Automatic Grapevine Trunk Detection on UAV-Based Point Cloud
Abstract
1. Introduction
2. Materials and Methods
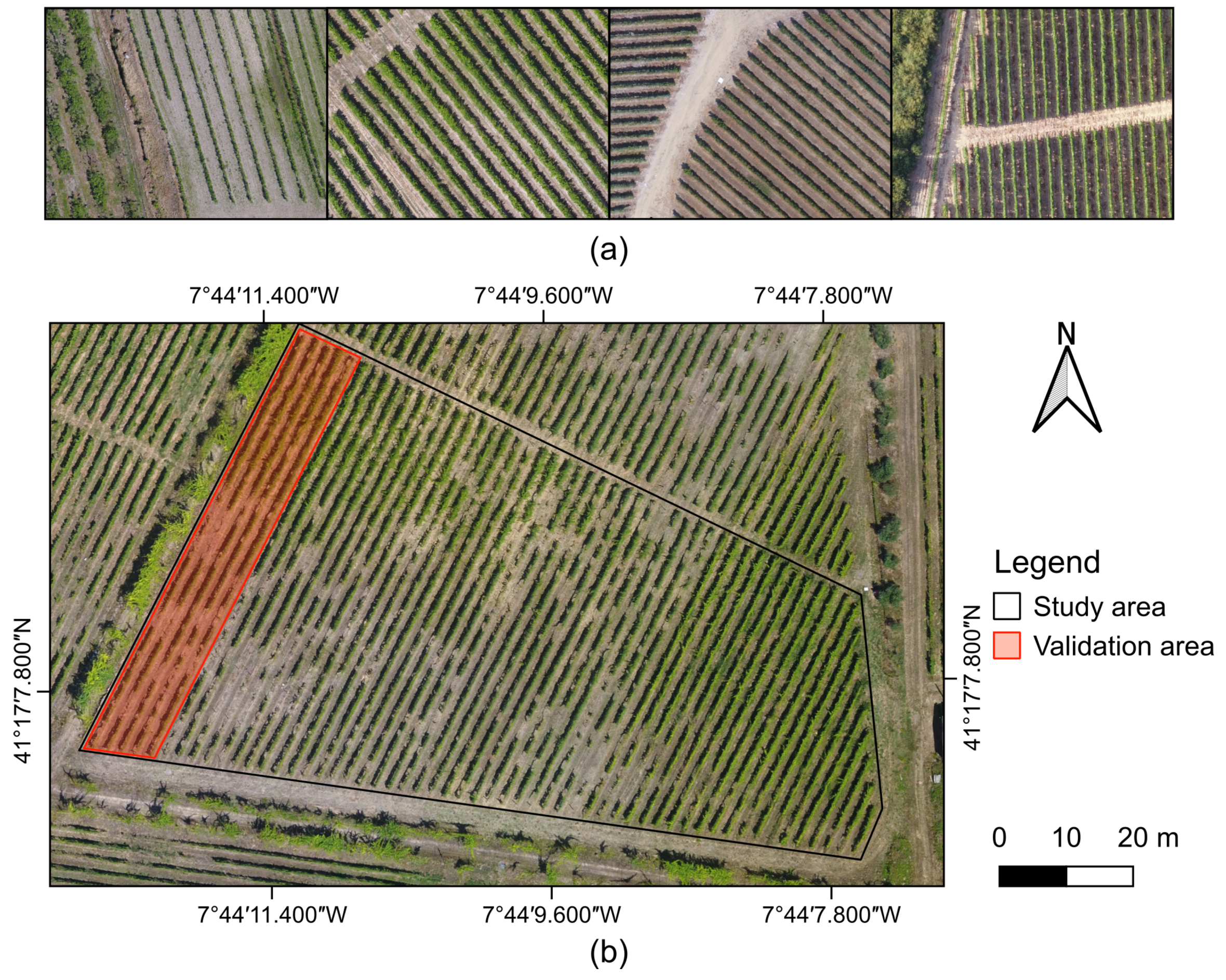
2.1. Study Area
2.2. UAV-Based Data Acquisition
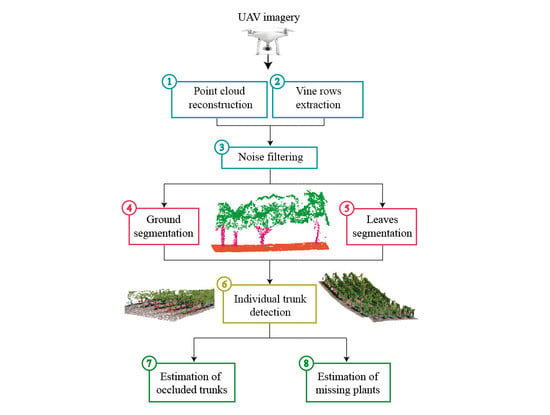
2.3. Proposed Method
2.3.1. SFM Reconstruction and Noise Removal
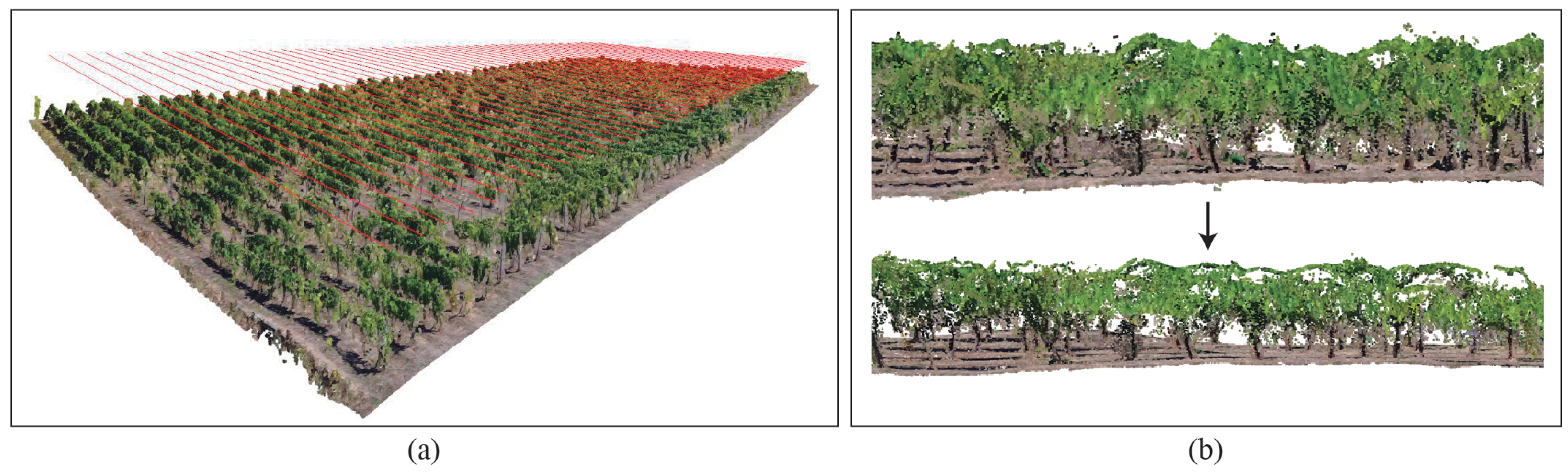
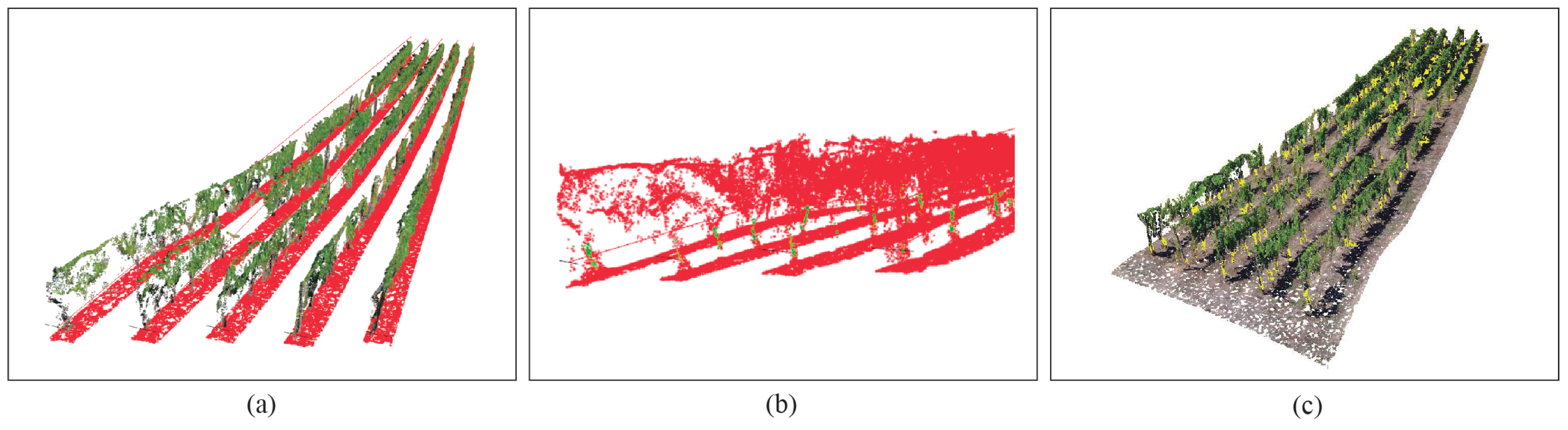
2.3.2. Vine Rows Extraction
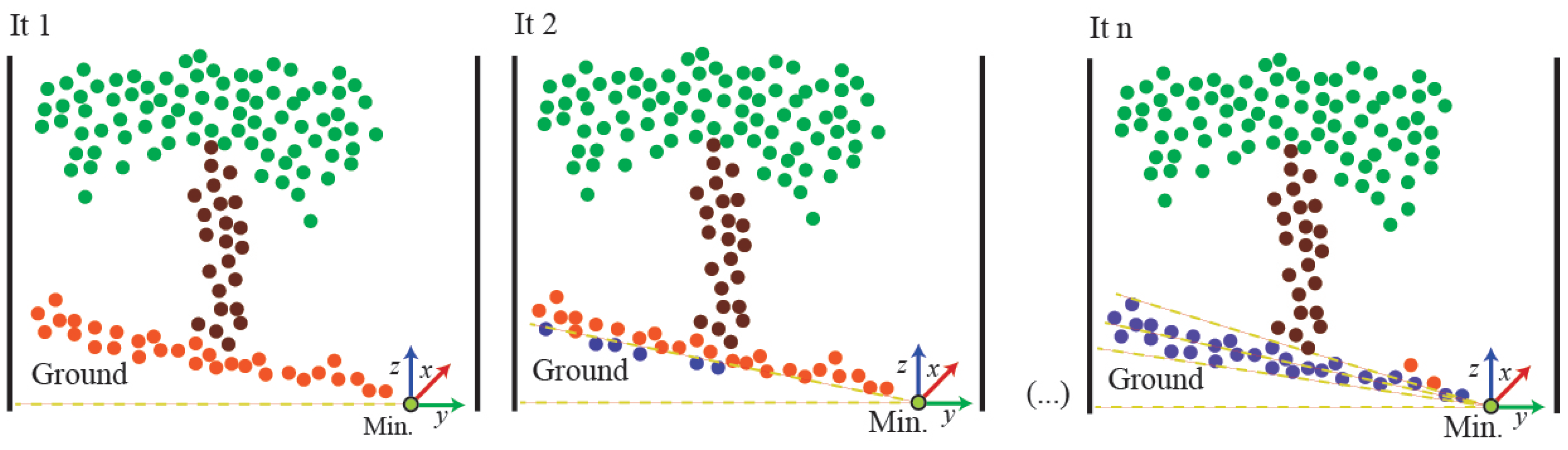
2.3.3. Ground and Leaves Segmentation
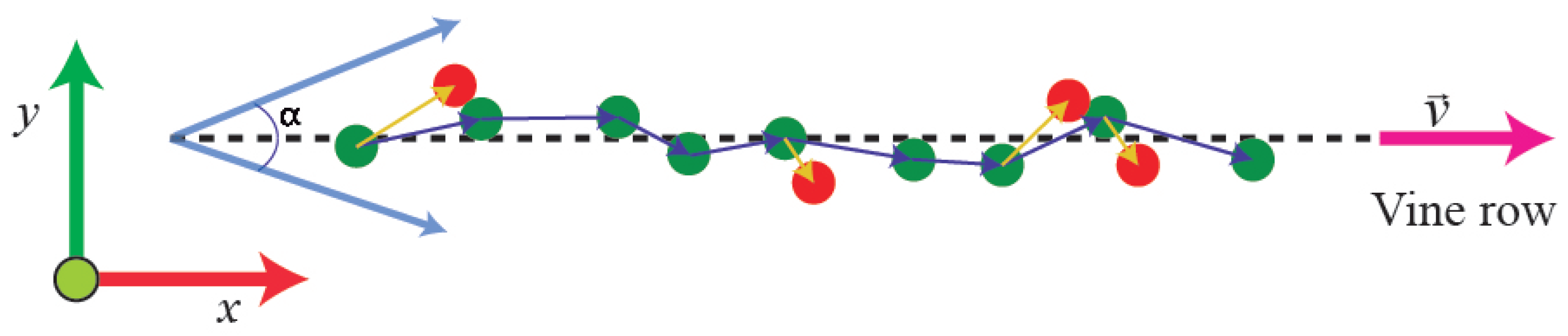
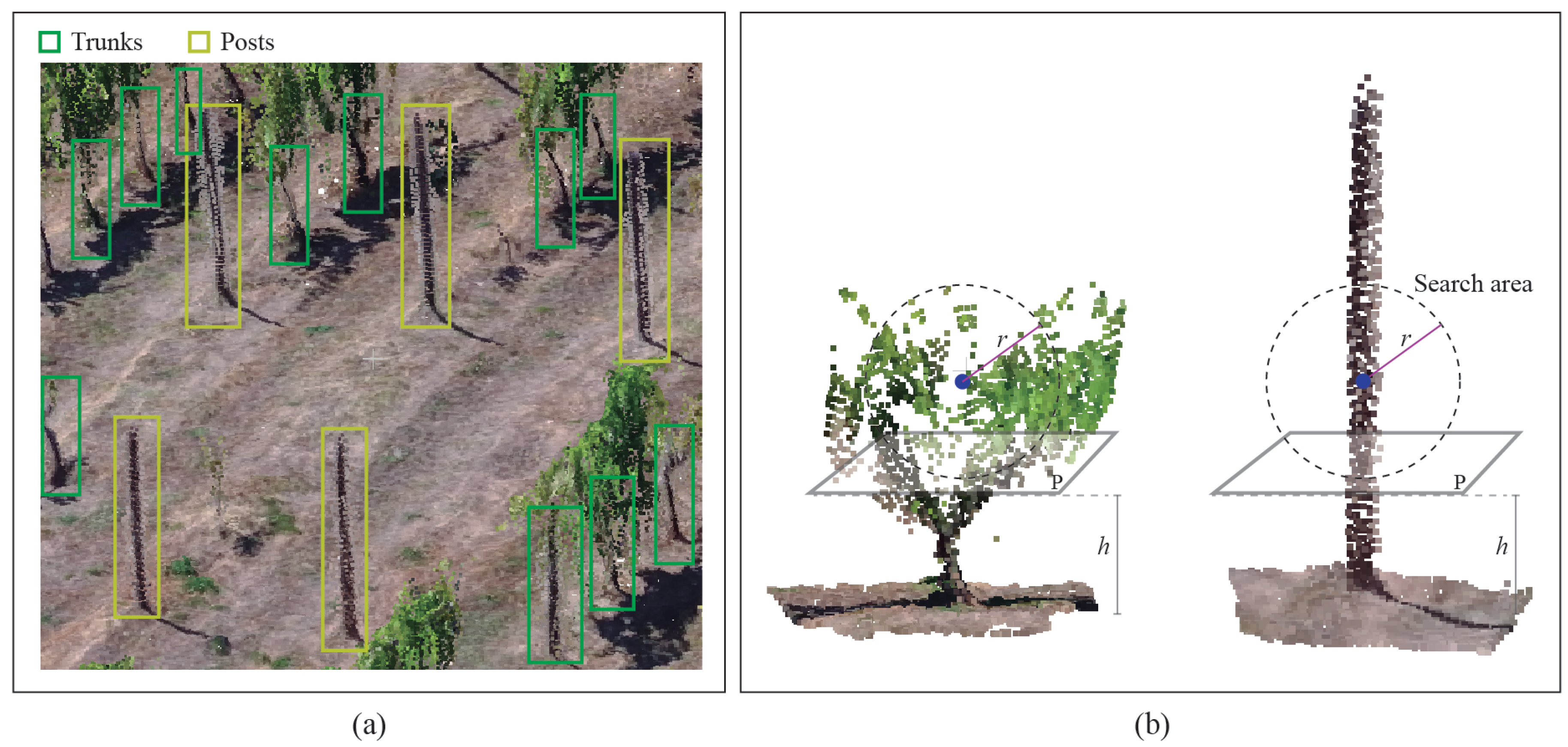
2.3.4. Trunk Detection
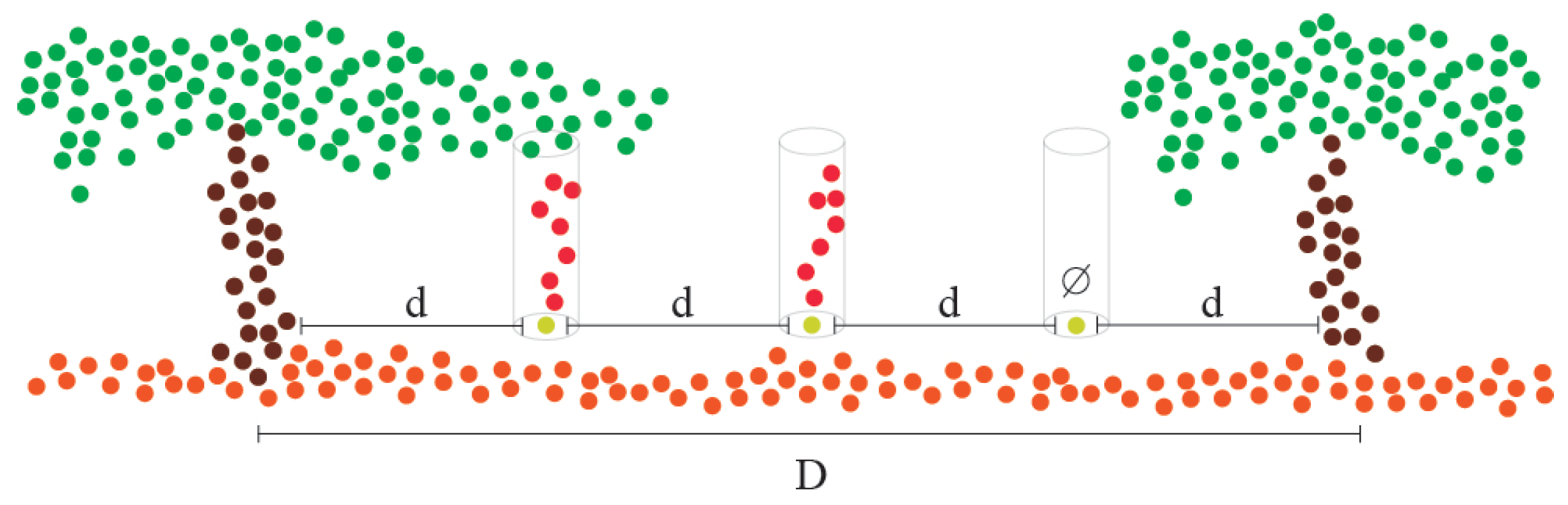
2.3.5. Estimation of Missing Plants and Occluded Trunks
2.4. Validation Process
3. Results
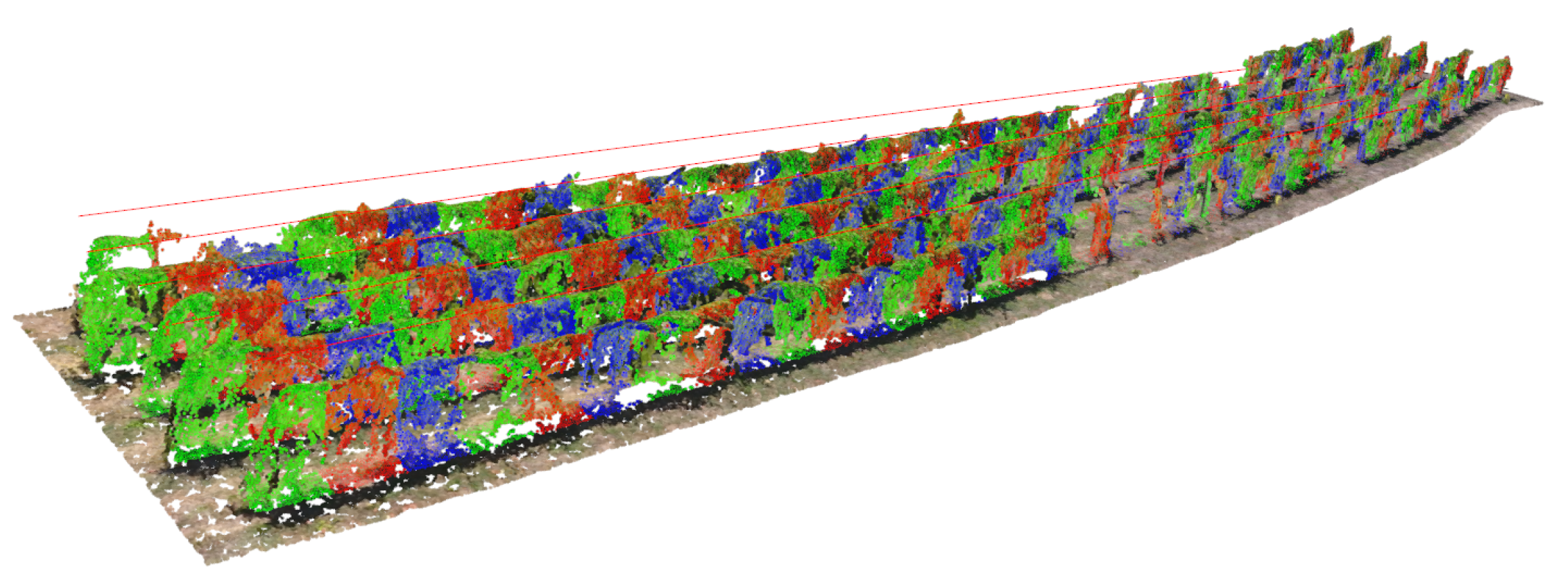
3.1. Point Cloud Reconstruction and Processing
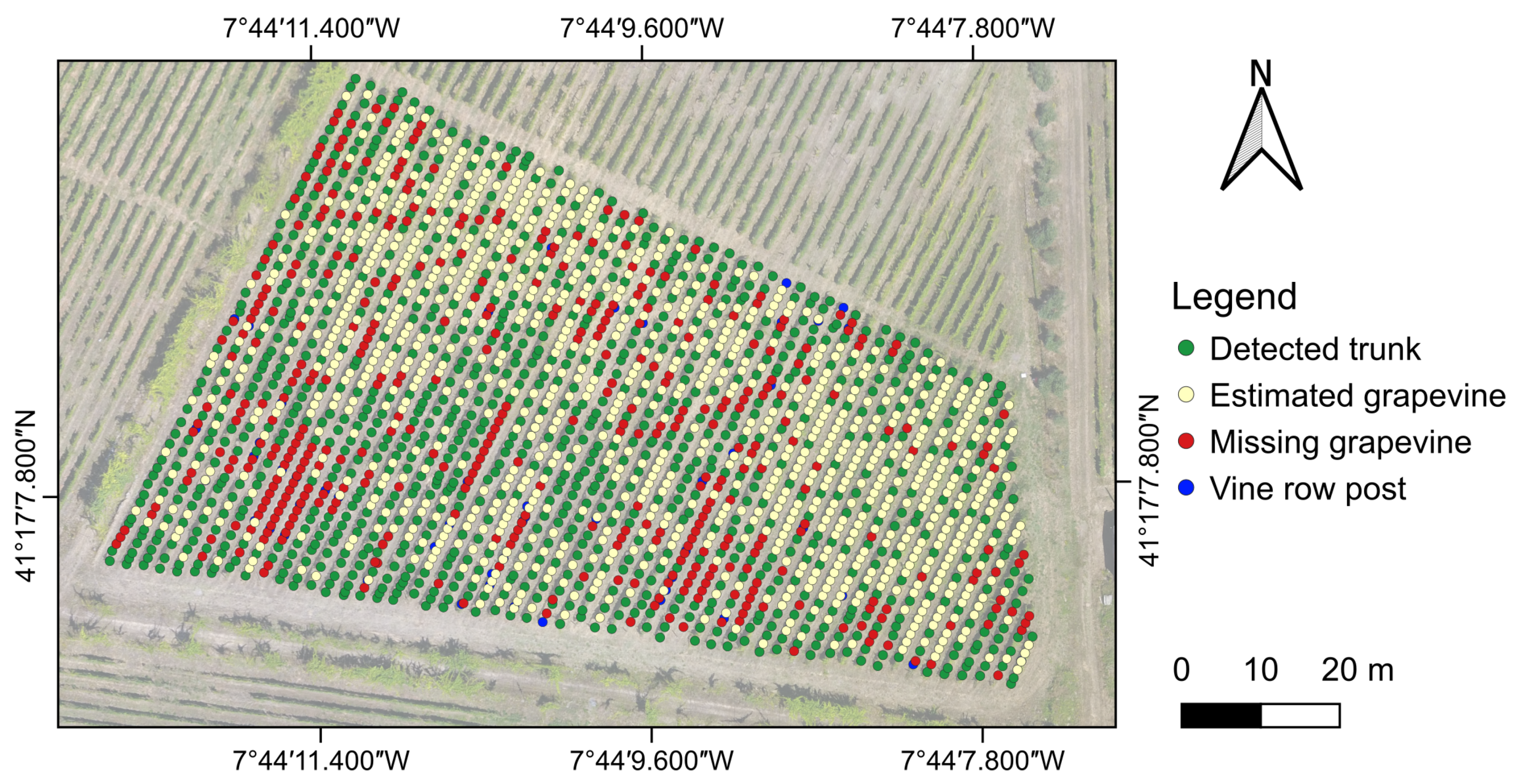
3.2. Individual Grapevine Detection
3.3. Grapevine Estimation Accuracy
4. Discussion
4.1. Point Cloud Reconstruction and Processing
4.2. Individual Grapevine Detection
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Mogili, U.R.; Deepak, B. Review on application of drone systems in precision agriculture. Procedia Comput. Sci. 2018, 133, 502–509. [Google Scholar] [CrossRef]
- Bernardes, M.F.F.; Pazin, M.; Pereira, L.C.; Dorta, D.J. Impact of pesticides on environmental and human health. Toxicol. Stud. Cells Drugs Environ. 2015, 195–233. [Google Scholar]
- Scott, G.; Rajabifard, A. Sustainable development and geospatial information: A strategic framework for integrating a global policy agenda into national geospatial capabilities. Geo-Spat. Inf. Sci. 2017, 20, 59–76. [Google Scholar] [CrossRef]
