and Quercus spp. groups. The discriminant score centroid and distribution calculated by canonical discriminant functions is shown in Figure S3. The classification agreement was excellent as indicated by a kappa value greater than 0.90 [38] (Table S3).
4.2. Plot Volume
4.2.1. Explanatory Variables for Plot Volume
In the case of the plots dominated by Larix kaempferi, five variables were selected using the stepwise selection method at a significance level of 0.05: 90th percentile height (HEI,90), standard deviation of height (HEI,std), mode of the intensity (INT,mode), standard error of the mean of the intensity (INT,se), and the sum of the intensity (INT,TSum). These variables were shown to have low multicollinearity by a VIF of approximately 1 (Table 4). Then, the selected variables were examined by comparing Pearson’s correlation coefficients derived from correlation analysis between candidate variables. From the results, HEI,std was highly correlated with HEI,90 with a coefficient higher than 0.5, so this variable was eliminated to reduce multicollinearity (Table 5). Therefore, the explanatory variables to regress the plot volume model for Larix kaempferi were HEI,90, INT,mode, INT,se, and INT,TSum.

Table 4.
Results of variable selection to each species by variance inflation.

Table 5.
Results of variable selection to each species by correlation coefficient.
In the case of Pinus densiflora, the mean of the height (HEI,mean), the mode of the height (HEI,mode), the standard deviation of intensity (INT,std), and the range of intensity (INT,range) were selected. The multicollinearity between the selected variables was low compared to their VIFs (below 10), as shown in Table 4. An assessment of the one-to-one correlations between candidate variables was completed; however, all four variables were included in the regression analysis due their relatively low correlation coefficients (below 0.5) (Table 5).
To extract explanatory variables, acquired LiDAR parameters for Quercus spp. plots were reduced by the same procedure. Consequently, the candidate independent variables, shown in Table 4, are the 80th percentile of height (HEI,80), the 90th percentile of height (HEI,90), the mode of the intensity (INT,mode), and the kurtosis of the intensity distribution (INT,kurt). The multicollinearity between the selected variables was weak, as each VIF had a value below 10 (Table 4). Furthermore, as a result of the correlation analysis between the selected variables, HEI,80 and HEI,90 were highly correlated (r > 0.5) (Table 5). Therefore, HEI,90 was rejected as an explanatory independent variable for regressing the plot volume model because both the probability value from a t-test and the VIF of HEI,80 were lower than those for HEI,90. Eventually, HEI,80, INT,mode, and INT,kurt were adopted for the multiple linear regression analysis for predicting the plot volume.
4.2.2. Evaluation of Plot-Volume Models
The four independent variables selected for Larix kaempferi were used in developing regression models. The predictable equation was estimated by adopting the optimal regression model for estimating the plot volume represented by ΔAICc less than 2 (Table S4). The two optimal regression models found with ΔAICc ≤ 2 were model no. 1, which estimates the plot volume using HEI,90, INT,mode, and IT, and model no. 2, in which the explanatory variables were HEI,90, INT,mode, INT,se, and INT, when AICc was employed as the first criterion for selecting the best model. In addition, models 3 to 7 were rejected because their ΔAICc values were higher than 2, and the significance of their statistics was reduced with increasing ΔAICc. Among these two models, I chose the model no. 2 as it shows a better performance in RMSE, SEE, R2, and adjusted R2.
For Pinus densiflora plots, the combinable regression model was estimated with optimal regression models having ΔAICc less than 2 (Table S5). However, the best model and the only one with ΔAICc ≤ 2 was model no. 1, by which the plot volume could be estimated using HEI,mean, HEI,mode, INT,std, and INT,range. Models 2 to 7 were eliminated as candidate regression models because their ΔAICc was higher than 2 and because they lacked statistical significance with increasing ΔAICc.
In the case of Quercus spp. plots, three explanatory variables (HEI,80, HEI,mode, and INT,kurt) were selected and applied to the regression analysis. As a result of the regression procedure, one regression model, no. 1, with ΔAICc ≤ 2 was recommended. Models 2 to 7 were eliminated as candidate regression models for estimating plot volume of Quercus spp. stands because their ΔAICc was greater than 2 and because they lacked statistical significance with increasing ΔAICc (Table S6).
