Shape Adaptive Neighborhood Information-Based Semi-Supervised Learning for Hyperspectral Image Classification
Abstract
1. Introduction
- (1)
- Compared with the fixed size window-based and superpixel based spatial neighborhood information that most existing research methods adopted, the SA based method can represent the neighborhood information in a pointwise adaptive manner. We exploit the SANI in our proposed SSL method in order to select new unlabeled samples, which can make the training samples more representative and valuable and, thus, achieve better classification accuracy.
- (2)
- The unlabeled samples’ selection makes full use of SANI of the whole image which avoids the restriction of limited labeled samples. In addition, an adaptive interval strategy is proposed in our method to ensure the informativeness of unlabeled samples selected from unlabeled samples’ neighborhoods. The proposed strategy utilizes the uncertainty information of available training samples to select more diverse unlabeled samples.
2. Methodology
2.1. Spatial Information Extraction
2.1.1. Extended Morphological Attribute Profile
2.1.2. Shape Adaptive Neighborhood Information
2.2. Classification Method
2.2.1. Sparse Multinomial Logistic Regression
2.2.2. Adaptive Sparse Representation
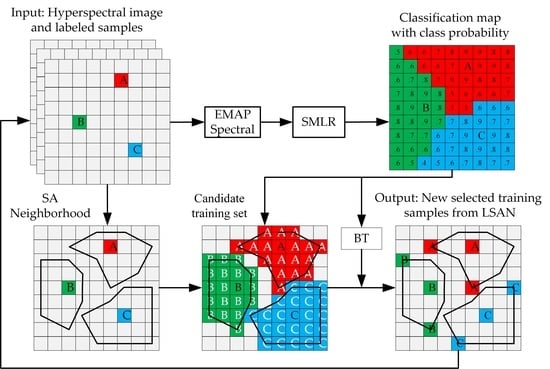
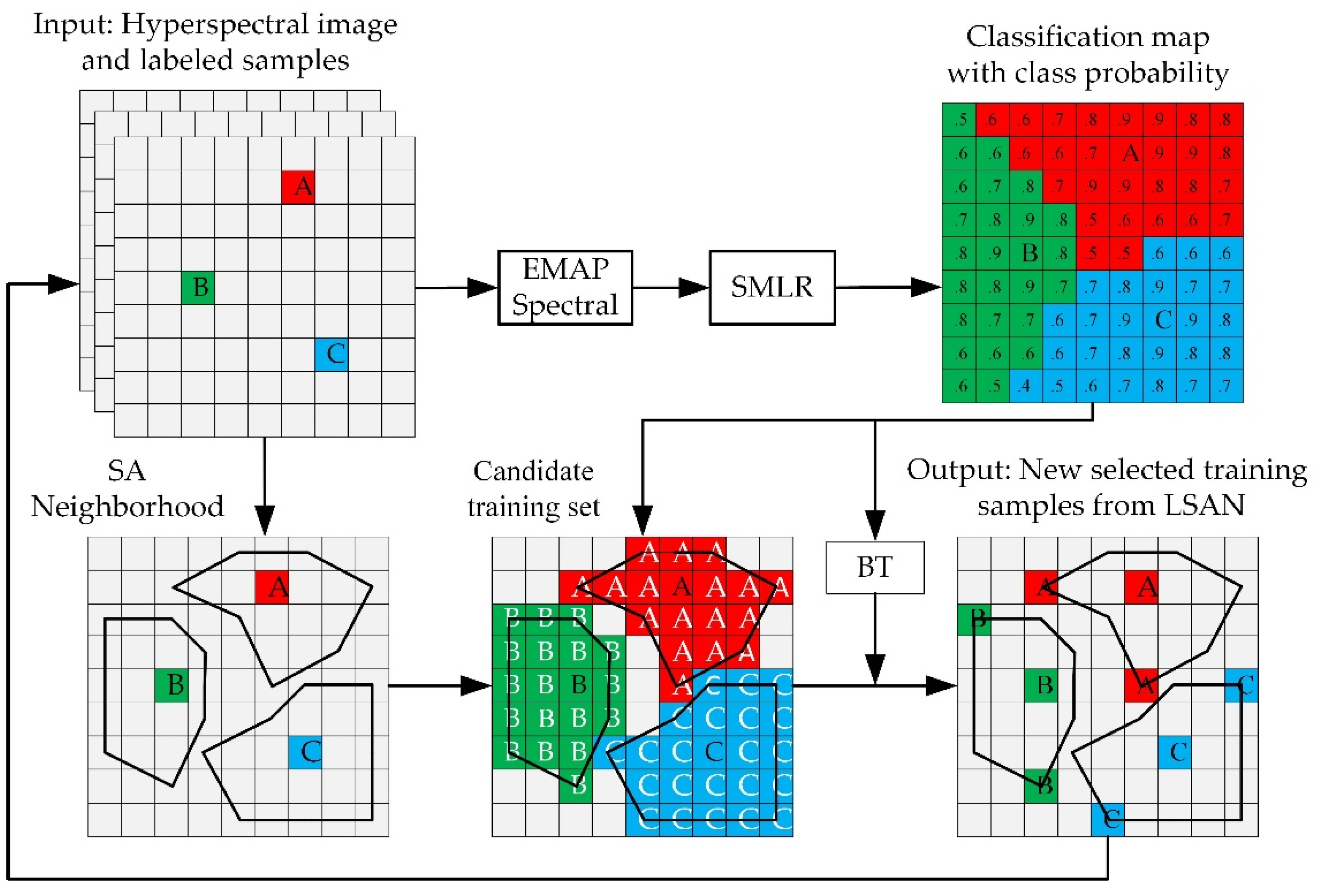
2.3. Unlabeled Samples Selection Method in Proposed SANI-SSL
2.3.1. Selecting Unlabeled Samples from LSAN
- (1)
- The construction of candidate training samples
- (2)
- Active learning
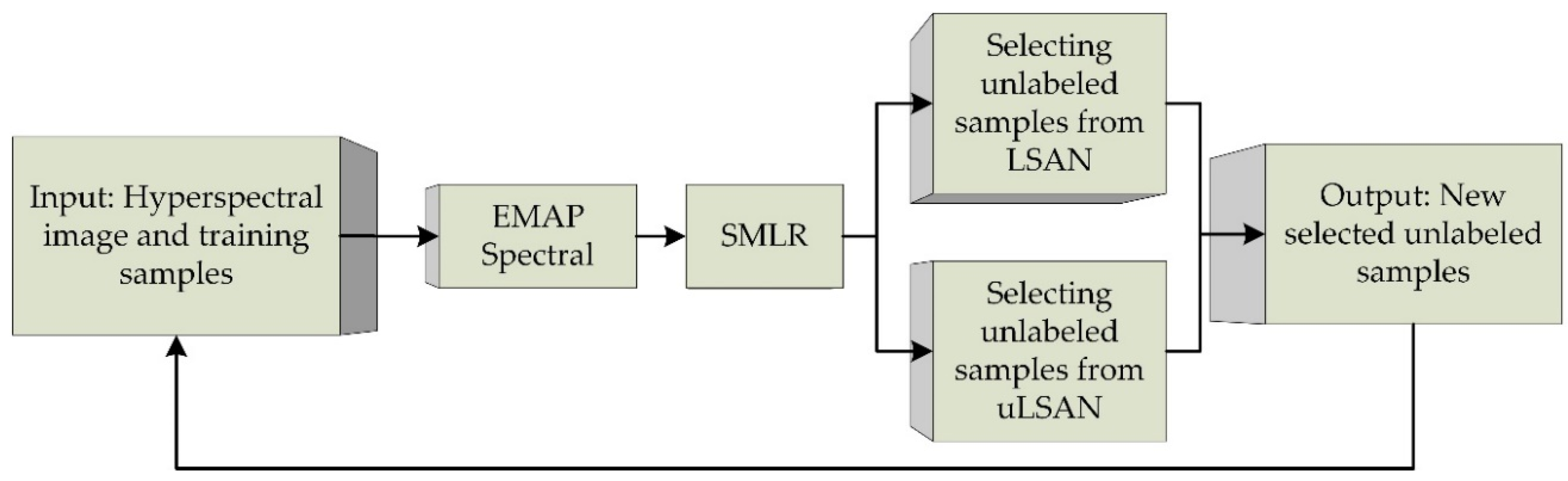
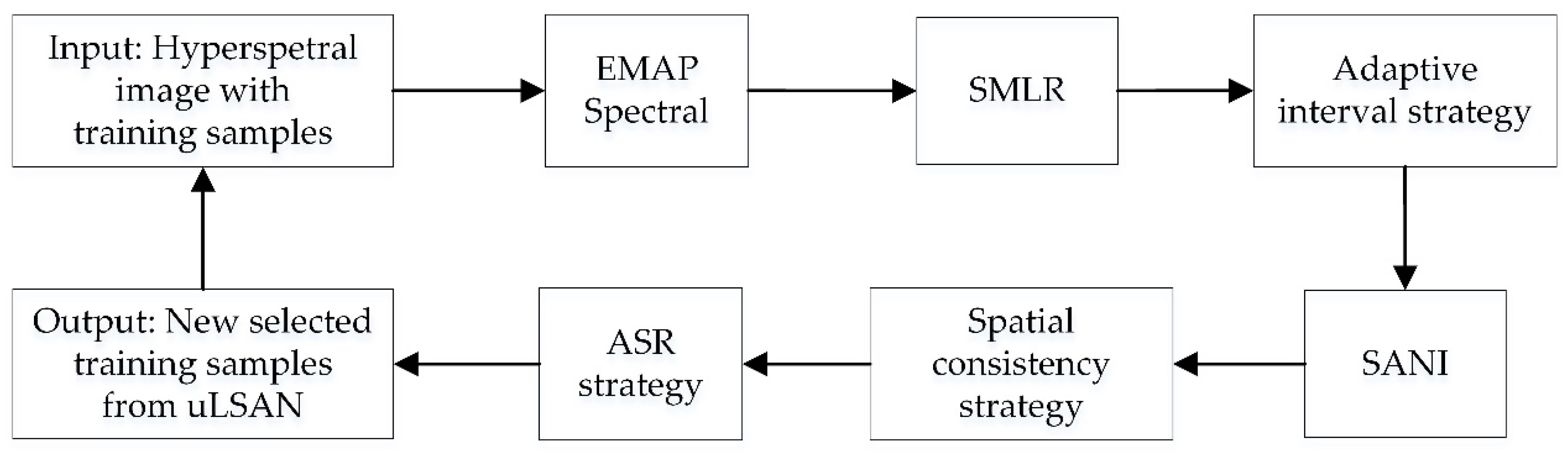
2.3.2. Selecting Unlabeled Samples from uLSAN
- (1)
- Strategy to ensure informativeness
- (2)
- Strategies to ensure confidence
3. Experiments
3.1. Datasets Used in the Experiments
- (1)
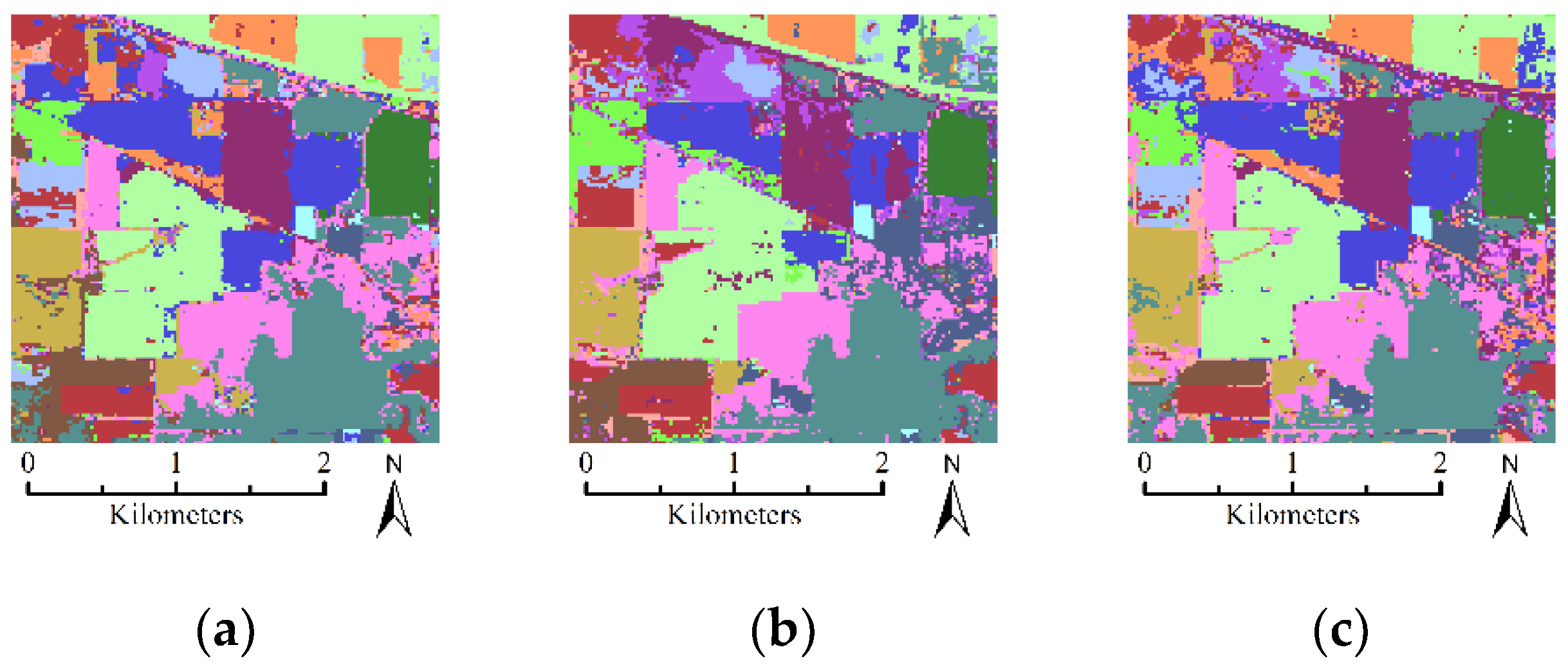
- The first hyperspectral dataset used in this paper was acquired by the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) sensor over a mixed agricultural/forest area in northwestern Indiana in 1992. The dataset contains 145 lines by 145 samples with a spatial resolution of 20 m. It is composed of 224 spectral reflectance bands in the wavelength range from 0.4 um–2.5 um at 10 nm intervals. After an initial screening, the spectral bands were reduced to 200 by removing the bands that cover the region of water absorption. The available ground-truth map contains 10,366 labeled samples with 16 different classes. Figure 5 shows the false color composition image of Indian Pines datasets and ground-truth map.