- Pádua, L.; Vanko, J.; Hruška, J.; Adão, T.; Sousa, J.J.; Peres, E.; Morais, R. UAS, sensors, and data processing in agroforestry: A review towards practical applications. Int. J. Remote Sens. 2017, 38, 2349–2391. [Google Scholar] [CrossRef]
- Ezenne, G.; Jupp, L.; Mantel, S.; Tanner, J. Current and potential capabilities of UAS for crop water productivity in precision agriculture. Agric. Water Manag. 2019, 218, 158–164. [Google Scholar] [CrossRef]
- Shi, X.; Han, W.; Zhao, T.; Tang, J. Decision support system for variable rate irrigation based on UAV multispectral remote sensing. Sensors 2019, 19, 2880. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, J.; Sudduth, K.A.; Kitchen, N.R. Estimation of maize yield and effects of variable-rate nitrogen application using UAV-based RGB imagery. Biosyst. Eng. 2020, 189, 24–35. [Google Scholar] [CrossRef]
- Mendes, J.; Pinho, T.M.; Neves dos Santos, F.; Sousa, J.J.; Peres, E.; Boaventura-Cunha, J.; Cunha, M.; Morais, R. Smartphone Applications Targeting Precision Agriculture Practices—A Systematic Review. Agronomy 2020, 10, 855. [Google Scholar] [CrossRef]
- Matese, A.; Di Gennaro, S.F. Technology in precision viticulture: A state of the art review. Int. J. Wine Res. 2015, 7, 69–81. [Google Scholar] [CrossRef]
- Proffitt, A.P.B.; Bramley, R.; Lamb, D.; Winter, E. Precision Viticulture: A New Era in Vineyard Management and Wine Production; Winetitles Pty Ltd.: Ashford, SA, USA, 2006. [Google Scholar]