The developed plot volume models for each species were evaluated by implementing the models to independent 30 testing plots (10 plots for each dominant species) (Table 6; Figure S4). The model for Larix kaempferi shows high performance (R2 = 0.71, RMSE = 2.8 m3). In the case of Pinus densiflora, validation results showed that R2 and RMSE values of the model were 0.74 and 2.59 m3, respectively. When compared with plot volume predictions for Larix kaempferi, the best model of Pinus densiflora showed slightly better validation performance. Prediction by the model for Pinus densiflora was slightly overestimated, while estimation by the model for Larix kaempferi was underestimated. For Quercus spp., the developed model predicted plot volume with relatively a lower accuracy of R2 (0.56) and RMSE (3.01 m3) than those of other species (Figure S4d). The best plot-volume model of Quercus spp. produced overestimated plot volume predictions. The significance of the t-test for the developed and selected optimal plot volume models for each dominant species was statistically satisfactory.

Table 6.
Selected optimal regression models for each species.
5. Discussion
5.1. Plot-Dominant Species Classification
The stepwise technique selected nine explanatory variables: the 80th and 90th percentiles and standard deviation of height (HEI,80, HEI,90, and HEI,std), the mean, mode, standard deviation, coefficient of variation, skewness of intensity (INT,mean, INT,mode, INT,std, INT,cv, and INT,skew), and the canopy return ratio (CRR) (Table 2). For the three plot-dominant species, each species showed significant differences in height percentile parameters (such as HEI,80 and HEI,90) (Figure S5). Næsset and Bjerknes [41] and Holmgren and Persson [23] showed that the 90 percentile statistic could be used as a determining factor for species identification according to the close relationship between the height of the dominant trees in the plot and crown shape. This study determined that the HEI,80 and HEI,90 parameters could represent the dominant height of trees within a plot and showed high potential for being powerful discriminant variables. The height standard deviation of lidar returns which shows how much variation or dispersion exists from the average of returns was closely related with crown depth and corresponded to crown base height. The standard deviation of height was usually higher for Larix kaempferi than Quercus spp. and Pinus densiflora because Larix kaempferi usually has deeper tree crowns than those of the other species. This distinct aspect of standard deviation of height could be a potential indicator for differentiating plot-dominant species [23].
Descriptive statistics of lidar near infrared intensity returns provide useful discriminators for species identification through recording the different characteristics of the near-infrared radiation reflected from forest canopies (Figure S6). In this study, five intensity statistics were effectively used to classify plot-dominant species. The mean of intensity showed the highest value among the three dominant species for Quercus spp. because reflectivity characteristics of broadleaf and dense foliage produce higher intensity values than needle-like leaves and sparse foliage [42]. Coniferous trees generally show significantly lower reflection values than broadleaf trees [43], so our result supports his finding of species-specific intensity differences (Figure S6). In addition, due to densely covered leaves or other components, this study also found that the mean of intensity of Pinus densiflora was higher than that of Larix kaempferi. This is likely due to higher reflectivity of Pinus densiflora and denser canopy structure (Figure S1) [44].
The statistics related to dispersion of lidar intensity, such as standard deviation and coefficient of variation, were considered to be explanatory variables for identifying individual trees or dominant species in previous studies [20,24,42]. The coefficient of variation of intensity, a normalized measure of dispersion, was closely related to the standard deviation of intensity. Generally, intensity was affected not only by canopy closure, but also by specific reflectivity characteristics that depend on species. Consequently, dense forest canopies were associated with low lidar penetration rates and therefore such forests had low coefficients of variation and standard deviation [33]. When this study examined intensity dispersion and the influences of canopy closure, Pinus densiflora showed the lowest standard deviation and coefficient of variation. These lowest values originated from its permeability, which is determined by the densely covered canopy. The intensity dispersion of Quercus spp. showed higher standard deviation and higher variability of normalized standard deviation. Considering the variability of field measured tree density, the intensity dispersions of Quercus spp. might be crucially influenced by the intermingled effects of tree density and degree of canopy closure.
The mode of intensity might be expected to describe the concentrativeness of the returned intensity distribution. The mode could be strongly influenced by the shape and structure of the canopy and the degree of canopy closure. Species-specific reflective characteristics were shown in three plot-dominant species; however, the dense canopy closure of Pinus densiflora and variability of tree density of Quercus spp. affected those distributions. Highly dense needle-like leaves increased the returned intensity value, and the severe variation of tree density in Quercus spp. dispersed its mode of the intensity distribution. The skewness of intensity is related to the asymmetry of the recorded intensity distribution. Positive skewness of intensity would be associated with a minimally skewed distribution and negative skewness would be relevant to a highly skewed distribution. INT,skew shown in box-whisker plots indicated that larger laser pulses were reflected from branches or bark in plots of Larix kaempferi, while highly recorded intensities were from leaves in plots of Pinus densiflora considering the structural and canopy closure characteristics of each species. Because Roberts et al. [45] found that intensities from branch and bark had lower spectral reflectance than that of leaves, this skewness of intensity suggested that the asymmetry of intensity might be an explainable indicator of reflective objects.
