Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model
Abstract
1. Introduction
- Varying the training data on which a neural network model is trained;
- Varying the initial set of random weights from which each neural network is trained, but keeping the training data constant;
- Varying the topology or the architecture of the hidden layers within the same algorithm;
- Varying the algorithm used for training the same data.
2. Materials and Methods
2.1. Study Sites and Materials
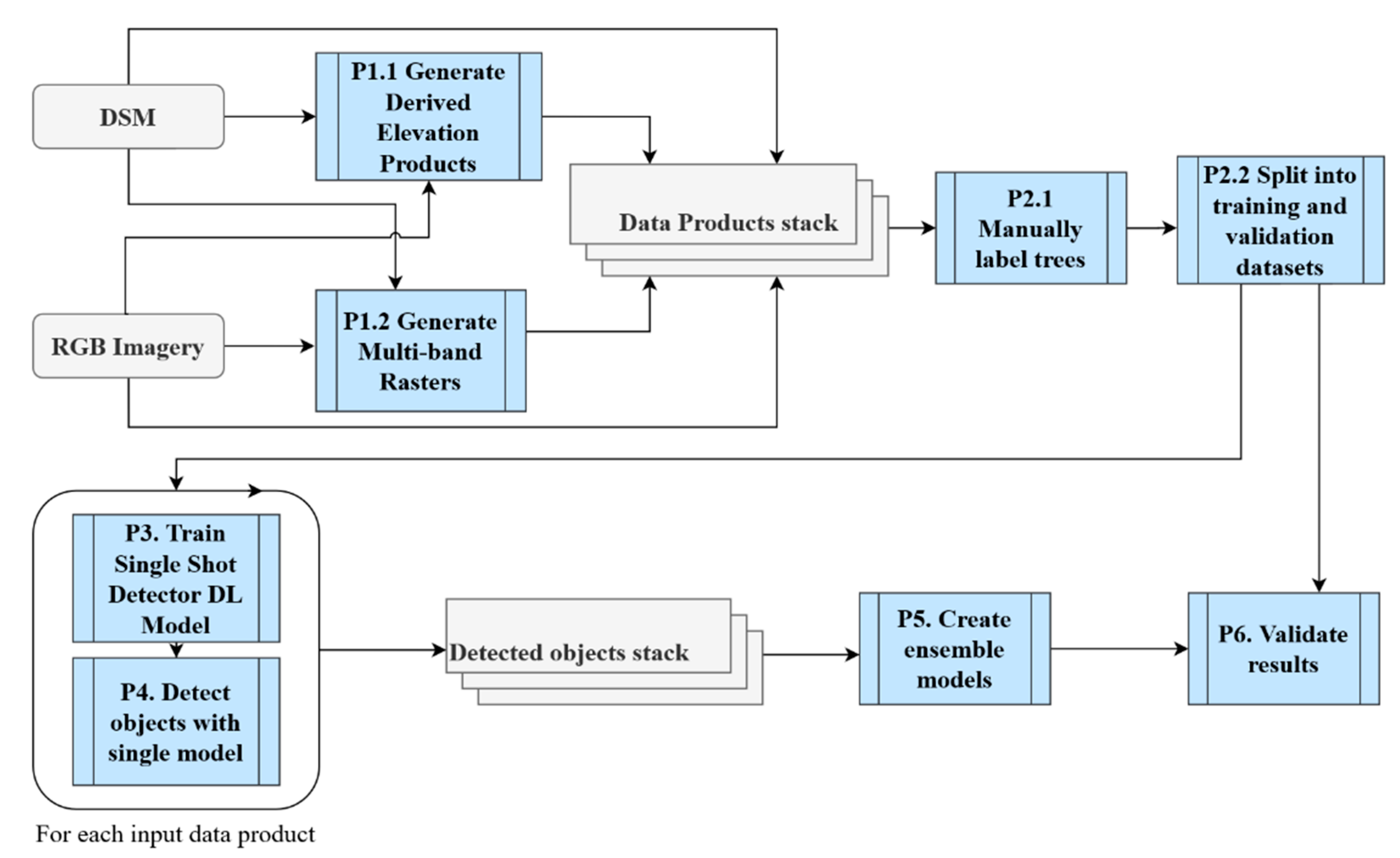
2.2. Technical Approach
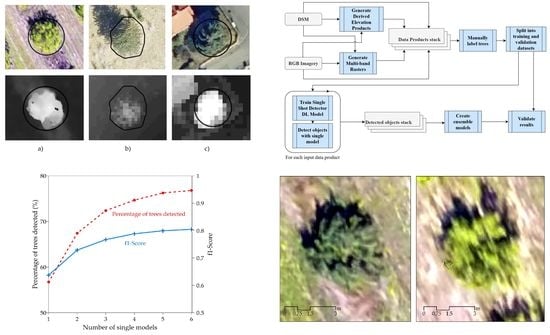
2.2.1. Overview
2.2.2. Data Pre-Processing
2.2.3. Preparing the Training and Validation Data
2.2.4. Training Single Shot Detector Deep Learning Models
2.2.5. Detecting with Single Models
2.2.6. Ensemble Learning
- Majority: the majority of single models must agree the output species detected;
- Unison: all single models must agree the output species detected;
- Confidence: the model gives the output species with the highest SSD confidence value;