- Campos, J.; Llop, J.; Gallart, M.; García-Ruiz, F.; Gras, A.; Salcedo, R.; Gil, E. Development of canopy vigour maps using UAV for site-specific management during vineyard spraying process. Precis. Agric. 2019, 20, 1136–1156. [Google Scholar] [CrossRef]
- Pádua, L.; Adão, T.; Sousa, A.; Peres, E.; Sousa, J.J. Individual Grapevine Analysis in a Multi-Temporal Context Using UAV-Based Multi-Sensor Imagery. Remote Sens. 2020, 12, 139. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Hruška, J.; Adão, T.; Bessa, J.; Sousa, A.; Peres, E.; Morais, R.; Sousa, J.J. Vineyard properties extraction combining UAS-based RGB imagery with elevation data. Int. J. Remote Sens. 2018, 39, 5377–5401. [Google Scholar] [CrossRef]
- Comba, L.; Gay, P.; Primicerio, J.; Aimonino, D.R. Vineyard detection from unmanned aerial systems images. Comput. Electron. Agric. 2015, 114, 78–87. [Google Scholar] [CrossRef]
- Mathews, A.J.; Jensen, J.L. Visualizing and quantifying vineyard canopy LAI using an unmanned aerial vehicle (UAV) collected high density structure from motion point cloud. Remote Sens. 2013, 5, 2164–2183. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Using 3D point clouds derived from UAV RGB imagery to describe vineyard 3D macro-structure. Remote Sens. 2017, 9, 111. [Google Scholar] [CrossRef]
- Poblete-Echeverría, C.; Olmedo, G.F.; Ingram, B.; Bardeen, M. Detection and segmentation of vine canopy in ultra-high spatial resolution RGB imagery obtained from unmanned aerial vehicle (UAV): A case study in a commercial vineyard. Remote Sens. 2017, 9, 268. [Google Scholar] [CrossRef]
- Caruso, G.; Tozzini, L.; Rallo, G.; Primicerio, J.; Moriondo, M.; Palai, G.; Gucci, R. Estimating biophysical and geometrical parameters of grapevine canopies (‘Sangiovese’) by an unmanned aerial vehicle (UAV) and VIS-NIR cameras. Vitis 2017, 56, 63–70. [Google Scholar]