The canopy return ratio was generally used to measure the degree of laser penetration (i.e., gap probability) against canopy components such as leaves, branches, and stem. The canopy return ratio showed a different pattern compared to other lidar height and intensity parameters revealing its potential for describing canopy structures (Figures S5 and S6). This is because the canopy return ratio was derived from calculating the ratio between the number of total returns and the number canopy returns while the other parameters were calculated from only canopy returns. Higher ratio value means that laser pulses are dominantly interrupted by dense canopy components. To the contrary, lower canopy return ratios can be considered as open canopy forests. Comparing these ratios between species, Quecus spp. showed the densest canopy cover with the highest canopy return ratio. This ratio based variable can be an important explanatory indicator for differentiating stand species.
5.2. Plot-Volume Estimation
For the plot volume model, this study identified different sets of explanatory variables for different plot-dominant species. This is likely because of different canopy structural conditions such as canopy cover ratio, crown depth, tree density, and others of the plot-dominant species. Across all cases, HEI,90, HEI,80, HEI,mean, HEI,mode, and HEI,kurt variables were chosen for plot volume estimation. These key structural parameters were corroborated by findings of previous studies that were able to estimate plot or stand volume using only canopy height distributional parameters [13,14]. The characteristics of these parameters were closely related to volumetric canopy structures [14,19]. The higher percentiles of height (80th and 90th) and mean of height were highly correlated with actual canopy height which has been used to calculate volume based on allometric relationships. In addition, the mode of height and kurtosis of the height distribution might help explain crown geometric volume [9,13]. Because crown geometric volume correlated with stem volume and was derived from 3-dimensional crown structures, these crown structures might be represented by variables based on their meanings. Therefore, these two variables also have a meaningful explanatory ability for plot volume estimation through expanding the linear relationship [9,13].
Intensity data were also included in plot volume estimation. The intensity parameters used here were INT,mode, INT,se, INT,TSum, INT,std, INT,range, INT,mode, and INT,kurt. According to van Aardt et al. [18], various statistics of lidar intensity data, such as median, standard deviation, minimum, and others, were adopted to predict stand volume under specified species information. As described in Section 5.1, lidar intensity at the near-infrared region contains canopy structure and reflectivity information which may improve the volume estimation model [46].
In multiple linear regression analysis, this study could distinguish the explanatory ability of each variable for plot volume estimation. For the case of Larix kaempferi, HEI,90 and INT,TSum acted as key explanatory variables at the highest significance. The plot volume dominated by Pinus densiflora took HEI,mean and INT,range as highly significant variables among the selected four variables. In the case of the Quercus spp., INT,mode largely explained plot volume of this species along with the HEI,80 and INT,kurt parameters. Different key explanatory variables could be attributed by the characteristics of canopy structures of each species. In general, Larix kaempferi showed a fat corn shape crown with deeper crown depth, whereas the other two species had a horizontally flattened and rounded shape in the study area. Particularly, the canopy depth of Quercus spp. was relatively deeper than that of Pinus densiflora.
Canopy structural characteristics of each species may determine performance of lidar based volume estimation model. Several studies have reported that plot- or stand-volume estimation using lidar data is highly feasible but most of them examined over coniferous forests. For instance, Næsset noted a range of the coefficient of determination of 0.80–0.93 for stand volume estimation in coniferous forest and age and site-quality classes. Means et al. [16] conducted volume modeling for coniferous forest with R2 values of 0.95 (including mature plots) and 0.97 (not including mature plots). By including coniferous, mixed and broadleaf forests, van Aardt et al. [18] attempted to develop a species-specific stand volume model and showed a limited performance ranging from R2 of 0.40–0.70. The performance of this study is relatively better, at 0.71, 0.74, and 0.56 for Pinus densiflora, L. kaempferi, and Quercus spp., respectively. Most studies including this study confirms that lidar based volume estimation performs better in coniferous forest than broadleaf or mixed forests. In this study, the plots dominated by Quercus spp. had especially large variability from the forest condition, so its plot volume model suffered. These accuracy shortcomings can be also attributed to unexplainable variability in the forest condition, such as stand density, species mixed ratio, etc. Therefore, further study is required to consider various forest conditions (age, density, ratio of mixture, site quality, etc.) and to precisely survey unbiased samples.