- Weighted: the output species is given by a weighted sum that applies weights based on the single models’ accuracy in terms of the f1-Score.
3. Results
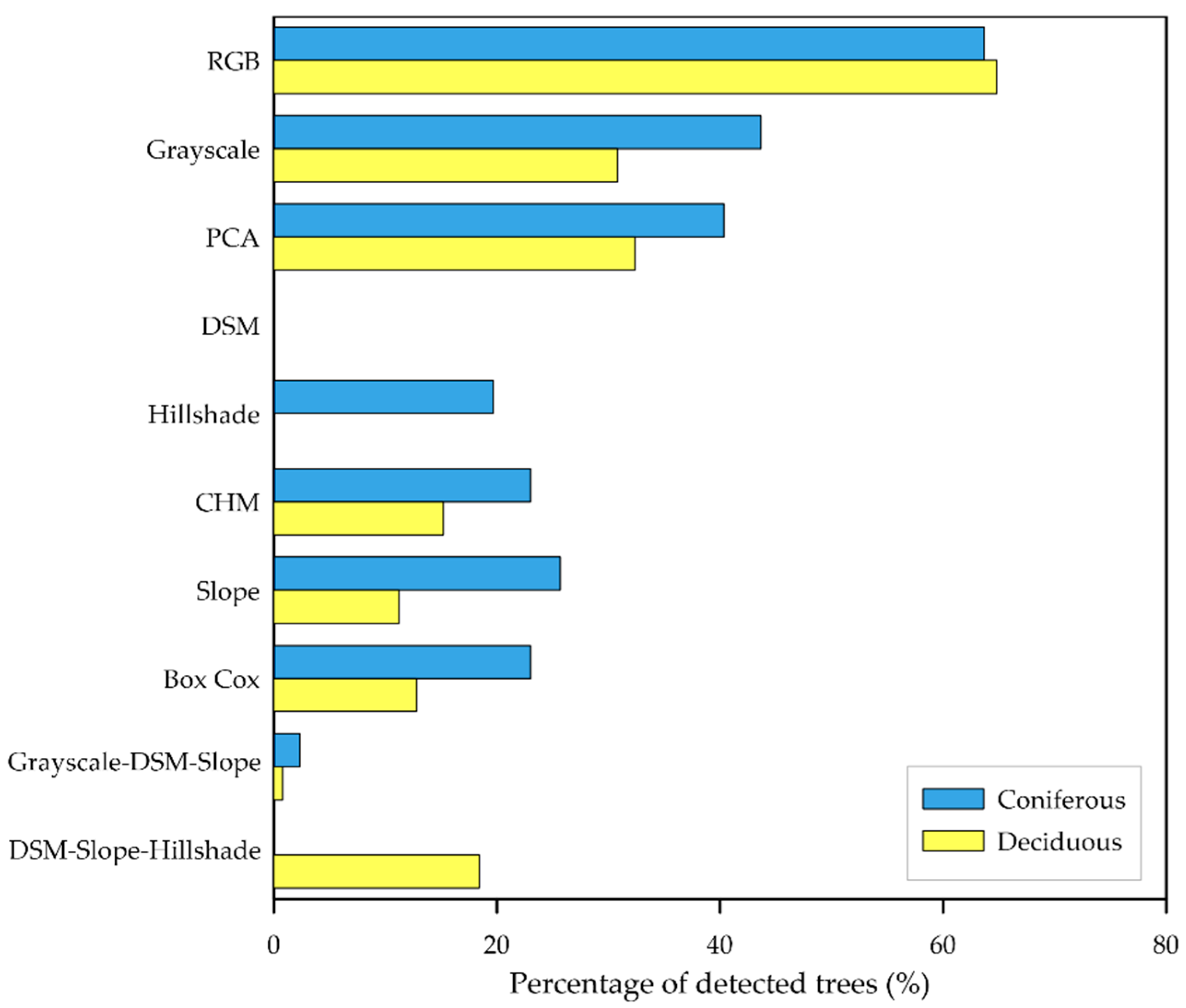
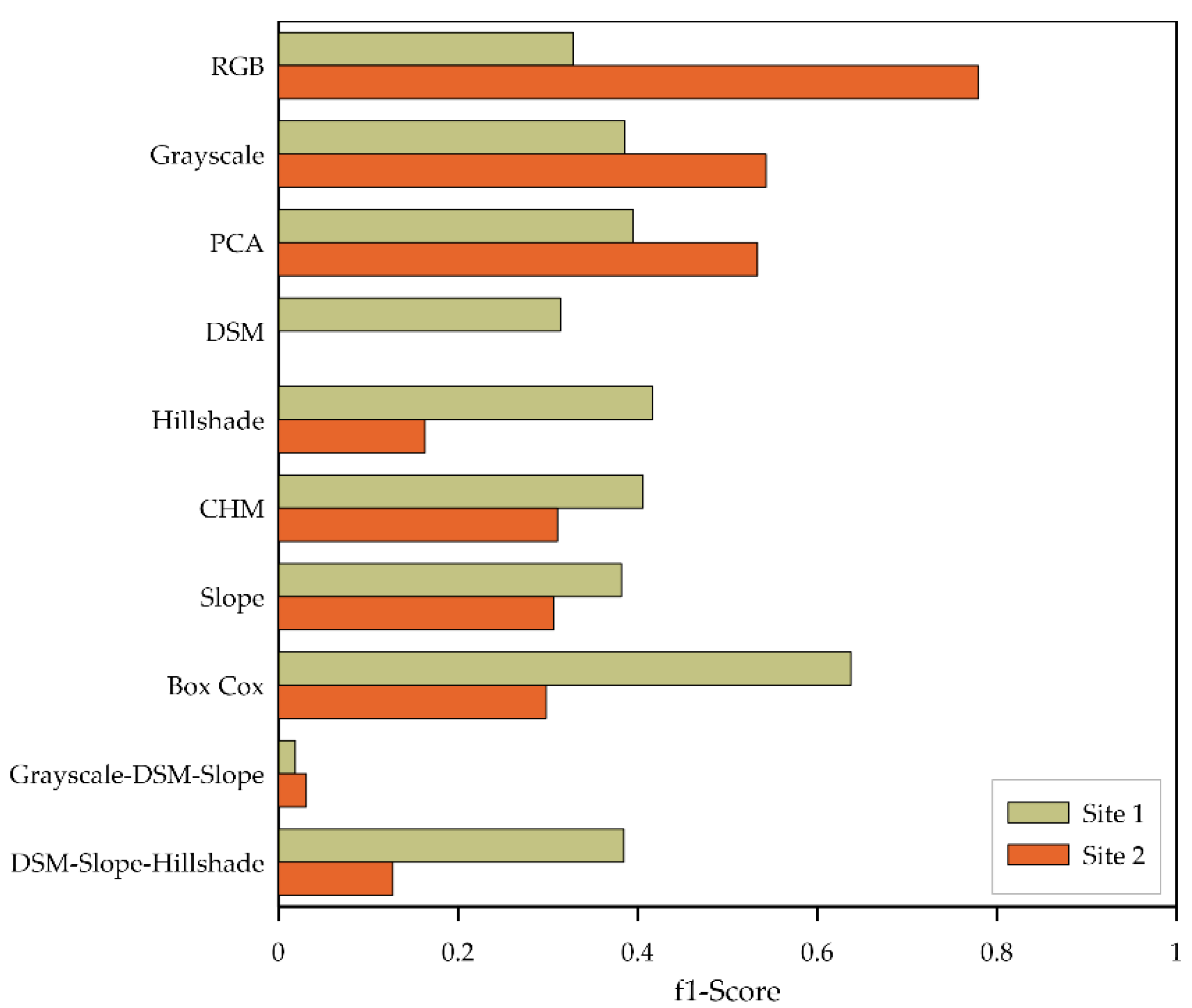
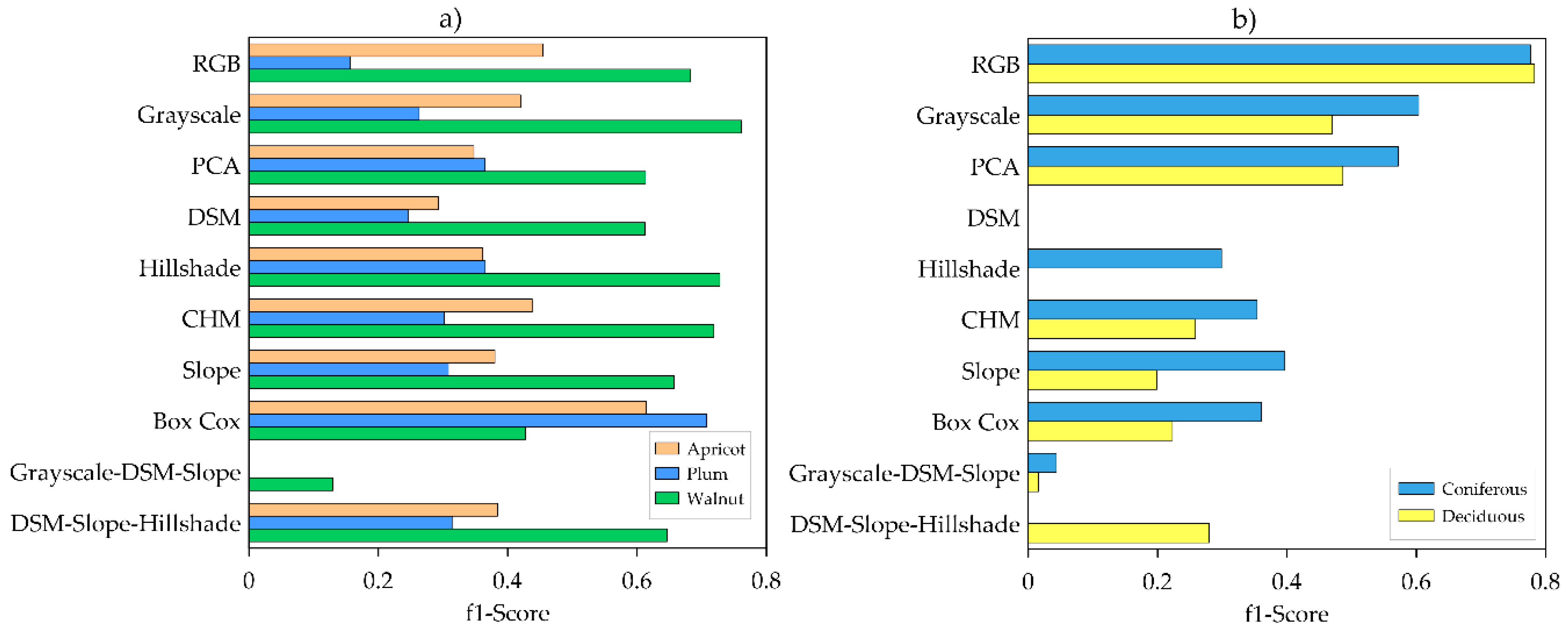
3.1. Single Product Models
3.2. Ensemble Models
4. Discussion
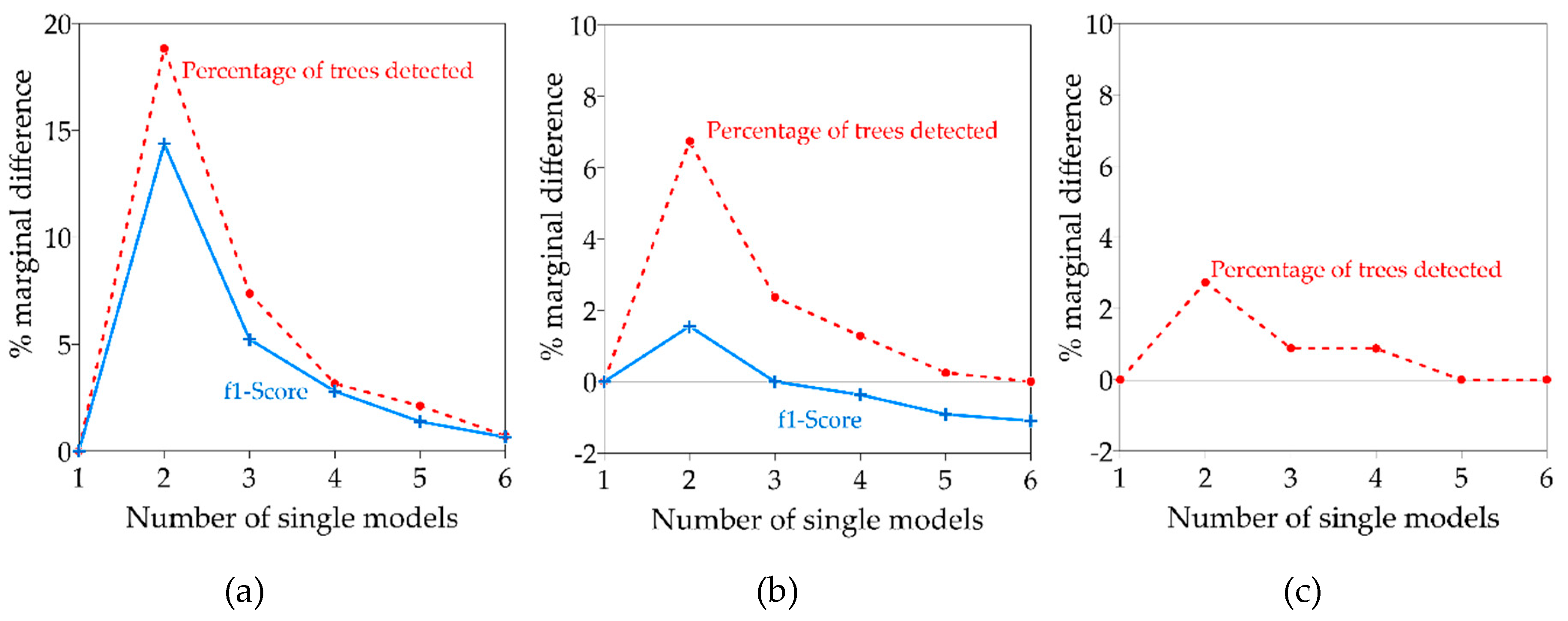
4.1. Single vs. Ensemble Models
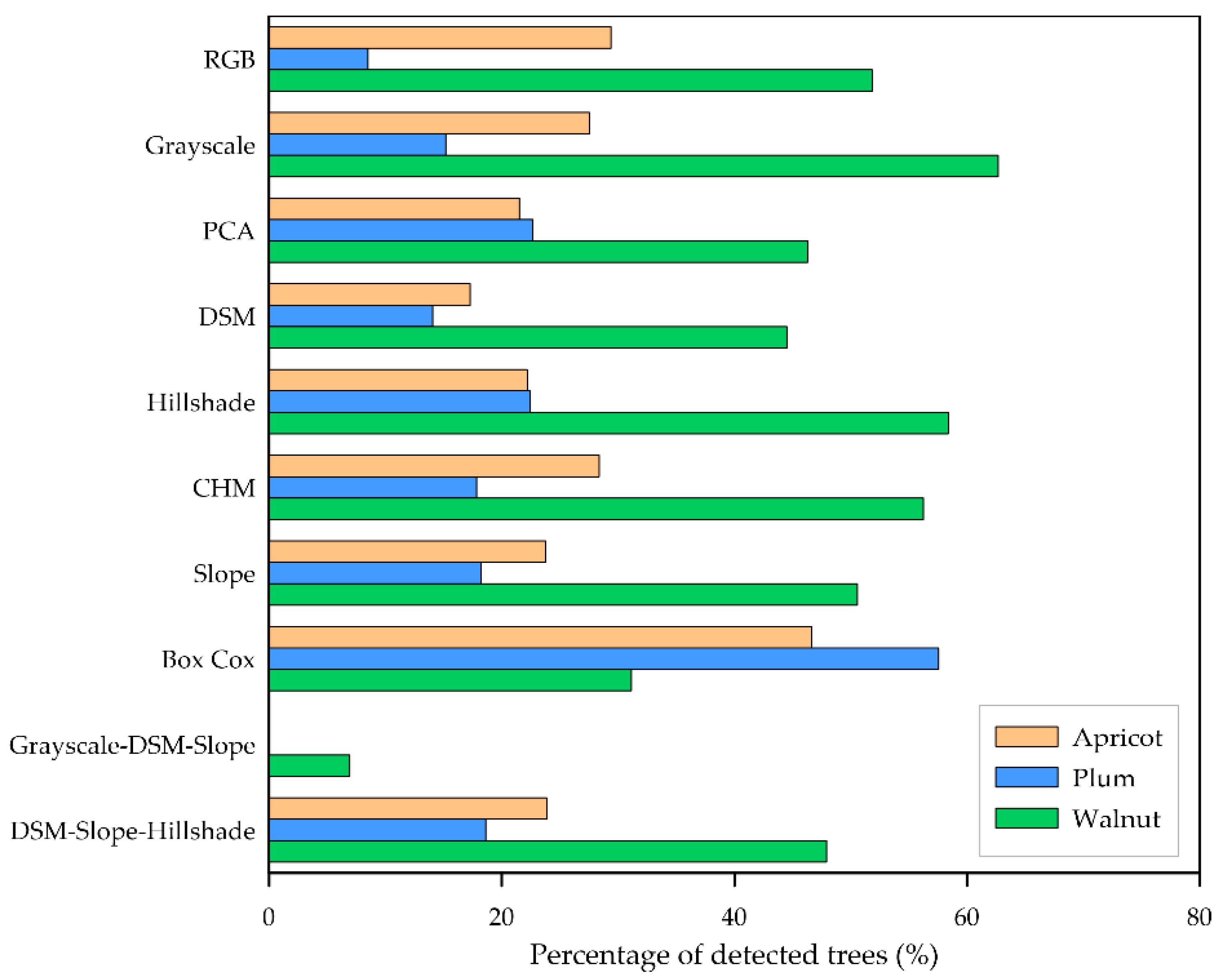
4.2. Input Data Products Performance vs. Image Resolution
4.3. Performance of Voting Strategies
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Parameter | Value |
|---|---|
| Batch size | 30 |
| Stochastic optimization method | Adam |
| Number of training epochs | 20 |
| Learning rate (lr) | lr ∈ (0.001, 0.01) |
| Early stopping condition | Valid—loss does not reduce after 5 epochs |
| Layer (Type) | Output Shape | Param # | Trainable |
|---|---|---|---|
| Conv2d | (64, 60, 60) | 9408 | FALSE |
| BatchNorm2d | (64, 60, 60) | 128 | TRUE |
| ReLU | (64, 60, 60) | 0 | FALSE |