- De Castro, A.I.; Jimenez-Brenes, F.M.; Torres-Sánchez, J.; Peña, J.M.; Borra-Serrano, I.; López-Granados, F. 3-D characterization of vineyards using a novel UAV imagery-based OBIA procedure for precision viticulture applications. Remote Sens. 2018, 10, 584. [Google Scholar] [CrossRef]
- Matese, A.; Di Gennaro, S.F. Practical applications of a multisensor uav platform based on multispectral, thermal and rgb high resolution images in precision viticulture. Agriculture 2018, 8, 116. [Google Scholar] [CrossRef]
- Primicerio, J.; Caruso, G.; Comba, L.; Crisci, A.; Gay, P.; Guidoni, S.; Genesio, L.; Ricauda Aimonino, D.; Vaccari, F.P. Individual plant definition and missing plant characterization in vineyards from high-resolution UAV imagery. Eur. J. Remote Sens. 2017, 50, 179–186. [Google Scholar] [CrossRef]
- Comba, L.; Biglia, A.; Aimonino, D.R.; Gay, P. Unsupervised detection of vineyards by 3D point-cloud UAV photogrammetry for precision agriculture. Comput. Electron. Agric. 2018, 155, 84–95. [Google Scholar] [CrossRef]
- Comba, L.; Biglia, A.; Aimonino, D.R.; Tortia, C.; Mania, E.; Guidoni, S.; Gay, P. Leaf Area Index evaluation in vineyards using 3D point clouds from UAV imagery. Precis. Agric. 2020, 21, 881–896. [Google Scholar] [CrossRef]
- Mesas-Carrascosa, F.J.; de Castro, A.I.; Torres-Sánchez, J.; Triviño-Tarradas, P.; Jiménez-Brenes, F.M.; García-Ferrer, A.; López-Granados, F. Classification of 3D point clouds using color vegetation indices for precision viticulture and digitizing applications. Remote Sens. 2020, 12, 317. [Google Scholar] [CrossRef]