- (2)
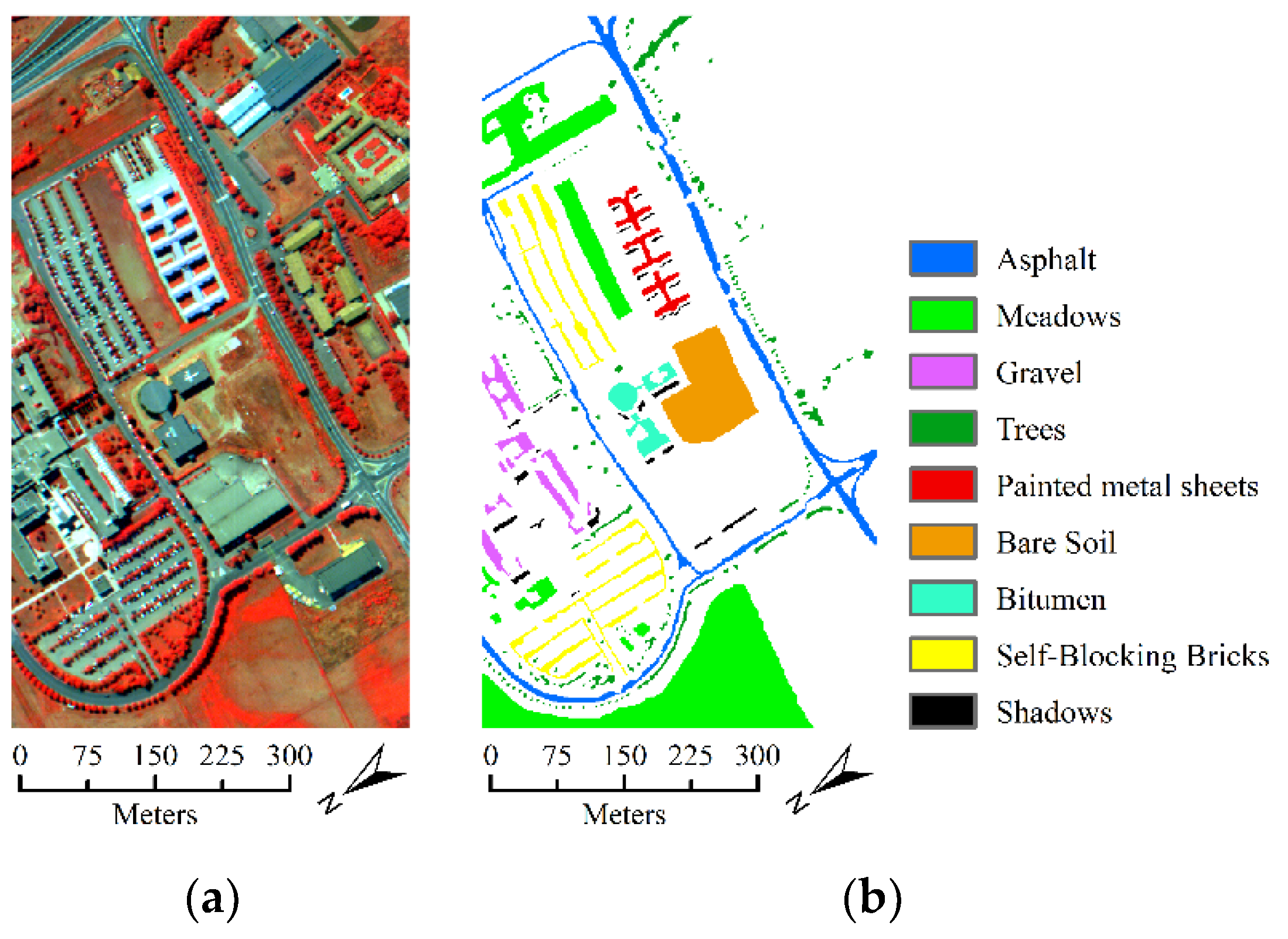
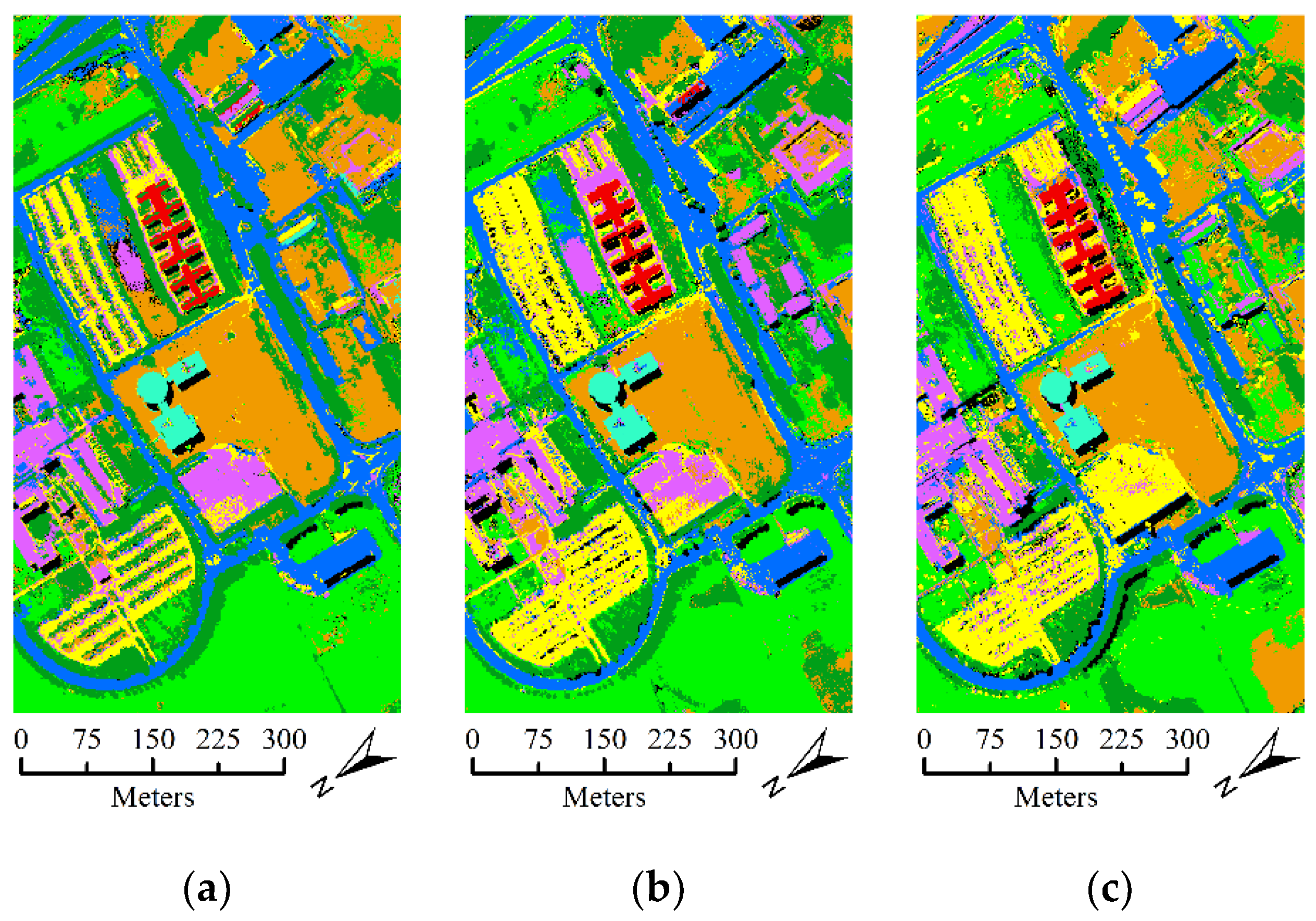
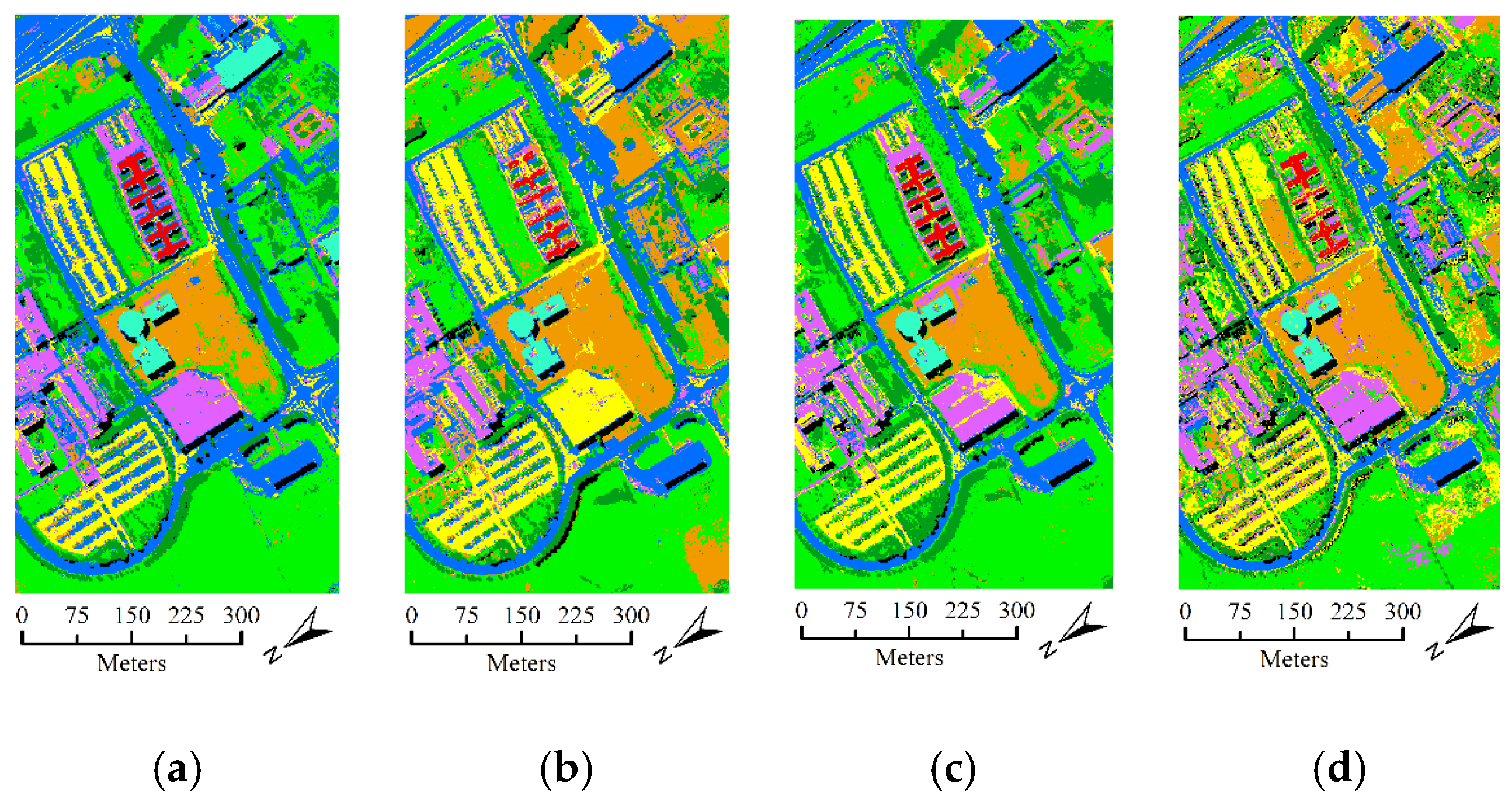
- The second hyperspectral dataset was acquired by the ROSIS optical sensor over the urban area of the Pavia University, Italy. The dataset contains 610 lines by 310 samples with a spatial resolution of 1.3 m. It comprises 115 spectral channels in the wavelength range of 0.43 um–0.68 µm and 103 spectral bands are used in the experiment after the noisy and water absorption channels were removed. For this dataset, the available ground-truth map contains 42,776 labeled samples with nine classes. Figure 6 shows the false color composition image of Pavia University data and ground-truth map.
- (3)
- The third hyperspectral dataset was acquired by the AVIRIS sensor over Salinas Valley, California in 1998. The dataset contains 512 lines by 217 samples with a spatial resolution of 3.7 m. It is composed of 224 spectral bands in the wavelength range from 0.4 um–2.5 um, and 204 bands are used in the experiment after noisy and water absorption channels were removed. There are 54,129 samples in total available in the scene which contains 16 classes. Figure 7 shows the false color composition image of Salinas Valley dataset and ground-truth map.