| MaxPool2d | (64, 30, 30) | 0 | FALSE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| ReLU | (64, 30, 30) | 0 | FALSE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| ReLU | (64, 30, 30) | 0 | FALSE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| ReLU | (64, 30, 30) | 0 | FALSE |
| Conv2d | (64, 30, 30) | 36,864 | FALSE |
| BatchNorm2d | (64, 30, 30) | 128 | TRUE |
| Conv2d | (128, 15, 15) | 73,728 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| ReLU | (128, 15, 15) | 0 | FALSE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| Conv2d | (128, 15, 15) | 8192 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| ReLU | (128, 15, 15) | 0 | FALSE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| ReLU | (128, 15, 15) | 0 | FALSE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| ReLU | (128, 15, 15) | 0 | FALSE |
| Conv2d | (128, 15, 15) | 147,456 | FALSE |
| BatchNorm2d | (128, 15, 15) | 256 | TRUE |
| Conv2d | (256, 8, 8) | 294,912 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 32,768 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| ReLU | (256, 8, 8) | 0 | FALSE |
| Conv2d | (256, 8, 8) | 589,824 | FALSE |
| BatchNorm2d | (256, 8, 8) | 512 | TRUE |
| Conv2d | (512, 4, 4) | 1,179,648 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| ReLU | (512, 4, 4) | 0 | FALSE |
| Conv2d | (512, 4, 4) | 2,359,296 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| Conv2d | (512, 4, 4) | 131,072 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| Conv2d | (512, 4, 4) | 2,359,296 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| ReLU | (512, 4, 4) | 0 | FALSE |
| Conv2d | (512, 4, 4) | 2,359,296 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| Conv2d | (512, 4, 4) | 2,359,296 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| ReLU | (512, 4, 4) | 0 | FALSE |
| Conv2d | (512, 4, 4) | 2,359,296 | FALSE |
| BatchNorm2d | (512, 4, 4) | 1024 | TRUE |
| Dropout | (512, 4, 4) | 0 | FALSE |
| Conv2d | (256, 4, 4) | 1,179,904 | TRUE |
| BatchNorm2d | (256, 4, 4) | 512 | TRUE |
| Dropout | (256, 4, 4) | 0 | FALSE |
| Conv2d | (256, 1, 1) | 1,048,832 | TRUE |
| BatchNorm2d | (256, 1, 1) | 512 | TRUE |
| Dropout | (256, 1, 1) | 0 | FALSE |
| Conv2d | (256, 3, 3) | 590,080 | TRUE |
| Upsample | (256, 3, 3) | 0 | FALSE |
| BatchNorm2d | (256, 3, 3) | 512 | TRUE |