- Aboutalebi, M.; Torres-Rua, A.F.; McKee, M.; Kustas, W.P.; Nieto, H.; Alsina, M.M.; White, A.; Prueger, J.H.; McKee, L.; Alfieri, J.; et al. Incorporation of Unmanned Aerial Vehicle (UAV) Point Cloud Products into Remote Sensing Evapotranspiration Models. Remote Sens. 2020, 12, 50. [Google Scholar] [CrossRef]
- Moreno, H.; Valero, C.; Bengochea-Guevara, J.M.; Ribeiro, Á.; Garrido-Izard, M.; Andújar, D. On-Ground Vineyard Reconstruction Using a LiDAR-Based Automated System. Sensors 2020, 20, 1102. [Google Scholar] [CrossRef]
- Rusu, R.B.; Cousins, S. 3d is here: Point cloud library (pcl). In Proceedings of the 2011 IEEE International Conference on Robotics and Automation, Shanghai, China, 9–13 May 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 1–4. [Google Scholar]
- Schönberger, J.L.; Frahm, J. Structure-from-Motion Revisited. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 4104–4113. [Google Scholar] [CrossRef]
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef]
- Han, X.F.; Jin, J.S.; Wang, M.J.; Jiang, W.; Gao, L.; Xiao, L. A review of algorithms for filtering the 3D point cloud. Signal Process. Image Commun. 2017, 57, 103–112. [Google Scholar] [CrossRef]
- Richardson, A.D.; Jenkins, J.P.; Braswell, B.H.; Hollinger, D.Y.; Ollinger, S.V.; Smith, M.L. Use of digital webcam images to track spring green-up in a deciduous broadleaf forest. Oecologia 2007, 152, 323–334. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Chen, S.; McDermid, G.J.; Castilla, G.; Linke, J. Measuring vegetation height in linear disturbances in the boreal forest with UAV photogrammetry. Remote Sens. 2017, 9, 1257. [Google Scholar] [CrossRef]