6. Conclusions
This study shows that lidar height, intensity, and ratio parameters are applicable for discriminating plot-dominant species (Pinus densiflora, Larix kaempferi, and Quercus spp.) and for estimating plot volume sequentially. A kappa accuracy of 0.75 was achieved in plot-dominant species classification, and species-specific optimal plot volume models were developed and evaluated by coefficients of determination of 0.71, 0.74, and 0.56, respectively. Further investigation found that dispersion and mean of lidar intensity within a plot are key classifiers of identifying three species while height related lidar variables play a key role in modeling forest plot volume. Selected explanatory variables are closely correlated to vertical and horizontal canopy structures and are enough to represent species-specific characteristics in both discriminative analysis and volume estimation. Additionally, observed different variable combinations for two important applications imply that future studies should use proper variable combinations for each purpose. This study only investigated over homogeneous forest stands without considering characteristics of mixed forest stands, such as species mixture, age class mixture, etc. Considering the characteristics of mixed forest stands can help provide an unbiased implementation for discriminating species and estimating volume.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-4292/12/19/3266/s1, Figure S1. Box-whisker plots for visualizing the distributional characteristics of selected parameters by three plot dominant species. Figure S2. Original grouped- and Cross validated-accuracy of discriminant analysis by number of selected variables in combinations. Figure S3. Distribution of discriminant score and its centroid by first and second canonical discriminant function. Figure S4. Evaluation of the developed plot volume models using independent testing data. Figure S5. Three dimensional view of the lidar height distribution of each species. X and Y axes are a spatial coordinate of the plot (meter scale). Figure S6. Three dimensional view of the lidar intensity distribution of each species. X and Y axes are a spatial coordinate of the plot (meter scale). Table S1. Descriptive statistics of the field measurements. Table S2. Sorted the highest accuracy results of linear discriminant analysis by number of variables. Table S3. Error matrix of plot dominant species classification results by discriminant analysis (case on the highest performance of 93.3%). Table S4. Result of plot volume parameters estimated multiple regression analysis to Larix kaempferi. Table S5. Result of plot volume parameters estimated multiple regression analysis to Pinus densiflora. Table S6. Result of plot volume parameters estimated multiple regression analysis to Quercus spp.
Funding
This study is partially supported by NASA’s Carbon Monitoring System program (80NSSC18K0173-CMS).
Conflicts of Interest
The author declares no conflict of interest.
References
- Jung, S.-E.; Kwak, D.-A.; Park, T.; Lee, W.-K.; Yoo, S. Estimating crown variables of individual trees using airborne and terrestrial laser scanners. Remote Sens. 2011, 3, 2346–2363. [Google Scholar] [CrossRef]
- Matasci, G.; Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W.; Zald, H.S. Large-area mapping of Canadian boreal forest cover, height, biomass and other structural attributes using Landsat composites and lidar plots. Remote Sens. Environ. 2018, 209, 90–106. [Google Scholar] [CrossRef]
- Næsset, E.; McRoberts, R.E.; Pekkarinen, A.; Saatchi, S.; Santoro, M.; Trier, Ø.D.; Zahabu, E.; Gobakken, T. Use of local and global maps of forest canopy height and aboveground biomass to enhance local estimates of biomass in miombo woodlands in Tanzania. Int. J. Appl. Earth Obs. Geoinf. 2020. [Google Scholar] [CrossRef]
- Choi, S.; Kempes, C.P.; Park, T.; Ganguly, S.; Wang, W.; Xu, L.; Basu, S.; Dungan, J.L.; Simard, M.; Saatchi, S.S. Application of the metabolic scaling theory and water–energy balance equation to model large-scale patterns of maximum forest canopy height. Glob. Ecol. Biogeogr. 2016, 25, 1428–1442. [Google Scholar] [CrossRef]
