Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance
Abstract
1. Introduction
2. Materials and Methods
2.1. Measurements and Datasets
2.2. Pre-Processing of the Raw Reflectance Data
2.3. Regression in Machine Learning
2.4. Statistical Criteria
3. Results
3.1. Photosynthetic Pigments Contents of Each Treatment
3.2. Spectral Patterns
3.3. Accuracy Assessment
3.4. Sensitivity Analysis
4. Discussion
4.1. Characteristics of the Samples Used in This Study
4.2. Performance of Different Pre-Processing and Machine Learning Algorithms
4.3. Direct Estimation vs. Indirect Estimation for Pigment Ratio
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gitelson, A.A.; Vina, A.; Verma, S.B.; Rundquist, D.C.; Arkebauer, T.J.; Keydan, G.; Leavitt, B.; Ciganda, V.; Burba, G.G.; Suyker, A.E. Relationship between gross primary production and chlorophyll content in crops: Implications for the synoptic monitoring of vegetation productivity. J. Geophys. Res. Atmos. 2006, 111. [Google Scholar] [CrossRef]
- Leong, T.Y.; Anderson, J.M. Adaptation of the thylakoid membranes of pea chloroplasts to light intensities. I. Study on the distribution of chlorophyll-protein complexes. Photosynth. Res. 1984, 5, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Jin, N.; Shi, Y.; Su, Y.Q.; Fei, B.J.; Li, W.; Qiao, D.R.; Cao, Y. Coordinate expression of light-harvesting chlorophyll a/b gene family of photosystem II and chlorophyll a oxygenase gene regulated by salt-induced phosphorylation in Dunaliella salina. Photosynthetica 2010, 48, 355–360. [Google Scholar] [CrossRef]
- Hobe, S.; Fey, H.; Rogl, H.; Paulsen, H. Determination of relative chlorophyll binding affinities in the major light-harvesting chlorophyll a/b complex. J. Biol. Chem. 2003, 278, 5912–5919. [Google Scholar] [CrossRef]
- Datt, B. Visible/near infrared reflectance and chlorophyll content in Eucalyptus leaves. Int. J. Remote Sens. 1999, 20, 2741–2759. [Google Scholar] [CrossRef]
- Demmig Adams, B.; Gilmore, A.M.; Adams, W.W. Carotenoids. In vivo functions of carotenoids in higher plants. Faseb. J. 1996, 10, 403–412. [Google Scholar] [CrossRef]
- Edge, R.; McGarvey, D.J.; Truscott, T.G. The carotenoids as anti-oxidants—A review. J. Photochem. Photobiol. B Biol. 1997, 41, 189–200. [Google Scholar] [CrossRef]
- Hendry, G.A.F.; Price, A.H. Stress Indicators: Chlorophylls and Carotenoids; Chapman Hall: London, UK, 1993. [Google Scholar]
- Féret, J.-B.; Francois, C.; Asner, G.P.; Gitelson, A.A.; Martin, R.E.; Bidel, L.P.R.; Ustin, S.L.; le Maire, G.; Jacquemoud, S. PROSPECT-4 and 5: Advances in the leaf optical properties model separating photosynthetic pigments. Remote Sens. Environ. 2008, 112, 3030–3043. [Google Scholar] [CrossRef]
- Sonobe, R.; Hirono, Y.; Oi, A. Non-Destructive Detection of Tea Leaf Chlorophyll Content Using Hyperspectral Reflectance and Machine Learning Algorithms. Plants 2020, 9, 368. [Google Scholar] [CrossRef]
- Sonobe, R.; Hirono, Y.; Oi, A. Quantifying chlorophyll-a and b content in tea leaves using hyperspectral reflectance and deep learning. Remote Sens. Lett. 2020, 11, 933–942. [Google Scholar] [CrossRef]
- Sonobe, R.; Yamashita, H.; Mihara, H.; Morita, A.; Ikka, T. Hyperspectral reflectance sensing for quantifying leaf chlorophyll content in wasabi leaves using spectral pre-processing techniques and machine learning algorithms. Int. J. Remote Sens. 2020. [Google Scholar] [CrossRef]
- Sonobe, R.; Miura, Y.; Sano, T.; Horie, H. Monitoring Photosynthetic Pigments of Shade-Grown Tea from Hyperspectral Reflectance. Can. J. Remote Sens. 2018, 44, 104–112. [Google Scholar] [CrossRef]
- Hege, N.; Kobayashi, M.; Michiki, N.; Takano, T.; Baba, F.; Kobayashi, K.; Ohyanagi, H.; Ohgane, J.; Yano, K.; Yamane, K. Complete chloroplast genome sequence and phylogenetic analysis of wasabi (Eutrema japonicum) and its relatives. Sci. Rep. 2019, 9, 14377. [Google Scholar]
- Li, S.J.; Gao, L.H.; Zhou, S.P.; Liu, G.Q.; Liu, W. Diversification of vegetable growing in the middle and lower reaches of the Yangtze River. Third Int. Symp. Diversif. Veg. Crop. 1998, 467, 253–258. [Google Scholar]
- Prado-Cabrero, A.; Beatty, S.; Howard, A.; Stack, J.; Bettin, P.; Nolan, J.M. Assessment of lutein, zeaxanthin and meso-zeaxanthin concentrations in dietary supplements by chiral high-performance liquid chromatography. Eur. Food Res. Technol. 2016, 242, 599–608. [Google Scholar] [CrossRef]
- Bilger, W.; Bjorkman, O.; Thayer, S.S. Light-induced spectral absorbance changes in relation to photosynthesis and the epoxidation state of xanthophyll cycle components in cotton leaves. Plant Physiol. 1989, 91, 542–551. [Google Scholar] [CrossRef]
- Yang, X.; Tang, J.W.; Mustard, J.F.; Wu, J.; Zhao, K.G.; Serbin, S.; Lee, J.E. Seasonal variability of multiple leaf traits captured by leaf spectroscopy at two temperate deciduous forests. Remote Sens. Environ. 2016, 179, 1–12. [Google Scholar] [CrossRef]
- Jacquemoud, S.É.P.; Ustin, S. Leaf Optical Properties; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar]