- Lisein, J.; Pierrot-Deseilligny, M.; Bonnet, S.; Lejeune, P. A photogrammetric workflow for the creation of a forest canopy height model from small unmanned aerial system imagery. Forests 2013, 4, 922–944. [Google Scholar] [CrossRef]
- Guimarães, N.; Pádua, L.; Marques, P.; Silva, N.; Peres, E.; Sousa, J.J. Forestry Remote Sensing from Unmanned Aerial Vehicles: A Review Focusing on the Data, Processing and Potentialities. Remote Sens. 2020, 12, 1046. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surovỳ, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Tian, J.; Dai, T.; Li, H.; Liao, C.; Teng, W.; Hu, Q.; Ma, W.; Xu, Y. A novel tree height extraction approach for individual trees by combining TLS and UAV image-based point cloud integration. Forests 2019, 10, 537. [Google Scholar] [CrossRef]
- Jurado, J.M.; Ramos, M.; Enríquez, C.; Feito, F. The Impact of Canopy Reflectance on the 3D Structure of Individual Trees in a Mediterranean Forest. Remote Sens. 2020, 12, 1430. [Google Scholar] [CrossRef]
- Jurado, J.M.; Ortega, L.; Cubillas, J.J.; Feito, F. Multispectral mapping on 3D models and multi-temporal monitoring for individual characterization of olive trees. Remote Sens. 2020, 12, 1106. [Google Scholar] [CrossRef]
- Cao, L.; Liu, H.; Fu, X.; Zhang, Z.; Shen, X.; Ruan, H. Comparison of UAV LiDAR and digital aerial photogrammetry point clouds for estimating forest structural attributes in subtropical planted forests. Forests 2019, 10, 145. [Google Scholar] [CrossRef]