| Dropout | (256, 3, 3) | 0 | FALSE |
| Conv2d | (4, 1, 1) | 9220 | TRUE |
| Conv2d | (4, 1, 1) | 9220 | TRUE |
| Conv2d | (4, 3, 3) | 9220 | TRUE |
| Conv2d | (4, 3, 3) | 9220 | TRUE |
References
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation-evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Disney, M. Terrestrial LiDAR: A three-dimensional revolution in how we look at trees. New Phytol. 2019, 222, 1736–1741. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Williams, J.; Schonlieb, C.B.; Swinfield, T.; Lee, J.; Cai, X.; Qie, L.; Coomes, D.A. 3D Segmentation of Trees through a Flexible Multiclass Graph Cut Algorithm. IEEE Trans. Geosci. Remote Sens. 2019, 58, 754–776. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Vastaranta, M.; Coops, N.C.; Pitt, D.; Woods, M. The utility of image-based point clouds for forest inventory: A comparison with airborne laser scanning. Forests 2013, 4, 518–536. [Google Scholar] [CrossRef]
- Goodbody, T.R.H.; Coops, N.C.; White, J.C. Digital Aerial Photogrammetry for Updating Area-Based Forest Inventories: A Review of Opportunities, Challenges, and Future Directions. Curr. For. Rep. 2019, 5, 55–75. [Google Scholar] [CrossRef]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from Motion Photogrammetry in Forestry: A Review. Curr. For. Rep. 2019, 5, 155–168. [Google Scholar] [CrossRef]
- Larsen, M.; Eriksson, M.; Descombes, X.; Perrin, G.; Brandtberg, T.; Gougeon, F.A. Comparison of six individual tree crown detection algorithms evaluated under varying forest conditions. Int. J. Remote Sens. 2011, 32, 5827–5852. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Forest understory trees can be segmented accurately within sufficiently dense airborne laser scanning point clouds. Sci. Rep. 2017, 7, 6770. [Google Scholar] [CrossRef]
- Li, L.; Chen, J.; Mu, X.; Li, W.; Yan, G.; Xie, D.; Zhang, W. Quantifying Understory and Overstory Vegetation Cover Using UAV-Based RGB Imagery in Forest Plantation. Remote Sens. 2020, 12, 298. [Google Scholar] [CrossRef]