- Yamamoto, A.; Nakamura, T.; Adu-Gyamfi, J.J.; Saigusa, M. Relationship between chlorophyll content in leaves of sorghum and pigeonpea determined by extraction method and by chlorophyll meter (SPAD-502). J. Plant Nutr. 2002, 25, 2295–2301. [Google Scholar] [CrossRef]
- Sonobe, R.; Wang, Q. Towards a Universal Hyperspectral Index to Assess Chlorophyll Content in Deciduous Forests. Remote Sens. 2017, 9, 191. [Google Scholar] [CrossRef]
- Lu, B.; He, Y.H. Evaluating Empirical Regression, Machine Learning, and Radiative Transfer Modelling for Estimating Vegetation Chlorophyll Content Using Bi-Seasonal Hyperspectral Images. Remote Sens. 2019, 11, 1979. [Google Scholar] [CrossRef]
- Shah, S.H.; Angel, Y.; Houborg, R.; Ali, S.; McCabe, M.F. A Random Forest Machine Learning Approach for the Retrieval of Leaf Chlorophyll Content in Wheat. Remote Sens. 2019, 11, 920. [Google Scholar] [CrossRef]
- Vanbrabant, Y.; Tits, L.; Delalieux, S.; Pauly, K.; Verjans, W.; Somers, B. Multitemporal Chlorophyll Mapping in Pome Fruit Orchards from Remotely Piloted Aircraft Systems. Remote Sens. 2019, 11, 1468. [Google Scholar] [CrossRef]
- Sonobe, R.; Wang, Q. Assessing the xanthophyll cycle in natural beech leaves with hyperspectral reflectance. Funct. Plant Biol. 2016, 43, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Gamon, J.A.; Penuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gamon, J.A.; Field, C.B.; Bilger, W.; Bjorkman, O.; Fredeen, A.L.; Penuelas, J. Remote sensing of the xanthophyll cycle and chlorophyll fluorescence in sunflower leaves and canopies. Oecologia 1990, 85, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Skoneczny, H.; Kubiak, K.; Spiralski, M.; Kotlarz, J. Fire Blight Disease Detection for Apple Trees: Hyperspectral Analysis of Healthy, Infected and Dry Leaves. Remote Sens. 2020, 12, 2101. [Google Scholar] [CrossRef]
- Li, Z.H.; Jin, X.L.; Wang, J.H.; Yang, G.J.; Nie, C.W.; Xu, X.G.; Feng, H.K. Estimating winter wheat (Triticum aestivum) LAI and leaf chlorophyll content from canopy reflectance data by integrating agronomic prior knowledge with the PROSAIL model. Int. J. Remote Sens. 2015, 36, 2634–2653. [Google Scholar] [CrossRef]
- Masemola, C.; Cho, M.A.; Ramoelo, A. Comparison of Landsat 8 OLI and Landsat 7 ETM + for estimating grassland LAI using model inversion and spectral indices: Case study of Mpumalanga, South Africa. Int. J. Remote Sens. 2016, 37, 4401–4419. [Google Scholar] [CrossRef]
- Izzuddin, M.A.; Idris, A.S.; Nisfariza, M.N.; Nordiana, A.A.; Shafri, H.Z.M.; Ezzati, B. The development of spectral indices for early detection of Ganoderma disease in oil palm seedlings. Int. J. Remote Sens. 2017, 38, 6505–6527. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A Model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Féret, J.-B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Hosgood, B.; Jacquemoud, S.; Andreoli, G.; Verdebout, J.; Pedrini, G.; Schmuck, G. Leaf Optical Properties EXperiment 93; Office for Official Publications of the European Communities: Luxembourg, 1994; p. 20. [Google Scholar]
- Sonobe, R.; Wang, Q. Hyperspectral indices for quantifying leaf chlorophyll concentrations performed differently with different leaf types in deciduous forests. Ecol. Inform. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Doktor, D.; Lausch, A.; Spengler, D.; Thurner, M. Extraction of Plant Physiological Status from Hyperspectral Signatures Using Machine Learning Methods. Remote Sens. 2014, 6, 12247–12274. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Nasi, R.; Niemelainen, O.; Nyholm, L.; Alhonoja, K.; Kaivosoja, J.; Jauhiainen, L.; Viljanen, N.; Nezami, S.; Markelin, L.; et al. Machine learning estimators for the quantity and quality of grass swards used for silage production using drone-based imaging spectrometry and photogrammetry. Remote Sens. Environ. 2020, 246, 20. [Google Scholar] [CrossRef]
- Burges, C.J.C. A tutorial on Support Vector Machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Houborg, R.; McCabe, M.F. A hybrid training approach for leaf area index estimation via Cubist and random forests machine-learning. Isprs. J. Photogramm. Remote Sens. 2018, 135, 173–188. [Google Scholar] [CrossRef]
- Wijesingha, J.; Astor, T.; Schulze-Bruninghoff, D.; Wengert, M.; Wachendorf, M. Predicting Forage Quality of Grasslands Using UAV-Borne Imaging Spectroscopy. Remote Sens. 2020, 12, 126. [Google Scholar] [CrossRef]
- Breunig, F.M.; Galvao, L.S.; Dalagnol, R.; Dauve, C.E.; Parraga, A.; Santi, A.L.; Della Flora, D.P.; Chen, S.S. Delineation of management zones in agricultural fields using cover crop biomass estimates from PlanetScope data. Int. J. Appl. Earth Obs. Geoinf. 2020, 85. [Google Scholar] [CrossRef]
- Maliha, A.; Yusof, R.; Shapiai, M.I. Extreme learning machine for structured output spaces. Neural Comput. Appl. 2018, 30, 1251–1264. [Google Scholar] [CrossRef]
- Sonobe, R.; Sano, T.; Horie, H. Using spectral reflectance to estimate leaf chlorophyll content of tea with shading treatments. Biosyst. Eng. 2018, 175, 168–182. [Google Scholar] [CrossRef]