- Ni, X.; Park, T.; Choi, S.; Shi, Y.; Cao, C.; Wang, X.; Lefsky, M.A.; Simard, M.; Myneni, R.B. Allometric scaling and resource limitations model of tree heights: Part 3. Model optimization and testing over continental China. Remote Sens. 2014, 6, 3533–3553. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef]
- Park, T.; Lee, W.-K.; Lee, J.-Y.; Byun, W.-H.; Kwak, D.-A.; Cui, G.; Kim, M.-I.; Jung, R.; Pujiono, E.; Oh, S. Forest plot volume estimation using national forest inventory, forest type map and airborne LiDAR data. For. Sci. Technol. 2012, 8, 89–98. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.; Acker, S.; Parker, G.G.; Spies, T.; Harding, D. Lidar remote sensing of the canopy structure and biophysical properties of Douglas-fir western hemlock forests. Remote Sens. Environ. 1999, 70, 339–361. [Google Scholar] [CrossRef]
- Kwak, D.-A.; Lee, W.-K.; Cho, H.-K.; Lee, S.-H.; Son, Y.; Kafatos, M.; Kim, S.-R. Estimating stem volume and biomass of Pinus koraiensis using LiDAR data. J. Plant Res. 2010, 123, 421–432. [Google Scholar] [CrossRef]
- Harding, D.; Lefsky, M.; Parker, G.; Blair, J. Laser altimeter canopy height profiles: Methods and validation for closed-canopy, broadleaf forests. Remote Sens. Environ. 2001, 76, 283–297. [Google Scholar] [CrossRef]
- Parker, G.A.; Smith, J.M. Optimality theory in evolutionary biology. Nature 1990, 348, 27–33. [Google Scholar] [CrossRef]
- Park, T.; Kennedy, R.E.; Choi, S.; Wu, J.; Lefsky, M.A.; Bi, J.; Mantooth, J.A.; Myneni, R.B.; Knyazikhin, Y. Application of physically-based slope correction for maximum forest canopy height estimation using waveform lidar across different footprint sizes and locations: Tests on LVIS and GLAS. Remote Sens. 2014, 6, 6566–6586. [Google Scholar] [CrossRef]
- Chen, Q.; Gong, P.; Baldocchi, D.; Tian, Y.Q. Estimating basal area and stem volume for individual trees from lidar data. Photogramm. Eng. Remote Sens. 2007, 73, 1355–1365. [Google Scholar] [CrossRef]
- Maltamo, M.; Suvanto, A.; Packalén, P. Comparison of basal area and stem frequency diameter distribution modelling using airborne laser scanner data and calibration estimation. For. Ecol. Manag. 2007, 247, 26–34. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Clark, D.B.; Knox, R.G.; Blair, J.B.; Hofton, M.A.; Chazdon, R.L.; Weishampel, J.F.; Prince, S. Estimation of tropical forest structural characteristics using large-footprint lidar. Remote Sens. Environ. 2002, 79, 305–319. [Google Scholar] [CrossRef]
- Means, J.E.; Acker, S.A.; Fitt, B.J.; Renslow, M.; Emerson, L.; Hendrix, C.J. Predicting forest stand characteristics with airborne scanning lidar. Photogramm. Eng. Remote Sens. 2000, 66, 1367–1372. [Google Scholar]
- Dubayah, R.; Prince, S.; JaJa, J.; Blair, J.; Bufton, J.L.; Knox, R.; Luthcke, S.B.; Clarke, D.B.; Weishampel, J. The vegetation canopy lidar mission. In Land Satellite Information in the Next Decade II: Sources and Applications; ASPRS: Washington, DC, USA, 1997. [Google Scholar]
- Aardt, J.A.v.; Wynne, R.H.; Oderwald, R.G. Forest volume and biomass estimation using small-footprint lidar-distributional parameters on a per-segment basis. For. Sci. 2006, 52, 636–649. [Google Scholar]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Korpela, I.S.; Tokola, T.E. Potential of aerial image-based monoscopic and multiview single-tree forest inventory: A simulation approach. For. Sci. 2006, 52, 136–147. [Google Scholar]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne lidar. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Brandtberg, T.; Warner, T.A.; Landenberger, R.E.; McGraw, J.B. Detection and analysis of individual leaf-off tree crowns in small footprint, high sampling density lidar data from the eastern deciduous forest in North America. Remote Sens. Environ. 2003, 85, 290–303. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree species classification using airborne LiDAR–effects of stand and tree parameters, downsizing of training set, intensity normalization, and sensor type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Naesset, E. Estimating timber volume of forest stands using airborne laser scanner data. Remote Sens. Environ. 1997, 61, 246–253. [Google Scholar] [CrossRef]