- Comba, L.; Zaman, S.; Biglia, A.; Ricauda, A.D.; Dabbene, F.; Gay, P. Semantic interpretation and complexity reduction of 3D point clouds of vineyards. Biosyst. Eng. 2020, 197, 216–230. [Google Scholar] [CrossRef]
- Magalhães, N. Tratado de Viticultura: A Videira, a Vinha e o Terroir; Publicações Chaves Ferreira Lisboa: Lisboa, Portugal, 2008; 605p, ISBN 9789899820739. [Google Scholar]
- Di Gennaro, S.F.; Matese, A. Evaluation of novel precision viticulture tool for canopy biomass estimation and missing plant detection based on 2.5 D and 3D approaches using RGB images acquired by UAV platform. Plant Methods 2020, 16, 1–12. [Google Scholar] [CrossRef]
- Siebers, M.H.; Edwards, E.J.; Jimenez-Berni, J.A.; Thomas, M.R.; Salim, M.; Walker, R.R. Fast phenomics in vineyards: Development of GRover, the grapevine rover, and LiDAR for assessing grapevine traits in the field. Sensors 2018, 18, 2924. [Google Scholar] [CrossRef] [PubMed]
- Milella, A.; Marani, R.; Petitti, A.; Reina, G. In-field high throughput grapevine phenotyping with a consumer-grade depth camera. Comput. Electron. Agric. 2019, 156, 293–306. [Google Scholar] [CrossRef]
- Mendes, J.; Dos Santos, F.N.; Ferraz, N.; Couto, P.; Morais, R. Vine trunk detector for a reliable robot localization system. In Proceedings of the 2016 International Conference on Autonomous Robot Systems and Competitions (ICARSC), Bragança, Portugal, 4–6 May 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–6. [Google Scholar]






| Vine Row | Number of Grapevines | Missing Grapevines | Row Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Obs. | Est. | OA (%) | Obs. | Est. | OA (%) | Obs. | Est. | OA (%) | |