- Meng, X.T.; Bao, Y.L.; Liu, J.G.; Liu, H.J.; Zhang, X.L.; Zhang, Y.; Wang, P.; Tang, H.T.; Kong, F.C. Regional soil organic carbon prediction model based on a discrete wavelet analysis of hyperspectral satellite data. Int. J. Appl. Earth Obs. Geoinf. 2020, 89. [Google Scholar] [CrossRef]
- Inoue, Y.; Sakaiya, E.; Zhu, Y.; Takahashi, W. Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sens. Environ. 2012, 126, 210–221. [Google Scholar] [CrossRef]
- Roman, J.R.; Rodriguez-Caballero, E.; Rodriguez-Lozano, B.; Roncero-Ramos, B.; Chamizo, S.; Aguila-Carricondo, P.; Canton, Y. Spectral Response Analysis: An Indirect and Non-Destructive Methodology for the Chlorophyll Quantification of Biocrusts. Remote Sens. 2019, 11, 1350. [Google Scholar] [CrossRef]
- Miller, D.L.; Alonzo, M.; Roberts, D.A.; Tague, C.L.; McFadden, J.P. Drought response of urban trees and turfgrass using airborne imaging spectroscopy. Remote Sens. Environ. 2020, 240. [Google Scholar] [CrossRef]
- Liang, K.; Huang, J.N.; He, R.Y.; Wang, Q.J.; Chai, Y.Y.; Shen, M.X. Comparison of Vis-NIR and SWIR hyperspectral imaging for the non-destructive detection of DON levels in Fusarium head blight wheat kernels and wheat flour. Infrared Phys. Technol. 2020, 106, 9. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circular 1938, 347, 1884–1949. [Google Scholar]
- Sultana, T.; Savage, G.P.; McNeil, D.L.; Porter, N.G.; Martin, R.J.; Deo, B. Effects of fertilisation on the allyl isothiocyanate profile of above-ground tissues of New Zealand-grown wasabi. J. Sci. Food Agric. 2002, 82, 1477–1482. [Google Scholar] [CrossRef]
- Prasad, K.A.; Gnanappazham, L.; Selvam, V.; Ramasubramanian, R.; Kar, C.S. Developing a spectral library of mangrove species of Indian east coast using field spectroscopy. Geocarto Int. 2015, 30, 580–599. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009; p. 745. [Google Scholar]
- Nguyen, K.A.; Chen, W.; Lin, B.S.; Seeboonruang, U.; Thomas, K. Predicting Sheet and Rill Erosion of Shihmen Reservoir Watershed in Taiwan Using Machine Learning. Sustainability 2019, 11, 3615. [Google Scholar] [CrossRef]
- Yan, L.; Pang, L.; Wang, H.; Xiao, J. Recognition of different Longjing fresh tea varieties using hyperspectral imaging technology and chemometrics. J. Food Process Eng. 2020, 43, 9. [Google Scholar] [CrossRef]
- Gopinath, G.; Sasidharan, N.; Surendran, U. Landuse classification of hyperspectral data by spectral angle mapper and support vector machine in humid tropical region of India. Earth Sci. Inform. 2020, 13, 633–640. [Google Scholar] [CrossRef]
- Sun, X.; Subedi, P.; Walker, R.; Walsh, K.B. NIRS prediction of dry matter content of single olive fruit with consideration of variable sorting for normalisation pre-treatment. Postharvest Biol. Technol. 2020, 163, 10. [Google Scholar] [CrossRef]
- Tsai, F.; Philpot, W. Derivative analysis of hyperspectral data. Remote Sens. Environ. 1998, 66, 41–51. [Google Scholar] [CrossRef]
- Clark, R.N.; Roush, T.L. Reflectance spectroscopy: Quantitative analysis techniques for remote sensing applications. J. Geophys. Res. Solid Earth 1984, 89, 6329–6340. [Google Scholar] [CrossRef]
- Ren, G.X.; Sun, Y.M.; Li, M.H.; Ning, J.M.; Zhang, Z.Z. Cognitive spectroscopy for evaluating Chinese black tea grades (Camellia sinensis): Near-infrared spectroscopy and evolutionary algorithms. J. Sci. Food Agric. 2020, 100, 3950–3959. [Google Scholar] [CrossRef]
- R Core Team. R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 11 August 2020).
- Stevens, A.; Ramirez-Lopez, L. Package ‘prospectr’. Available online: https://cran.r-project.org/web/packages/prospectr/prospectr.pdf (accessed on 11 August 2020).
- Snoek, J.; Rippel, O.; Swersky, K.; Kiros, R.; Satish, N.; Sundaram, N.; Patwary, M.M.A.; Prabhat; Adams, R.P. Scalable Bayesian optimization using deep neural networks . In Proceedings of the 32nd International Conference on Machine Learning (ICML), Paris, Lille, 6–11 July 2015; pp. 2171–2180. [Google Scholar]
- Yan, Y. Bayesian Optimization of Hyperparameters. Available online: https://cran.r-project.org/web/packages/rBayesianOptimization/rBayesianOptimization.pdf (accessed on 11 August 2020).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Belgiu, M.; Csillik, O. Sentinel-2 cropland mapping using pixel-based and object-based time-weighted dynamic time warping analysis. Remote Sens. Environ. 2018, 204, 509–523. [Google Scholar] [CrossRef]
- Ishwaran, H. The effect of splitting on random forests. Mach. Learn. 2015, 99, 75–118. [Google Scholar] [CrossRef] [PubMed]
- Ishwaran, H.; Kogalur, U.B. Random survival forests for R. R News 2007, 7, 25–31. [Google Scholar]
- Ding, S.F.; Shi, Z.Z.; Tao, D.C.; An, B. Recent advances in Support Vector Machines. Neurocomputing 2016, 211, 1–3. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F.; Chang, C.-C.; Lin, C.-C. Misc Functions of the Department of Statistics, Probability. Available online: https://rdrr.io/rforge/e1071/e1071.pdf (accessed on 7 September 2020).