3.2. Experimental Setup
- (1)
- Spatial information extraction: in this paper, the parameters about EMAP are determined with reference to related work in [20,47]. The thresholds of area attribute are determined according to the scale of the objects, the thresholds of moment of inertia, and standard deviation are determined according to geometry of the objects and the homogeneity of the intensity values of the pixels. Although finer threshold division is conducive to extracting detailed spatial information, the amount of calculation will increase accordingly. For the Indian Pines dataset, the EMAP are extracted while using the attributes area a and moment of inertia i with thresholds ; for the Pavia University dataset, the EMAP are extracted using attributes area a and moment of inertia with thresholds ; and, for Salinas Valley dataset, the EMAP are extracted using attributes area and standard deviation with thresholds . The SANI is extracted with the predefined candidate length set for the three datasets.
- (2)
- Classifier parameter: the kernel width , and the degree of sparsity for SMLR.
- (3)
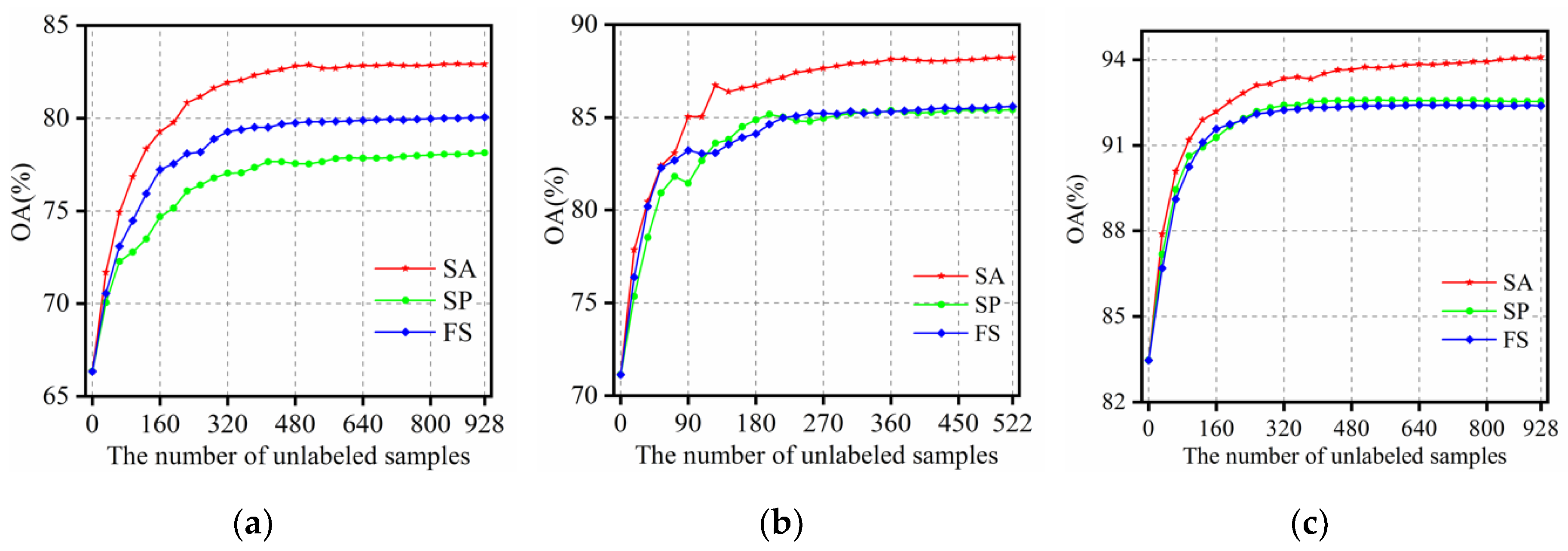
- Training sets: to evaluate performance of the proposed SANI-SSL method for scenarios with limited labeled samples, only five truly labeled samples per class were randomly chosen from the ground truth for all of the methods in this paper. The number of unlabeled samples is set to be , , for the three datasets, respectively.
- (4)
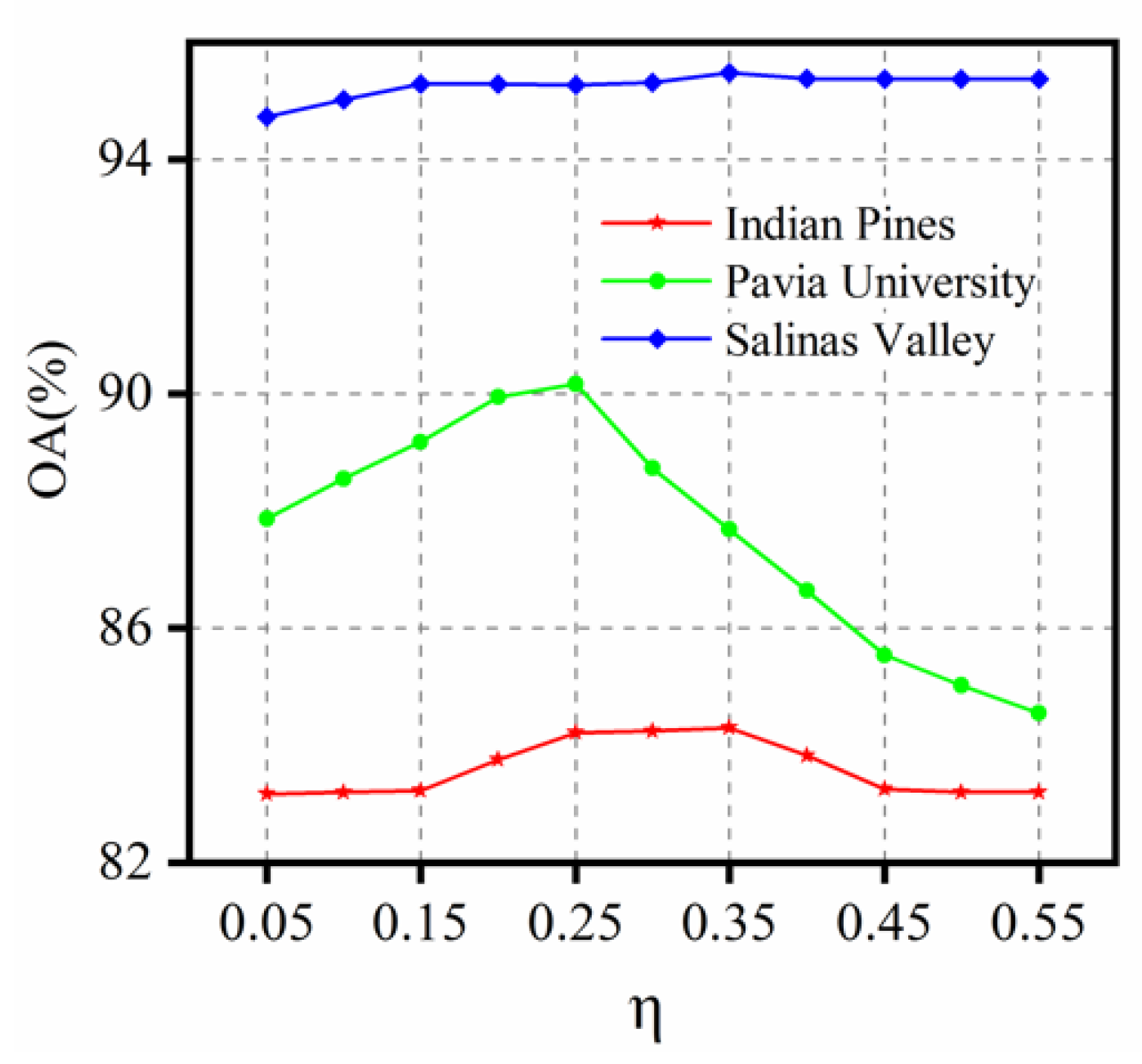
- Other settings: the parameter , which represents the length of the uncertainty interval is intuitively set to 0.25 because we have empirically found that this parameter setting provides better performance and more detailed discussion about is shown at 3.4.3. The SSL process is executed for 30 iterations. In order to evaluate the performance of our SANI-SSL method quantitatively, the overall accuracy (OA), average accuracy (AA), class-specific accuracies (CA), and kappa coefficient are computed by averaging ten Monte Carlo runs that correspond to independent initial labeled sets.