- Soininen, A. TerraScan User’s Guide; Terrasolid: Helsinki, Finland, 2004. [Google Scholar]
- Gholamy, A.; Kreinovich, V.; Kosheleva, O. Why 70/30 or 80/20 Relation Between Training and Testing Sets: A Pedagogical Explanation; ScholarWorks@UTEP: Berkeley, CA, USA, 2018. [Google Scholar]
- Korean Forest Research Institute. Guideline for 5th National Forest Inventory; KFRI: Seoul, Korea, 2008. [Google Scholar]
- Korean Forest Research Institute. Volume, Biomass, and Yield Table; KFRI: Seoul, Korea, 2014. [Google Scholar]
- Lee, J.-K.; Hwang, C.-S.; Jung, S.-H. Analysis of accuracy for the control points using the GPS continuous stations. J. Korean Soc. Civ. Eng. 2003, 23, 401–409. [Google Scholar]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Maltamo, M.; Mustonen, K.; Hyyppä, J.; Pitkänen, J.; Yu, X. The accuracy of estimating individual tree variables with airborne laser scanning in a boreal nature reserve. Can. J. For. Res. 2004, 34, 1791–1801. [Google Scholar] [CrossRef]
- Donoghue, D.N.; Watt, P.J.; Cox, N.J.; Wilson, J. Remote sensing of species mixtures in conifer plantations using LiDAR height and intensity data. Remote Sens. Environ. 2007, 110, 509–522. [Google Scholar] [CrossRef]
- Kwak, D.-A.; Cui, G.; Lee, W.-K.; Cho, H.-K.; Jeon, S.W.; Lee, S.-H. Estimating plot volume using LiDAR height and intensity distributional parameters. Int. J. Remote Sens. 2014, 35, 4601–4629. [Google Scholar] [CrossRef]
- Johnson, R.A.; Wichern, D.W. Applied Multivariate Statistical Analysis; Prentice Hall: Upper Saddle River, NJ, USA, 2002; Volume 5. [Google Scholar]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Lachenbruch, P.A.; Mickey, M.R. Estimation of error rates in discriminant analysis. Technometrics 1968, 10, 1–11. [Google Scholar] [CrossRef]
- Monserud, R.A.; Leemans, R. Comparing global vegetation maps with the Kappa statistic. Ecol. Model. 1992, 62, 275–293. [Google Scholar] [CrossRef]
- O’brien, R.M. A caution regarding rules of thumb for variance inflation factors. Qual. Quant. 2007, 41, 673–690. [Google Scholar] [CrossRef]
- Kutner, M.H.; Nachtsheim, C.J.; Neter, J.; Li, W. Applied Linear Statistical Models; McGraw-Hill Irwin: New York, NY, USA, 2005; Volume 5. [Google Scholar]
- Næsset, E.; Bjerknes, K.-O. Estimating tree heights and number of stems in young forest stands using airborne laser scanner data. Remote Sens. Environ. 2001, 78, 328–340. [Google Scholar] [CrossRef]
- Moffiet, T.; Mengersen, K.; Witte, C.; King, R.; Denham, R. Airborne laser scanning: Exploratory data analysis indicates potential variables for classification of individual trees or forest stands according to species. ISPRS J. Photogramm. Remote Sens. 2005, 59, 289–309. [Google Scholar] [CrossRef]
- Schreier, H.; Lougheed, J.; Tucker, C.; Leckie, D. Automated measurements of terrain reflection and height variations using an airborne infrared laser system. Int. J. Remote Sens. 1985, 6, 101–113. [Google Scholar] [CrossRef]
- Rautiainen, M.; Lukeš, P.; Homolova, L.; Hovi, A.; Pisek, J.; Mõttus, M. Spectral properties of coniferous forests: A review of in situ and laboratory measurements. Remote Sens. 2018, 10, 207. [Google Scholar] [CrossRef]
- Roberts, D.A.; Ustin, S.L.; Ogunjemiyo, S.; Greenberg, J.; Dobrowski, S.Z.; Chen, J.; Hinckley, T.M. Spectral and structural measures of northwest forest vegetation at leaf to landscape scales. Ecosystems 2004, 7, 545–562. [Google Scholar] [CrossRef]
- Knyazikhin, Y.; Schull, M.A.; Stenberg, P.; Mõttus, M.; Rautiainen, M.; Yang, Y.; Marshak, A.; Carmona, P.L.; Kaufmann, R.K.; Lewis, P. Hyperspectral remote sensing of foliar nitrogen content. Proc. Natl. Acad. Sci. USA 2013, 110, E185–E192. [Google Scholar] [CrossRef]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).