- Huang, G.B.; Zhu, Q.-Y.; Siew, C.-K. Extreme Learning Machine: A New Learning Scheme of Feedforward Neural Networks. In Proceedings of the International Joint Conference on Neural Networks (IJCNN2004), Budapest, Hungary, 25–29 July 2004; pp. 985–990. [Google Scholar]
- Quinlan, J.R. Learning with Continuous Classe AI ’92 . In Proceedings of the 5th Australian Joint Conference on Artificial Intelligence, Hobart, TAS, Australia, 16–18 November 1992; pp. 343–348. [Google Scholar]
- Kuhn, M.; Weston, S.; Keefer, C.; Coulter, N.; Quinlan, R.; Rulequest Research Pty Ltd. Package ‘Cubist’. Available online: https://cran.r-project.org/web/packages/Cubist/Cubist.pdf (accessed on 11 August 2020).
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Greenwell, B.; Boehmke, B.; Cunningham, J.; Developers, G. Package ‘GBM’. Available online: https://cran.r-project.org/web/packages/gbm/gbm.pdf (accessed on 11 August 2020).
- Burns, R.P.; Burns, R. Business Research Methods and Statistics Using SPSS; SAGE Publications: New York, NY, USA, 2008. [Google Scholar]
- Draper, N.H. Applied Regression Analysis; Wiley Series in Probability and Statistics; Wiley-Interscience: Hoboken, NJ, USA, 1998. [Google Scholar]
- Williams, P. Variables affecting near-infraredreflectance spectroscopic analysis. In Near-Infrared Technology in the Agricultural and Food Industries; Williams, P., Norris, K., Eds.; American Association of Cereal Chemists Inc.: Saint Paul, MN, USA, 1987; pp. 143–167. [Google Scholar]
- Ishwaran, H. Variable importance in binary regression trees and forests. Electron. J. Stat. 2007, 1, 519–537. [Google Scholar] [CrossRef]
- Sonobe, R. Parcel-Based Crop Classification Using Multi-Temporal TerraSAR-X Dual Polarimetric Data. Remote Sens. 2019, 11, 1148. [Google Scholar] [CrossRef]
- Cortez, P.; Embrechts, M.J. Using sensitivity analysis and visualization techniques to open black box data mining models. Inf. Sci. 2013, 225, 1–17. [Google Scholar] [CrossRef]
- Kewley, R.H.; Embrechts, M.J.; Breneman, C. Data strip mining for the virtual design of pharmaceuticals with neural networks. IEEE Trans. Neural Netw. 2000, 11, 668–679. [Google Scholar] [CrossRef]