| 1 | 46 | 43 | 93.5 | 12 | 15 | 75.0 | 58 | 58 | 100.0 |
| 2 | 39 | 37 | 94.9 | 18 | 21 | 83.3 | 57 | 58 | 98.2 |
| 3 | 42 | 45 | 92.9 | 15 | 12 | 80.0 | 57 | 57 | 100.0 |
| 4 | 40 | 46 | 85.0 | 17 | 9 | 52.9 | 57 | 55 | 96.5 |
| 5 | 49 | 50 | 98.0 | 7 | 7 | 100.0 | 56 | 57 | 98.2 |
| Total | 216 | 221 | 97.7 | 69 | 64 | 92.8 | 285 | 285 | 100.0 |
| Vine Row | TP | FP | TN | FN | Precision | Recall | F1score | O.A. (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 39 | 4 | 8 | 7 | 0.91 | 0.85 | 0.88 | 81.0 |
| 2 | 32 | 5 | 16 | 5 | 0.86 | 0.86 | 0.86 | 82.8 |
| 3 | 40 | 5 | 7 | 5 | 0.89 | 0.89 | 0.89 | 82.5 |
| 4 | 42 | 4 | 6 | 3 | 0.91 | 0.93 | 0.92 | 87.3 |
| 5 | 45 | 5 | 4 | 3 | 0.90 | 0.94 | 0.92 | 86.0 |
| Total | 198 | 23 | 41 | 23 | 0.90 | 0.90 | 0.90 | 83.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurado, J.M.; Pádua, L.; Feito, F.R.; Sousa, J.J. Automatic Grapevine Trunk Detection on UAV-Based Point Cloud. Remote Sens. 2020, 12, 3043. https://doi.org/10.3390/rs12183043
Jurado JM, Pádua L, Feito FR, Sousa JJ. Automatic Grapevine Trunk Detection on UAV-Based Point Cloud. Remote Sensing. 2020; 12(18):3043. https://doi.org/10.3390/rs12183043
Chicago/Turabian StyleJurado, Juan M., Luís Pádua, Francisco R. Feito, and Joaquim J. Sousa. 2020. "Automatic Grapevine Trunk Detection on UAV-Based Point Cloud" Remote Sensing 12, no. 18: 3043. https://doi.org/10.3390/rs12183043
APA StyleJurado, J. M., Pádua, L., Feito, F. R., & Sousa, J. J. (2020). Automatic Grapevine Trunk Detection on UAV-Based Point Cloud. Remote Sensing, 12(18), 3043. https://doi.org/10.3390/rs12183043