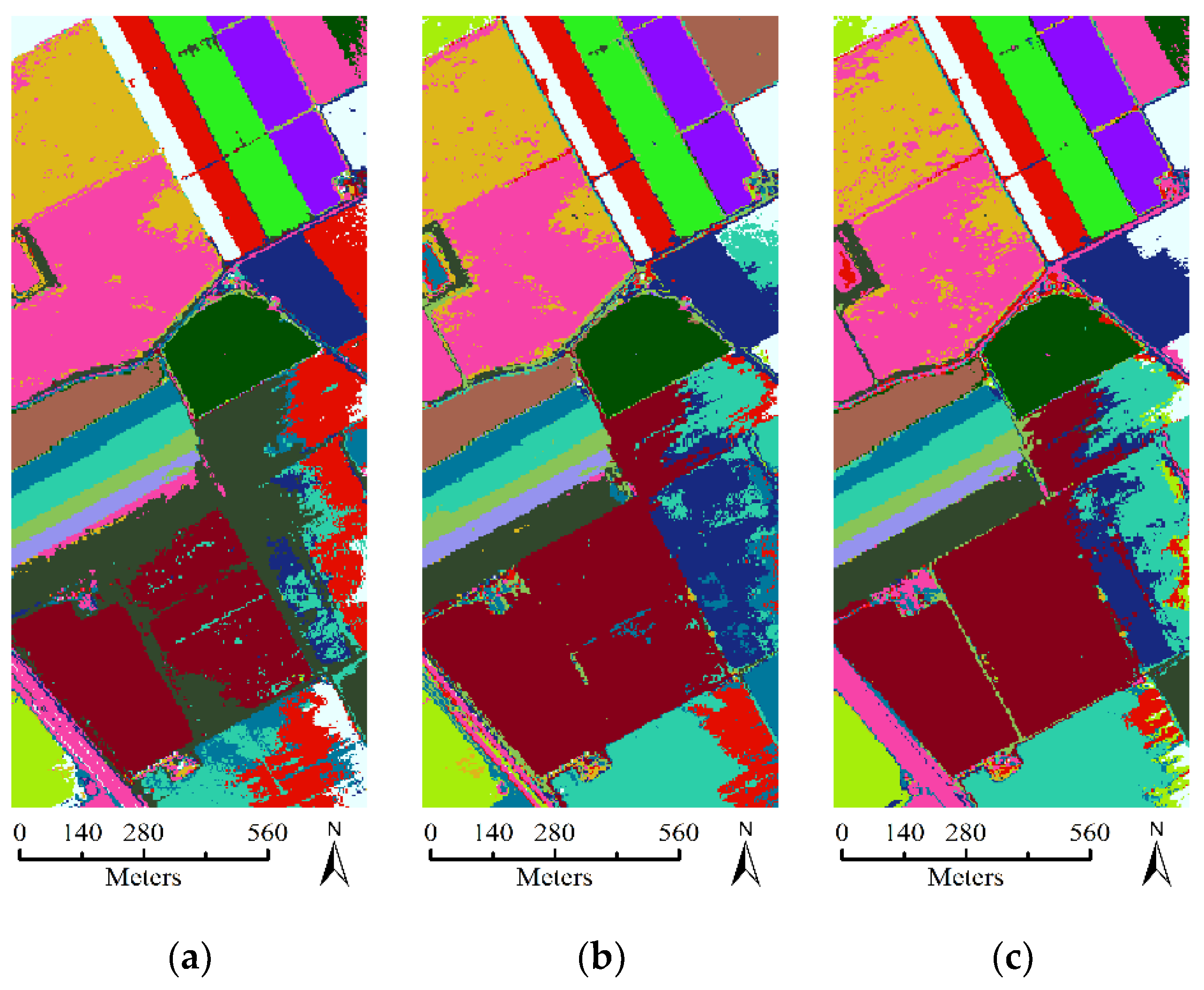
3.3. Experimental Results with Unlabeled Samples Selected from Only LSAN
3.4. Experimental Results with Unlabeled Samples Selected from LSAN and uLSAN
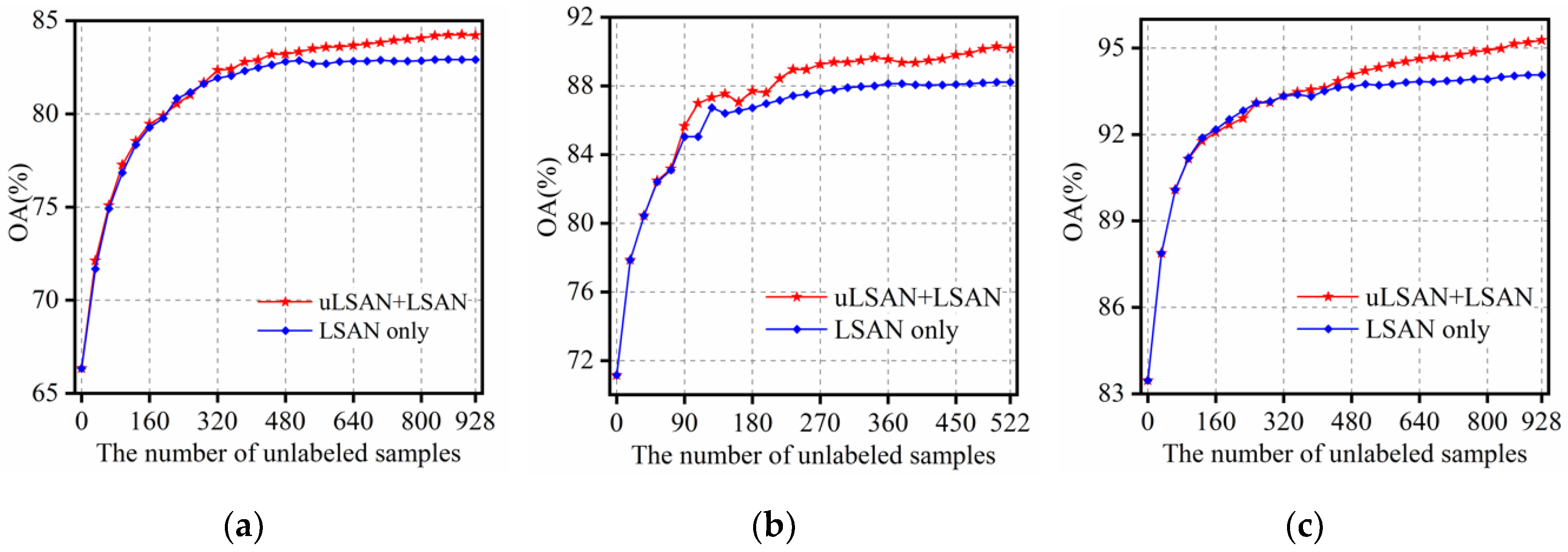
3.4.1. The Influence of Unlabeled Samples from uLSAN
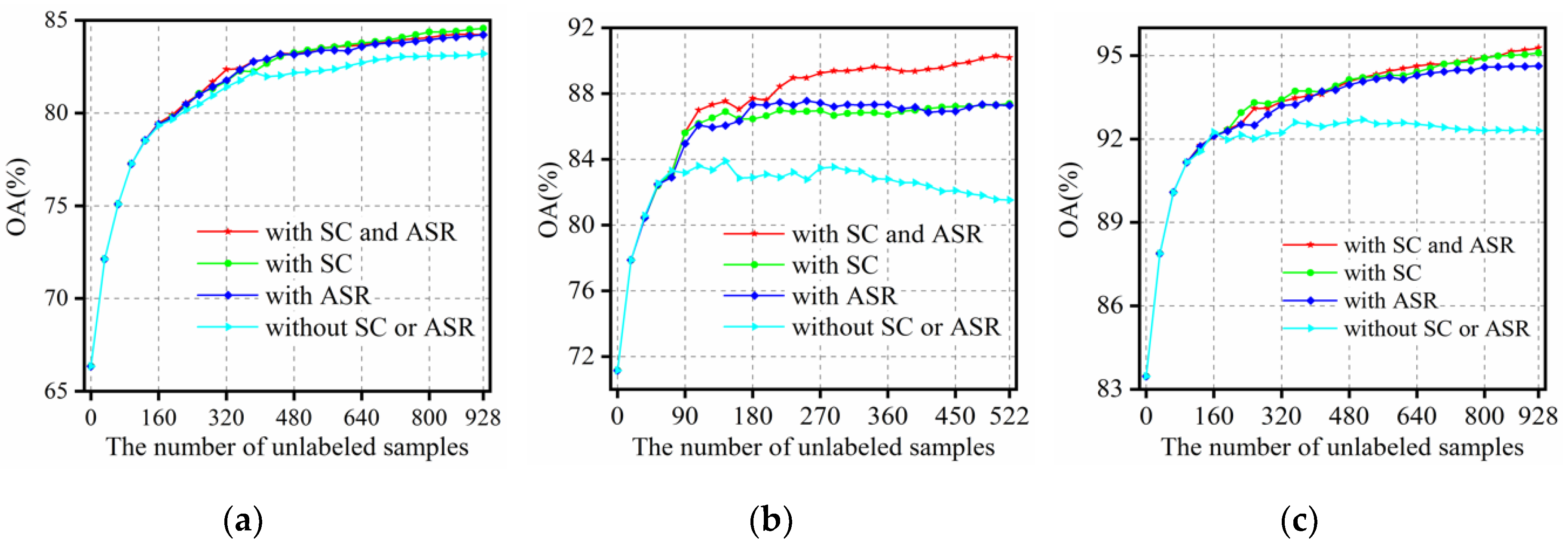
3.4.2. The Influence of the Strategy to Ensure Confidence
3.4.3. The Influence of the Strategy to Ensure Informativeness
4. Discussion
- (1)
- The comparison experiment between the SA-, SP-, and FS- based SSL methods shows that our proposed SA-based method performs better than the other two methods. This finding can be explained by the fact that the SA spatial information is beneficial to construct a more representative neighborhood for every pixel. In homogeneous regions of the image, SA has the advantage of representing the neighborhood information comprehensively, while in the heterogeneous region, SA has the advantage of more accurately representing the neighborhood information.