- Katayama, Y.; Shida, S. Studies on the Change of Chlorophyll a and b Contents Due to Projected Materials and Some Environmental Conditions. Cytologia 1970, 35, 171–180. [Google Scholar] [CrossRef]
- Terashima, I.; Hikosaka, K. Comparative ecophysiology of leaf and canopy photosynthesis. Plant Cell Environ. 1995, 18, 1111–1128. [Google Scholar] [CrossRef]
- Tang, Y.L.; Wen, X.G.; Lu, Q.T.; Yang, Z.P.; Cheng, Z.K.; Lu, C.M. Heat stress induces an aggregation of the light-harvesting complex of photosystem II in spinach plants. Plant Physiol. 2007, 143, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Embry, J.L.; Nothnagel, E.A. Leaf senescence of postproduction poinsettias in low-light stress. J. Am. Soc. Hortic. Sci. 1994, 119, 1006–1013. [Google Scholar] [CrossRef]
- Pinnola, A. The rise and fall of Light-Harvesting Complex Stress-Related proteins as photoprotection agents during evolution. J. Exp. Bot. 2019, 70, 5527–5535. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, A.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Bruning, B.; Berger, B.; Lewis, M.; Liu, H.; Garnett, T. Approaches, applications, and future directions for hyperspectral vegetation studies: An emphasis on yield-limiting factors in wheat. Plant Phenome J. 2020, 3. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey, J.E. Ratio analysis of reflectance spectra (RARS): An algorithm for the remote estimation of the concentrations of chlorophyll A, chlorophyll B, and carotenoids in soybean leaves. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of chlorophyll a, chlorophyll b, chlorophyll a + b, and total carotenoid content in eucalyptus leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing carotenoid content in plant leaves with reflectance spectroscopy. Photochem. Photobiol. 2002, 75, 272–281. [Google Scholar] [CrossRef]
- Hernandez-Clemente, R.; Navarro-Cerrillo, R.M.; Zarco-Tejada, P.J. Carotenoid content estimation in a heterogeneous conifer forest using narrow-band indices and PROSPECT plus DART simulations. Remote Sens. Environ. 2012, 127, 298–315. [Google Scholar] [CrossRef]