- Chianucci, F.; Cutini, A.; Corona, P.; Puletti, N. Estimation of leaf area index in understory deciduous trees using digital photography. Agric. For. Meteorol. 2014, 198, 259–264. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Guan, H.; Yu, Y.; Ji, Z.; Li, J.; Zhang, Q. Deep learning-based tree classification using mobile LiDAR data. Remote Sens. Lett. 2015, 6, 864–873. [Google Scholar] [CrossRef]
- Zou, X.; Cheng, M.; Wang, C.; Xia, Y.; Li, J. Tree Classification in Complex Forest Point Clouds Based on Deep Learning. IEEE Geosci. Remote Sens. Lett. 2017, 14, 2360–2364. [Google Scholar] [CrossRef]
- Li, W.; Fu, H.; Yu, L.; Cracknell, A. Deep Learning Based Oil Palm Tree Detection and Counting for High-Resolution Remote Sensing Images. Remote Sens. 2017, 9, 22. [Google Scholar] [CrossRef]
- Liao, W.; Van Coillie, F.; Gao, L.; Li, L.; Zhang, B.; Chanussot, J. Deep Learning for Fusion of APEX Hyperspectral and Full-Waveform LiDAR Remote Sensing Data for Tree Species Mapping. IEEE Access 2018, 6, 68716–68729. [Google Scholar] [CrossRef]
- Hartling, S.; Sagan, V.; Sidike, P.; Maimaitijiang, M.; Carron, J. Urban tree species classification using a worldview-2/3 and liDAR data fusion approach and deep learning. Sensors 2019, 19, 1284. [Google Scholar] [CrossRef] [PubMed]