- (2)
- The comparison experiment between the selection of unlabeled samples from LSAN and uLSAN shows that the uLSAN has great potential in finding valuable samples. This potential could be attributed to two reasons. First, when compared with the limited and high-related information in LSAN, the uLSAN contains abundant undiscovered and unused information, which is helpful in improving the classifier. Second, taking the characteristics of both LSAN and uLSAN into full consideration, the unlabeled samples are selected from these two regions at generally appropriate time by adjusting the number of unlabeled samples from both regions.
- (3)
- The influence of different strategies confidence to ensure on the classification accuracy is analyzed in Section 3.4.2, and the result shows that the two strategies can ensure that the selected samples are assigned by correct labels, and the SC strategy shows better performance in computational efficiency. The influence of the informativeness parameter on the classification accuracy is analyzed by conducting experiments using different value of , and the best performance is acquired by setting to 0.25 according to the results of three datasets. As for the difference in performance of the three datasets, the main reason is that the spatial characteristics in the datasets are different. The main ground objects of Indian Pines and Salinas Valley datasets are cropland, which have larger spatial scale and regular distribution. However, for the urban scene in Pavia University dataset, the spatial distribution is more complicated; therefore, more stringent requirements on the strategies need to be satisfied in order to achieve satisfactory results.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Luo, Y.H.; Tao, Z.P.; Ke, G.; Wang, M.Z. The Application Research of Hyperspectral Remote Sensing Technology in Tailing Mine Environment Pollution Supervise Management. In Proceedings of the 2012 2nd International Conference on Remote Sensing, Environment and Transportation Engineering, Nanjing, China, 1–3 June 2012; pp. 1–4. [Google Scholar]
- Tong, Q.; Zhang, B.; Zheng, L. Hyperspectral Remote Sensing: The Principle, Technology and Application; Higher Education Press: Beijing, China, 2006. [Google Scholar]
- Majdar, R.S.; Ghassemian, H. A probabilistic SVM approach for hyperspectral image classification using spectral and texture features. Int. J. Remote Sens. 2017, 38, 4265–4284. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Du, P.; Xia, J.; Xue, Z.; Tan, K.; Su, H.; Bao, R. Review of hyperspectral remote sensing image classification. J. Remote Sens. 2016, 20, 236–256. [Google Scholar]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree Species Classification Using Hyperspectral Imagery: A Comparison of Two Classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Yu, H.; Gao, L.; Li, J.; Li, S.S.; Zhang, B.; Benediktsson, J.A. Spectral-Spatial Hyperspectral Image Classification Using Subspace-Based Support Vector Machines and Adaptive Markov Random Fields. Remote Sens. 2016, 8, 355. [Google Scholar] [CrossRef]
- Cao, F.; Yang, Z.; Ren, J.; Ling, W.K.; Zhao, H.; Marshall, S. Extreme Sparse Multinomial Logistic Regression: A Fast and Robust Framework for Hyperspectral Image Classification. Remote Sens. 2017, 9, 1255. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Spectral-Spatial Hyperspectral Image Segmentation Using Subspace Multinomial Logistic Regression and Markov Random Fields. IEEE Trans. Geosci. Remote Sens. 2012, 50, 809–823. [Google Scholar] [CrossRef]
- Li, J.; Gamba, P.; Plaza, A. A Novel Semi-Supervised Method for Obtaining Finer Resolution Urban Extents Exploiting Coarser Resolution Maps. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 4276–4287. [Google Scholar] [CrossRef]
- Chen, C.; Li, W.; Su, H.; Liu, K. Spectral-Spatial Classification of Hyperspectral Image Based on Kernel Extreme Learning Machine. Remote Sens. 2014, 6, 5795–5814. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Shen, Q. Spectral-Spatial Classification of Hyperspectral Imagery with 3D Convolutional Neural Network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Mirzapour, F.; Ghassemian, H. Improving hyperspectral image classification by combining spectral, texture, and shape features. Int. J. Remote Sens. 2015, 36, 1070–1096. [Google Scholar] [CrossRef]
- Huang, X.; Liu, X.; Zhang, L. A Multichannel Gray Level Co-Occurrence Matrix for Multi/Hyperspectral Image Texture Representation. Remote Sens. 2014, 6, 8424–8445. [Google Scholar] [CrossRef]
- Li, J.; Xi, B.; Li, Y.; Du, Q.; Wang, K. Hyperspectral Classification Based on Texture Feature Enhancement and Deep Belief Networks. Remote Sens. 2018, 10, 396. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Song, H. A Spectral-Texture Kernel-Based Classification Method for Hyperspectral Images. Remote Sens. 2016, 8, 919. [Google Scholar] [CrossRef]
- Zhang, B.; Li, S.; Jia, X.; Gao, L.; Peng, M. Adaptive Markov Random Field Approach for Classification of Hyperspectral Imagery. IEEE Geosci. Remote Sens. Lett. 2011, 8, 973–977. [Google Scholar] [CrossRef]
- Andrejchenko, V.; Liao, W.; Philips, W.; Scheunders, P. Decision Fusion Framework for Hyperspectral Image Classification Based on Markov and Conditional Random Fields. Remote Sens. 2019, 11, 624. [Google Scholar] [CrossRef]
- Cao, X.; Xu, Z.; Meng, D. Spectral-Spatial Hyperspectral Image Classification via Robust Low-Rank Feature Extraction and Markov Random Field. Remote Sens. 2019, 11, 1565. [Google Scholar] [CrossRef]
- Dalla Mura, M.; Benediktsson, J.A.; Waske, B.; Bruzzone, L. Extended profiles with morphological attribute filters for the analysis of hyperspectral data. Int. J. Remote Sens. 2010, 31, 5975–5991. [Google Scholar] [CrossRef]
- Liang, H.; Li, Q. Hyperspectral Imagery Classification Using Sparse Representations of Convolutional Neural Network Features. Remote Sens. 2016, 8, 99. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, S.; Liu, Q.; Hang, R.; Liu, G. Hypergraph Embedding for Spatial-Spectral Joint Feature Extraction in Hyperspectral Images. Remote Sens. 2017, 9, 506–519. [Google Scholar]
- Andekah, Z.A.; Naderan, M.; Akbarizadeh, G.; IEEE. Semi-Supervised Hyperspectral Image Classification using Spatial-Spectral Features and Superpixel-Based Sparse Codes. In Proceedings of the 25th Iranian Conference on Electrical Engineering, Teheran, Iran, 2–4 May 2017; pp. 2229–2234. [Google Scholar]