- Sonobe, R.; Wang, Q. Nondestructive assessments of carotenoids content of broadleaved plant species using hyperspectral indices. Comput. Electron. Agric. 2018, 145, 18–26. [Google Scholar] [CrossRef]
- Horvath, G. CMAC neural network as an SVM with B-spline kernel functions. Proceedings of 20th IEEE Instrumentation and Measurement Technology Conference, Vail, CO, USA, 20–22 May 2003; pp. 1108–1113. [Google Scholar]
- Phienthrakul, T.; Kijsirikul, B. Evolutionary strategies for hyperparameters of support vector machines based on multi-scale radial basis function kernels. Soft Comput. 2010, 14, 681–699. [Google Scholar] [CrossRef]
- Trisasongko, B.H. Mapping stand age of rubber plantation using ALOS-2 polarimetric SAR data. Eur. J. Remote Sens. 2017, 50, 64–76. [Google Scholar] [CrossRef]
- Zhou, J.; Li, E.M.; Wei, H.X.; Li, C.Q.; Qiao, Q.Q.; Armaghani, D.J. Random Forests and Cubist Algorithms for Predicting Shear Strengths of Rockfill Materials. Appl. Sci. 2019, 9, 1621. [Google Scholar] [CrossRef]
- Walton, J.T. Subpixel urban land cover estimation: Comparing Cubist, Random Forests, and support vector regression. Photogramm. Eng. Remote Sens. 2008, 74, 1213–1222. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1491–1507. [Google Scholar] [CrossRef]
- Zhou, X.F.; Huang, W.J.; Zhang, J.C.; Kong, W.P.; Casa, R.; Huang, Y.B. A novel combined spectral index for estimating the ratio of carotenoid to chlorophyll content to monitor crop physiological and phenological status. Int. J. Appl. Earth Obs. Geoinf. 2019, 76, 128–142. [Google Scholar] [CrossRef]

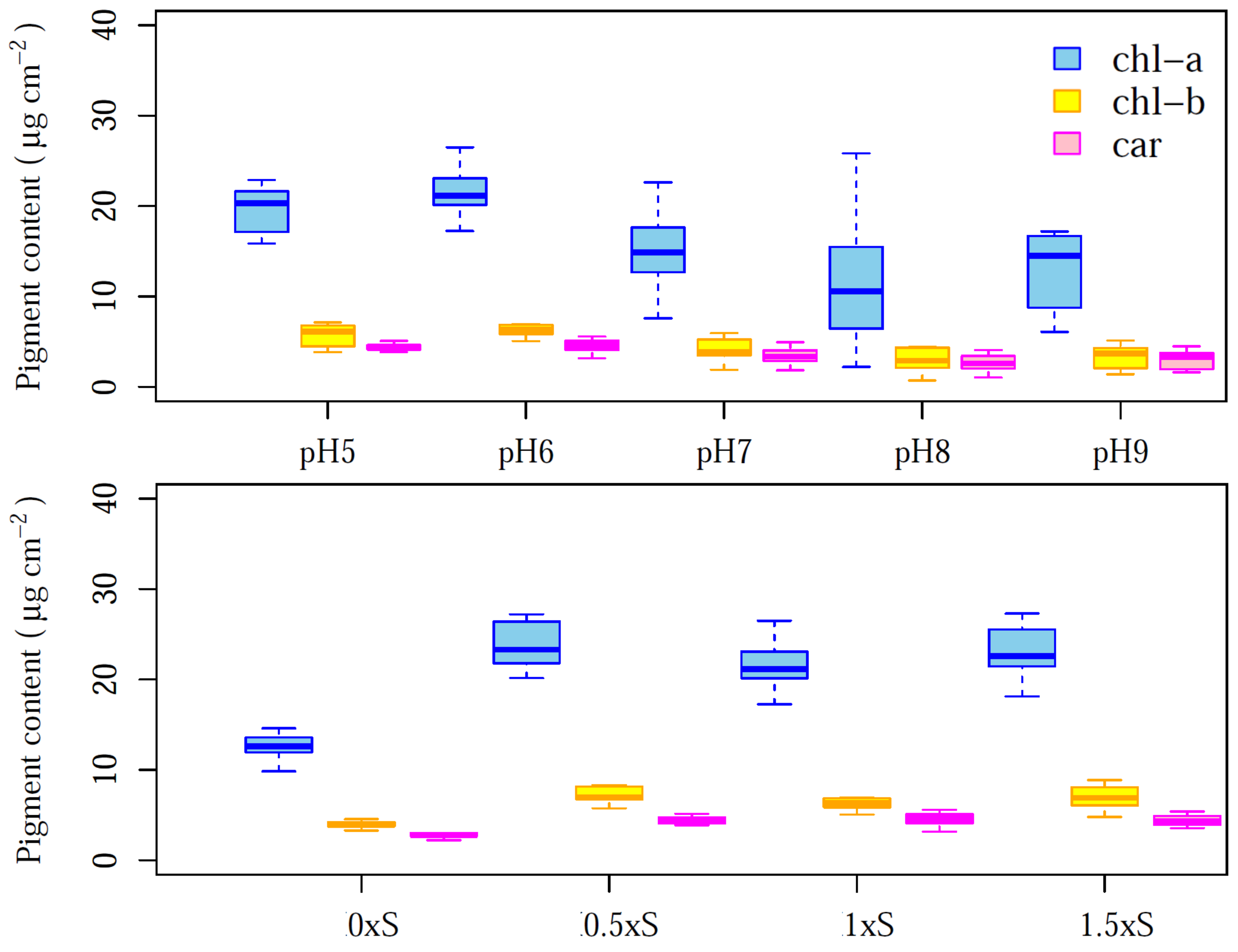
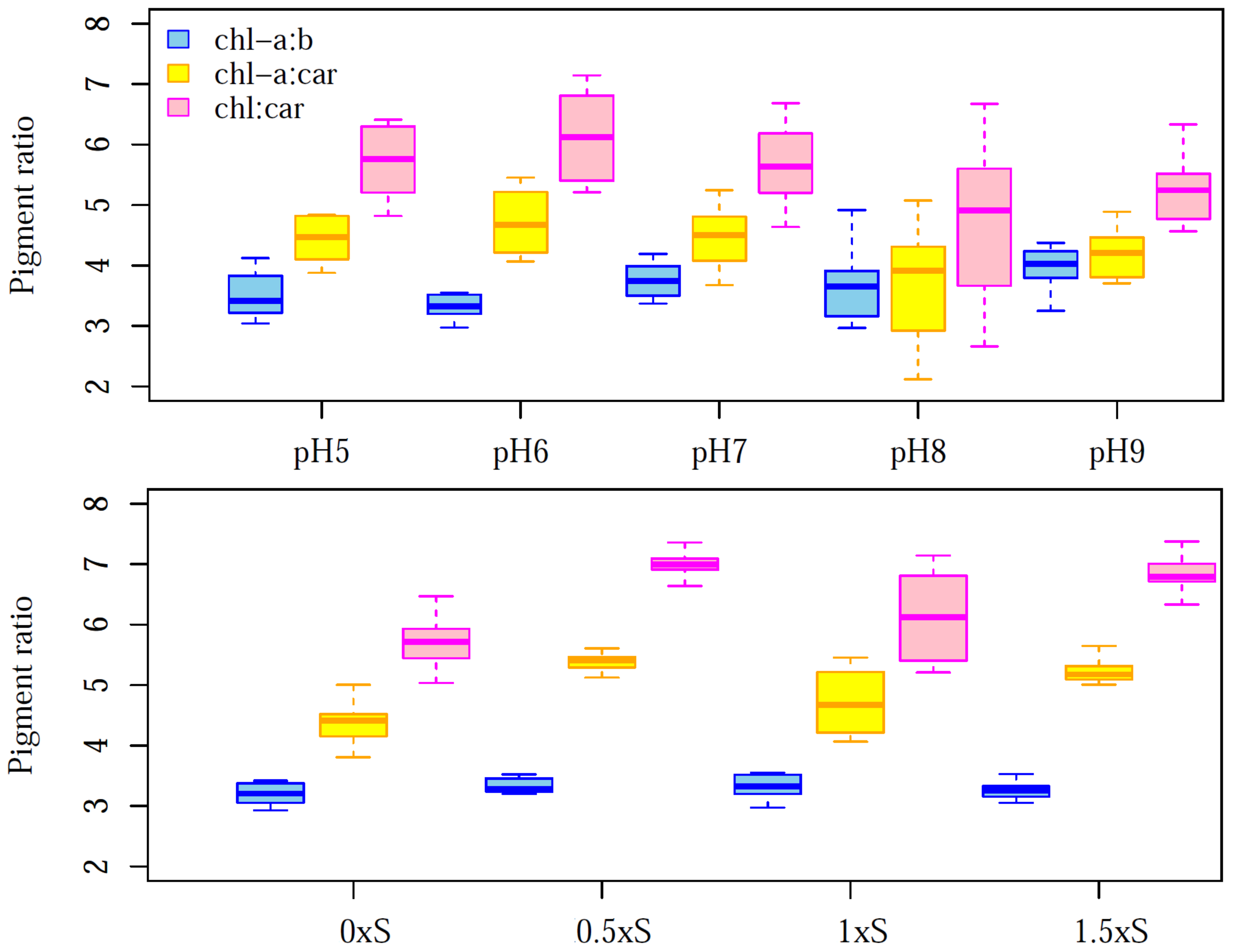
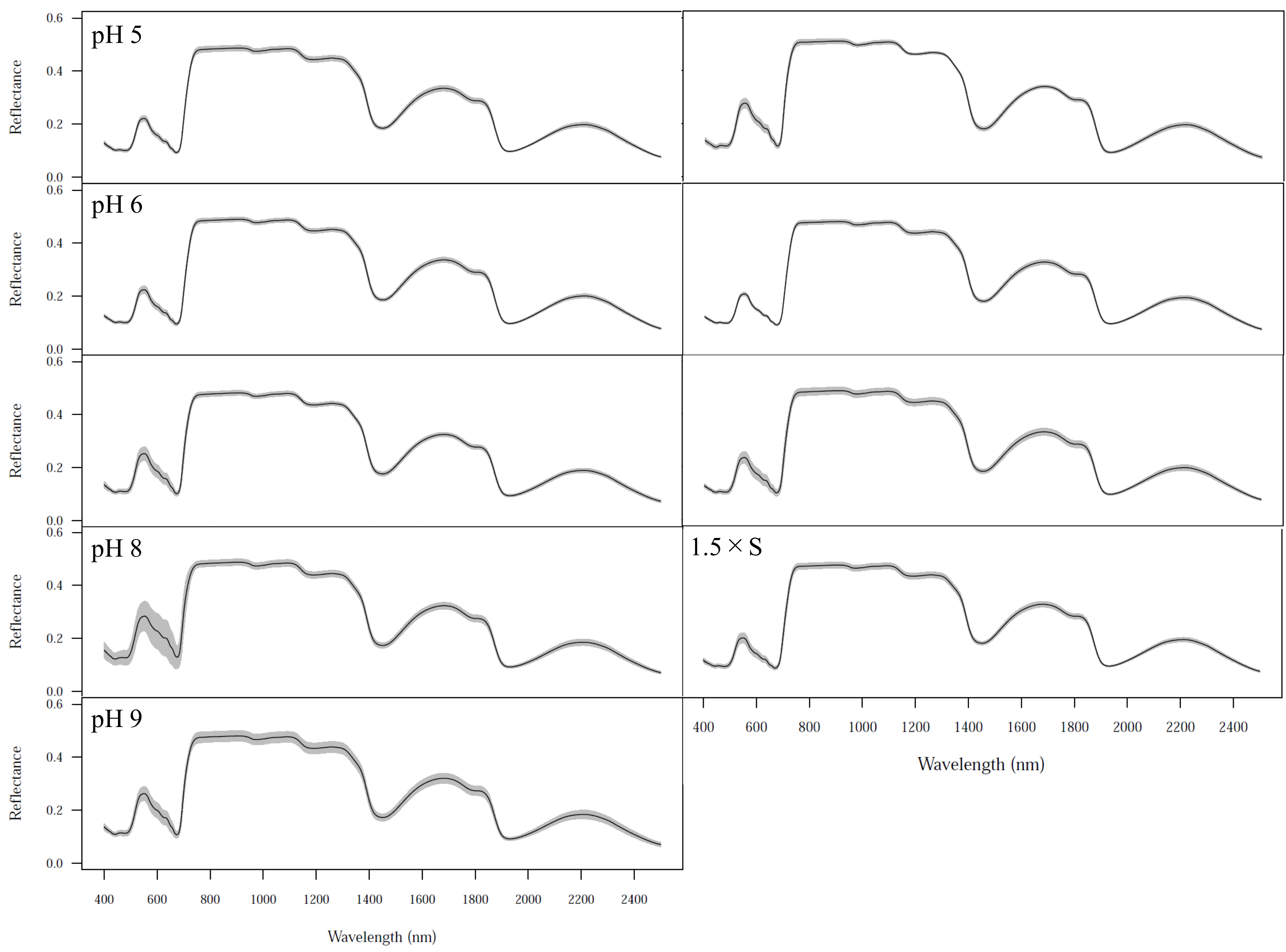
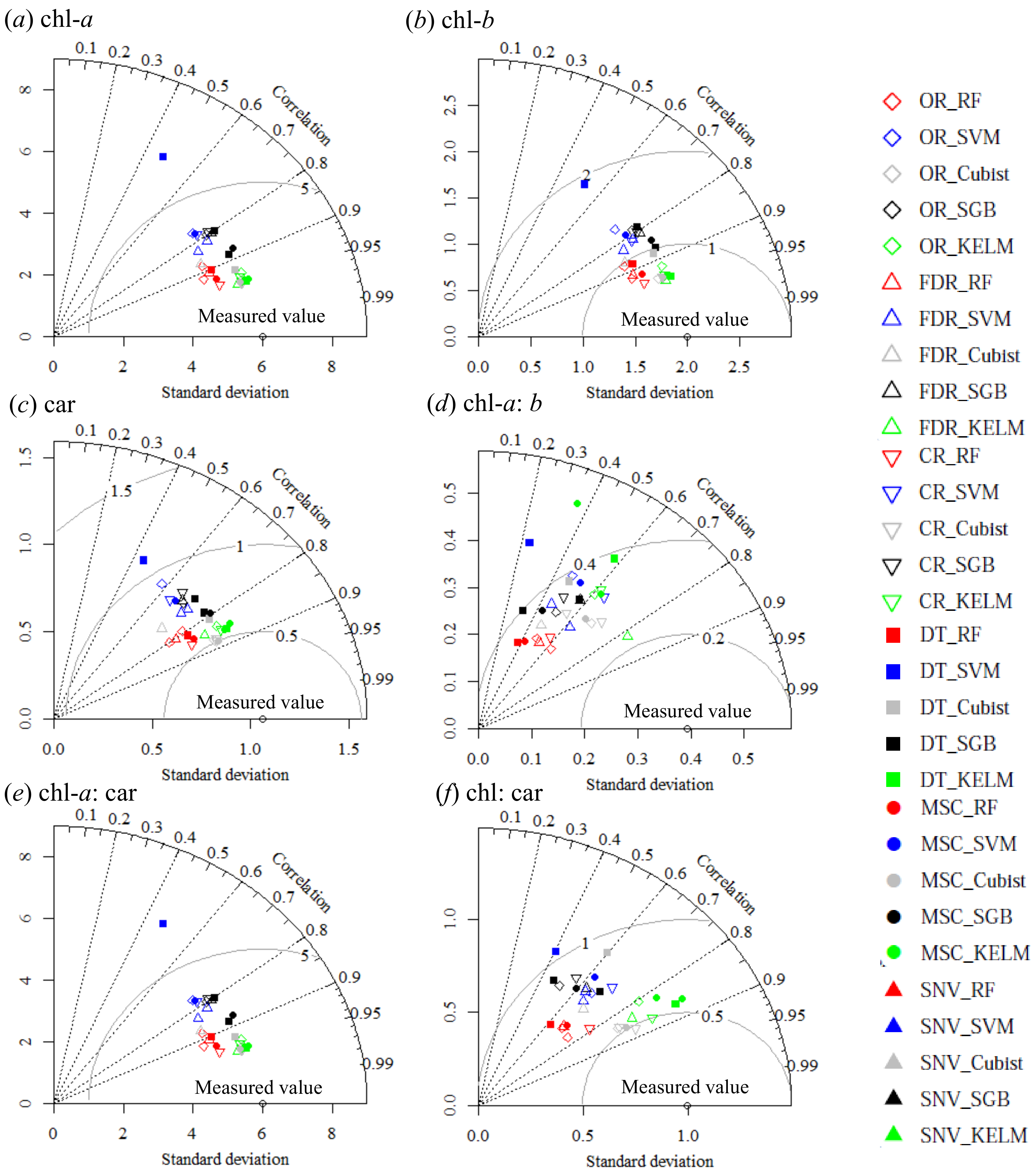
| Dataset | chl a | chl b | Car | |||
|---|---|---|---|---|---|---|
| pH | S strength | pH | S strength | pH | S strength | |
| All | −0.587 *** | 0.606 *** | −0.596 *** | 0.580 *** | −0.550 *** | 0.567 *** |
| Onimidori | −0.420 * | 0.575 * | −0.400 * | 0.518 * | −0.428 * | 0.526 * |
| Mazuma | −0.734 *** | 0.633 ** | −0.751 *** | 0.637 ** | −0.746 *** | 0.610 ** |