- Fu, Q.; Yu, X.; Zhang, P.; Wei, X. Semi-supervised ELM combined with spectral-spatial featuresfor hyperspectral imagery classification. J. Huazhong Univ. Sci. Technol. Nat. Sci. 2017, 45, 89–93. [Google Scholar]
- He, Z.; Liu, H.; Wang, Y.; Hu, J. Generative Adversarial Networks-Based Semi-Supervised Learning for Hyperspectral Image Classification. Remote Sens. 2017, 9, 1042. [Google Scholar] [CrossRef]
- Tan, K.; Li, E.; Du, Q.; Du, P. An efficient semi-supervised classification approach for hyperspectral imagery. ISPRS J. Photogramm. Remote Sens. 2014, 97, 36–45. [Google Scholar] [CrossRef]
- Dopido, I.; Li, J.; Marpu, P.R.; Plaza, A.; Bioucas Dias, J.M.; Benediktsson, J.A. Semisupervised Self-Learning for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2013, 51, 4032–4044. [Google Scholar] [CrossRef]
- Wu, Y.; Mu, G.F.; Qin, C.; Miao, Q.G.; Ma, W.P.; Zhang, X.R. Semi-Supervised Hyperspectral Image Classification via Spatial-Regulated Self-Training. Remote Sens. 2020, 12, 159. [Google Scholar] [CrossRef]
- Wang, L.; Hao, S.; Wang, Q.; Wang, Y. Semi-supervised classification for hyperspectral imagery based on spatial-spectral Label Propagation. ISPRS J. Photogramm. Remote Sens. 2014, 97, 123–137. [Google Scholar] [CrossRef]
- Liu, C.; Li, J.; He, L. Superpixel-Based Semisupervised Active Learning for Hyperspectral Image Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 357–370. [Google Scholar] [CrossRef]
- Balasubramaniam, R.; Namboodiri, S.; Nidamanuri, R.R.; Gorthi, R.K.S.S. Active Learning-Based Optimized Training Library Generation for Object-Oriented Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 575–585. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, F.; Yan, F. Novel Semi-Supervised Hyperspectral Image Classification Based on a Superpixel Graph and Discrete Potential Method. Remote Sens. 2020, 12, 1528–1547. [Google Scholar] [CrossRef]
- Tan, K.; Hu, J.; Li, J.; Du, P.J. A novel semi-supervised hyperspectral image classification approach based on spatial neighborhood information and classifier combination. ISPRS J. Photogramm. Remote Sens. 2015, 105, 19–29. [Google Scholar] [CrossRef]
- Luo, T.; Kramer, K.; Goldgof, D.B.; Hall, L.O.; Samson, S.; Remsen, A.; Hopkins, T. Active learning to recognize multiple types of plankton. J. Mach. Learn. Res. 2005, 6, 589–613. [Google Scholar]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Semisupervised Hyperspectral Image Segmentation Using Multinomial Logistic Regression with Active Learning. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4085–4098. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Hyperspectral Image Segmentation Using a New Bayesian Approach with Active Learning. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3947–3960. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Spectral-Spatial Classification of Hyperspectral Data Using Loopy Belief Propagation and Active Learning. IEEE Trans. Geosci. Remote Sens. 2013, 51, 844–856. [Google Scholar] [CrossRef]
- Wang, L.; Hao, S.; Wang, Y.; Lin, Y.; Wang, Q. Spatial-Spectral Information-Based Semisupervised Classification Algorithm for Hyperspectral Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3577–3585. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, X.; Huang, X. An Active Relearning Framework for Remote Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3468–3486. [Google Scholar] [CrossRef]
- Shu, W.; Liu, P.; He, G.; Wang, G. Hyperspectral Image Classification Using Spectral-Spatial Features with Informative Samples. IEEE Access 2019, 7, 20869–20878. [Google Scholar] [CrossRef]
- Pesaresi, M.; Benediktsson, J.A. A new approach for the morphological segmentation of high-resolution satellite imagery. IEEE Trans. Geosci. Remote Sens. 2001, 39, 309–320. [Google Scholar] [CrossRef]
- Foi, A.; Katkovnik, V.; Egiazarian, K. Pointwise shape-adaptive DCT for high-quality denoising and deblocking of grayscale and color images. IEEE Trans. Image Process. 2007, 16, 1395–1411. [Google Scholar] [CrossRef]
- Yang, J.; Qian, J. Joint Collaborative Representation with Shape Adaptive Region and Locally Adaptive Dictionary for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2020, 17, 671–675. [Google Scholar] [CrossRef]
- Xue, Z.; Du, P.; Li, J.; Su, H. Simultaneous Sparse Graph Embedding for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6114–6133. [Google Scholar] [CrossRef]
- Du, P.; Xue, Z.; Li, J.; Plaza, A. Learning Discriminative Sparse Representations for Hyperspectral Image Classification. IEEE J. Sel. Top. Signal. Process. 2015, 9, 1089–1104. [Google Scholar] [CrossRef]
- Kayabol, K. Approximate Sparse Multinomial Logistic Regression for Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 490–493. [Google Scholar] [CrossRef]
- Li, J.; Huang, X.; Gamba, P.; Bioucas-Dias, J.M.; Zhang, L.; Benediktsson, J.A.; Plaza, A. Multiple Feature Learning for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1592–1606. [Google Scholar] [CrossRef]
- Dopido, I.; Li, J.; Plaza, A.; Bioucas-Dias, J.M.; IEEE. A New Semi-supervised Approach for Hyperspectral Image Classification with Different Active Learnings Strategies. In Proceedings of the 2012 4th Workshop on Hyperspectral Image and Signal Processing, Shangai, China, 4–7 June 2012. [Google Scholar]
- Böhning, D. Multinomial logistic regression algorithm. Ann. Inst. Stat. Math. 1992, 44, 197–200. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Bruzzone, L. Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1351–1362. [Google Scholar] [CrossRef]
- Bioucasdias, B.J.; Figueiredo, M. Logistic Regression via Variable Splitting and Augmented Lagrangian Tools; Technical Report; Instituto Superior Técnico: Lisboa, Portugal, 2009. [Google Scholar]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spectral-Spatial Hyperspectral Image Classification via Multiscale Adaptive Sparse Representation. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7738–7749. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Liu, M.Y.; Tuzel, O.; Ramalingam, S.; Chellappa, R.; IEEE. Entropy Rate Superpixel Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 20–25 June 2011. [Google Scholar]
- Tan, K.; Zhu, J.; Du, Q.; Wu, L.; Du, P. A Novel Tri-Training Technique for Semi-Supervised Classification of Hyperspectral Images Based on Diversity Measurement. Remote Sens. 2016, 8, 749. [Google Scholar] [CrossRef]




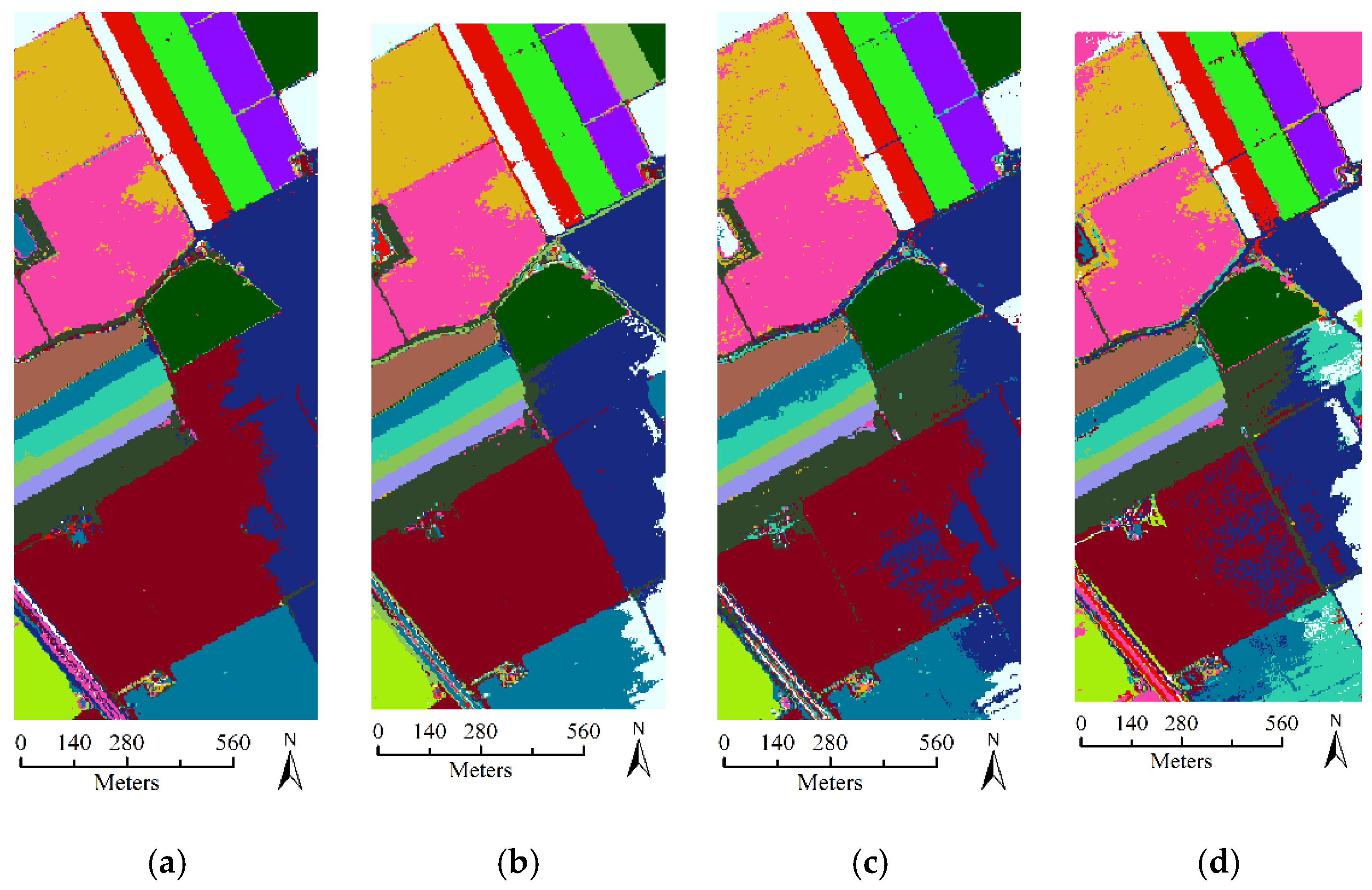
| Class | Number of Testing Samples | SA | SP | FS |
|---|---|---|---|---|
| Alfalfa | 49 | 91.63 ± (5.66) | 91.02 ± (5.49) | 92.45 ± (6.13) |
| Corn-Notill | 1429 | 69.66 ± (10.72) | 64.85 ± (8.90) | 68.01 ± (7.97) |
| Corn-Mintill | 829 | 72.98 ± (11.77) | 62.36 ± (11.51) | 65.11 ± (8.32) |
| Corn | 229 | 79.87 ± (11.77) | 93.49 ± (6.42) | 87.60 ± (7.80) |
| Grass-Pasture | 492 | 81.36 ± (6.16) | 78.01 ± (5.57) | 80.08 ± (5.53) |
| Grass-Trees | 742 | 96.73 ± (1.38) | 87.80 ± (8.78) | 95.22 ± (3.72) |
| Grass-Pasture-Mowed | 21 | 96.67 ± (3.05) | 98.57 ± (2.18) | 99.05 ± (1.90) |
| Hay-Windrowed | 484 | 100.00 ± (0.00) | 99.77 ± (0.22) | 99.67 ± (0.28) |
| Oats | 15 | 100.00 ± (0.00) | 100.00 ± (0.00) | 100.00 ± (0.00) |
| Soybean-Notill | 963 | 81.73 ± (5.00) | 80.82 ± (5.08) | 80.08 ± (4.60) |
| Soybean-Mintill | 2463 | 81.12 ± (6.08) | 75.54 ± (8.95) | 75.28 ± (10.36) |
| Soybean-Clean | 609 | 78.83 ± (13.92) | 60.31 ± (18.88) | 70.89 ± (17.34) |
| Wheat | 207 | 99.52 ± (2.22) | 99.23 ± (0.24) | 99.52 ± (0.22) |
| Woods | 1289 | 91.61 ± (9.32) | 89.34 ± (10.42) | 91.39 ± (8.91) |
| Buildings-Grass-Trees-Drives | 375 | 85.79 ± (10.16) | 87.76 ± (10.19) | 87.44 ± (10.34) |
| Stone-Steel-Towers | 90 | 97.22 ± (2.12) | 96.44 ± (4.38) | 98.78 ± (1.26) |
| OA | 82.90 ± (2.37) | 78.12 ± (3.39) | 80.05 ± (3.35) | |
| AA | 87.79 ± (1.80) | 85.33 ± (2.02) | 86.91 ± (1.79) | |
| Kappa | 80.67 ± (2.63) | 75.46 ± (3.69) | 77.59 ± (3.66) | |
| Time(s) | 242 | 236 | 237 | |
| Class | Number of Testing Samples | SA | SP | FS |
|---|---|---|---|---|
| Asphalt | 6626 | 92.79 ± (3.26) | 93.62 ± (6.50) | 96.02 ± (2.83) |
| Meadows | 18,644 | 81.53 ± (6.82) | 75.95 ± (6.35) | 73.91 ± (6.67) |
| Gravel | 2094 | 92.95 ± (7.12) | 90.21 ± (8.08) | 91.63 ± (4.71) |
| Trees | 3059 | 86.54 ± (6.14) | 86.90 ± (5.07) | 91.01 ± (5.07) |
| Painted Metal Sheets | 1340 | 94.31 ± (6.47) | 97.49 ± (3.71) | 98.07 ± (2.44) |
| Bare Soil | 5024 | 93.64 ± (5.09) | 92.87 ± (5.56) | 94.22 ± (5.10) |
| Bitumen | 1325 | 99.88 ± (0.08) | 99.55 ± (0.66) | 99.71 ± (0.35) |
| Self-Blocking Bricks | 3677 | 96.32 ± (2.90) | 96.69 ± (2.48) | 96.96 ± (3.03) |
| Shadows | 942 | 97.26 ± (4.28) | 78.94 ± (12.15) | 84.21 ± (11.38) |
| OA | 88.21 ± (2.55) | 85.42 ± (2.45) | 85.59 ± (2.80) | |
| AA | 92.80 ± (1.93) | 90.25 ± (1.37) | 91.75 ± (1.76) | |
| Kappa | 84.92 ± (3.07) | 81.59 ± (2.89) | 81.85 ± (3.33) | |
| Time(s) | 267 | 263 | 258 | |
| Class | Number of Testing Samples | SA | SP | FS |
|---|---|---|---|---|
| Brocoli_Green_Weeds_1 | 2004 | 99.89 ± (0.14) | 99.50 ± (0.46) | 99.31 ± (0.71) |
| Brocoli_Green_Weeds_2 | 3721 | 99.29 ± (0.26) | 99.38 ± (0.28) | 99.44 ± (0.26) |
| Fallow | 1971 | 93.28 ± (15.64) | 92.57 ± (16.59) | 93.19 ± (16.38) |
| Fallow_Rough_Plow | 1389 | 97.27 ± (4.41) | 98.60 ± (2.39) | 99.25 ± (0.90) |
| Fallow_Smooth | 2673 | 92.79 ± (4.81) | 97.01 ± (2.01) | 97.34 ± (0.94) |
| Stubble | 3954 | 98.21 ± (1.65) | 96.46 ± (2.22) | 96.78 ± (2.52) |
| Celery | 3574 | 98.82 ± (0.70) | 99.39 ± (0.30) | 99.39 ± (0.31) |
| Grapes_Untrained | 11,266 | 84.98 ± (3.21) | 78.83 ± (6.01) | 76.95 ± (8.55) |
| Soil_Vinyard_Develop | 6198 | 99.75 ± (0.32) | 99.07 ± (1.29) | 99.65 ± (0.61) |
| Corn_Senesced_Green_Weeds | 3273 | 96.02 ± (2.62) | 92.62 ± (7.43) | 92.06 ± (7.96) |
| Lettuce_Romaine_4 wk | 1063 | 95.50 ± (12.18) | 97.54 ± (1.60) | 97.48 ± (1.47) |
| Lettuce_Romaine_5 wk | 1922 | 96.15 ± (5.42) | 97.30 ± (6.42) | 99.99 ± (0.02) |
| Lettuce_Romaine_6 wk | 911 | 99.08 ± (0.50) | 99.13 ± (0.97) | 99.09 ± (0.74) |
| Lettuce_Romaine_7 wk | 1065 | 93.05 ± (7.05) | 89.92 ± (7.10) | 92.08 ± (6.32) |
| Vinyard_Untrained | 7263 | 91.91 ± (5.61) | 91.01 ± (4.46) | 91.47 ± (4.11) |
| Vinyard_Vertical_Trellis | 1802 | 96.03 ± (2.33) | 96.45 ± (2.97) | 94.51 ± (2.88) |
| OA | 94.07 ± (1.40) | 92.53± (1.78) | 92.38 ± (2.07) | |
| AA | 95.75 ± (1.45) | 95.30 ± (1.49) | 95.50 ± (1.28) | |
| Kappa | 93.41 ± (1.55) | 91.72 ± (1.96) | 91.55 ± (2.29) | |
| Time(s) | 425 | 430 | 401 | |
| Strategy to Ensure Confidence | with SC and ASR | with SC | with ASR | without SC or ASR | ||
|---|---|---|---|---|---|---|
| Indian Pines | OA (%) | 84.22 ± (3.20) | 84.56 ± (2.51) | 84.20 ± (3.12) | 83.19 ± (2.83) | |
| AA (%) | 88.55 ± (2.32) | 88.47 ± (2.00) | 88.29 ± (2.46) | 87.64 ± (2.04) | ||
| Kappa (%) | 82.14 ± (3.59) | 82.52 ± (2.80) | 82.12 ± (3.49) | 80.98 ± (3.16) | ||
| Time(s) | 289 | 244 | 367 | 269 | ||
| Pavia University | OA (%) | 90.17 ± (4.87) | 87.39 ± (3.27) | 87.27 ± (4.39) | 81.52 ± (3.63) | |
| AA (%) | 90.81 ± (3.64) | 88.30 ± (1.94) | 90.34 ± (1.80) | 83.00 ± (2.10) | ||
| Kappa (%) | 87.29 ± (6.03) | 83.76 ± (3.91) | 83.68 ± (5.34) | 76.42 ± (4.20) | ||
| Time(s) | 441 | 425 | 565 | 258 | ||
| Salinas Valley | OA (%) | 95.28 ± (1.51) | 95.10 ± (1.04) | 94.62 ± (1.48) | 92.30 ± (2.25) | |
| AA (%) | 96.76 ± (1.03) | 96.49 ± (1.14) | 96.52 ± (1.15) | 94.58 ± (1.72) | ||
| Kappa (%) | 94.75 ± (1.68) | 94.56 ± (1.15) | 94.02 ± (1.64) | 91.45 ± (2.50) | ||
| Time(s) | 571 | 367 | 640 | 394 | ||
| Datasets | TT-SSL | GANs-SSL | SDP-SSL | SANI-SSL | |
|---|---|---|---|---|---|
| Indian Pines | OA | 77.55 | 75.62 | 82.66 | 84.22 |
| AA | 85.16 | 81.05 | 88.12 | 88.55 | |
| Kappa | 74.79 | 72.23 | 80.27 | 82.14 | |
| Pavia University | OA | 82.55 | 77.94 | 84.20 | 90.17 |
| AA | 88.69 | 81.36 | 89.14 | 90.81 | |
| Kappa | 78.01 | 71.82 | 79.86 | 87.29 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; An, R.; Wang, B.; Xing, F.; Ju, F. Shape Adaptive Neighborhood Information-Based Semi-Supervised Learning for Hyperspectral Image Classification. Remote Sens. 2020, 12, 2976. https://doi.org/10.3390/rs12182976
Hu Y, An R, Wang B, Xing F, Ju F. Shape Adaptive Neighborhood Information-Based Semi-Supervised Learning for Hyperspectral Image Classification. Remote Sensing. 2020; 12(18):2976. https://doi.org/10.3390/rs12182976
Chicago/Turabian StyleHu, Yina, Ru An, Benlin Wang, Fei Xing, and Feng Ju. 2020. "Shape Adaptive Neighborhood Information-Based Semi-Supervised Learning for Hyperspectral Image Classification" Remote Sensing 12, no. 18: 2976. https://doi.org/10.3390/rs12182976
APA StyleHu, Y., An, R., Wang, B., Xing, F., & Ju, F. (2020). Shape Adaptive Neighborhood Information-Based Semi-Supervised Learning for Hyperspectral Image Classification. Remote Sensing, 12(18), 2976. https://doi.org/10.3390/rs12182976