| Dataset | chl a:b | chl a:car | chl:car | |||
| pH | S strength | pH | S strength | pH | S strength | |
| All | 0.402 ** | 0.137 | −0.298 * | 0.400 * | −0.335 * | 0.390 * |
| Onimidori | 0.309 | 0.457 * | −0.262 | 0.41 | −0.283 | 0.379 |
| Mazuma | 0.571 ** | −0.328 | −0.39 | 0.520 * | −0.461 * | 0.558 * |
| Pre-Processing | Selected Wavelength (nm) | Overall Accuracy |
|---|---|---|
| OR | 706 | 0.340 |
| FDR | 614, 619, 756 | 0.680 |
| CR | 536 | 0.340 |
| DT | 714 | 0.420 |
| MSC | 726 | 0.480 |
| SNV | 729 | 0.440 |
| Pre-Processing | Selected Wavelength (nm) | Overall Accuracy |
|---|---|---|
| OR | 514, 694 | 0.692 |
| FDR | 689, 702 | 0.795 |
| CR | 695, 893 | 0.718 |
| DT | 693 | 0.615 |
| MSC | 697 | 0.615 |
| SNV | 697 | 0.615 |
| Pro–Processing | Algorithm | chl a | chl b | car | chl a:b | chl a:car | chl:car |
|---|---|---|---|---|---|---|---|
| OR | SVM | 2 | 3 | 3 | |||
| OR | KELM | 6 | 4 | 6 | 2 | 4 | 1 |
| OR | Cubist | 3 | 6 | 13 | 4 | 4 | 4 |
| FDR | RF | 1 | |||||
| FDR | SVM | 1 | 1 | 2 | 2 | ||
| FDR | KELM | 22 | 19 | 2 | 36 | 4 | 6 |
| FDR | Cubist | 1 | 2 | ||||
| CR | RF | 1 | 1 | ||||
| CR | SVM | 1 | 1 | 2 | 1 | ||
| CR | KELM | 4 | 16 | 6 | 4 | 25 | 17 |
| CR | Cubist | 10 | 4 | 14 | 7 | 5 | |
| DT | RF | 5 | 3 | 2 | 1 | ||
| DT | SVM | 1 | 8 | 1 | |||
| DT | KELM | 9 | 11 | 4 | 17 | 19 | 7 |
| DT | Cubist | 14 | 7 | 17 | 7 | 10 | 26 |
| DT | SGB | 1 | |||||
| MSC | KELM | 9 | 12 | 8 | 5 | 9 | 5 |
| MSC | Cubist | 4 | 5 | 2 | 1 | 1 | |
| SNV | SVM | 1 | 5 | 1 | 3 | ||
| SNV | KELM | 6 | 5 | 1 | 6 | 3 | 7 |
| SNV | Cubist | 4 | 3 | 16 | 10 | 14 | |
| SNV | SGB | 1 |
| chl a:b | |||||
| RF | SVM | Cubist | SGB | KELM | |
| OR | 0.204 ± 0.235 | 0.825 ± 0.482 | 0.537 ± 0.474 | 0.566 ± 0.277 | 0.537 ± 0.545 |
| FDR | 0.315 ± 0.286 | 0.942 ± 0.363 | 0.564 ± 0.354 | 0.770 ± 0.254 | 1.200 ± 0.626 |
| CR | 0.202 ± 0.276 | 0.750 ± 0.297 | 0.323 ± 0.356 | 0.632 ± 0.218 | 0.552 ± 0.558 |
| DT | 0.127 ± 0.282 | 1.075 ± 0.597 | 0.731 ± 0.471 | 0.637 ± 0.249 | 0.884 ± 0.755 |
| MSC | 0.153 ± 0.207 | 0.642 ± 0.341 | 0.446 ± 0.456 | 0.470 ± 0.256 | 0.775 ± 0.530 |
| SNV | 0.084 ± 0.219 | 0.929 ± 0.547 | 0.519 ± 0.504 | 0.461 ± 0.255 | 0.452 ± 0.655 |
| chl a:car | |||||
| RF | SVM | Cubist | SGB | KELM | |
| OR | 0.156 ± 0.212 | 0.766 ± 0.533 | 0.350 ± 0.454 | 0.255 ± 0.212 | 0.392 ± 0.594 |
| FDR | 0.139 ± 0.282 | 0.567 ± 0.378 | 0.578 ± 0.453 | 0.488 ± 0.265 | 0.518 ± 0.453 |
| CR | 0.131 ± 0.227 | 0.701 ± 0.472 | 0.305 ± 0.433 | 0.424 ± 0.246 | 0.657 ± 0.594 |
| DT | 0.172 ± 0.256 | 0.810 ± 0.521 | 0.431 ± 0.439 | 0.398 ± 0.261 | 0.529 ± 0.575 |
| MSC | −0.013 ± 0.221 | 0.516 ± 0.437 | 0.173 ± 0.631 | 0.250 ± 0.248 | 0.304 ± 0.613 |
| SNV | 0.047 ± 0.207 | 0.703 ± 0.534 | 0.417 ± 0.503 | 0.264 ± 0.242 | 0.504 ± 0.550 |
| chl:car | |||||
| RF | SVM | Cubist | SGB | KELM | |
| OR | 0.003 ± 0.246 | 0.571 ± 0.464 | 0.908 ± 0.472 | 0.332 ± 0.268 | 0.333 ± 0.478 |
| FDR | 0.118 ± 0.283 | 0.545 ± 0.468 | 0.442 ± 0.400 | 0.476 ± 0.285 | 0.532 ± 0.473 |
| CR | −0.055 ± 0.251 | 0.562 ± 0.558 | 0.276 ± 0.489 | 0.159 ± 0.275 | 0.630 ± 0.652 |
| DT | 0.182 ± 0.268 | 0.716 ± 0.579 | 0.496 ± 0.469 | 0.360 ± 0.272 | 0.456 ± 0.614 |
| MSC | −0.069 ± 0.211 | 0.406 ± 0.483 | −0.231 ± 0.674 | 0.135 ± 0.259 | 0.416 ± 0.613 |
| SNV | 0.039 ± 0.210 | 0.717 ± 0.530 | 0.455 ± 0.531 | 0.255 ± 0.256 | 0.430 ± 0.569 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonobe, R.; Yamashita, H.; Mihara, H.; Morita, A.; Ikka, T. Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance. Remote Sens. 2020, 12, 3265. https://doi.org/10.3390/rs12193265
Sonobe R, Yamashita H, Mihara H, Morita A, Ikka T. Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance. Remote Sensing. 2020; 12(19):3265. https://doi.org/10.3390/rs12193265
Chicago/Turabian StyleSonobe, Rei, Hiroto Yamashita, Harumi Mihara, Akio Morita, and Takashi Ikka. 2020. "Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance" Remote Sensing 12, no. 19: 3265. https://doi.org/10.3390/rs12193265
APA StyleSonobe, R., Yamashita, H., Mihara, H., Morita, A., & Ikka, T. (2020). Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance. Remote Sensing, 12(19), 3265. https://doi.org/10.3390/rs12193265