- Tianyang, D.; Jian, Z.; Sibin, G.; Ying, S.; Jing, F. Single-Tree Detection in High-Resolution Remote-Sensing Images Based on a Cascade Neural Network. ISPRS Int. J. Geo-Inf. 2018, 7, 367. [Google Scholar] [CrossRef]
- Safonova, A.; Tabik, S.; Alcaraz-Segura, D.; Rubtsov, A.; Maglinets, Y.; Herrera, F. Detection of Fir Trees (Abies sibirica) Damaged by the Bark Beetle in Unmanned Aerial Vehicle Images with Deep Learning. Remote Sens. 2019, 11, 643. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.A.; Zare, A.; White, E.P. Cross-site learning in deep learning RGB tree crown detection. Ecol. Inform. 2020, 56, 101061. [Google Scholar] [CrossRef]
- Hamraz, H.; Jacobs, N.B.; Contreras, M.A.; Clark, C.H. Deep learning for conifer/deciduous classification of airborne LiDAR 3D point clouds representing individual trees. ISPRS J. Photogramm. Remote Sens. 2019, 158, 219–230. [Google Scholar] [CrossRef]
- Opitz, D.; Maclin, R. Popular Ensemble Methods: An Empirical Study. J. Artif. Intell. Res. 1999, 11, 169–198. [Google Scholar] [CrossRef]
- Polikar, R. Ensemble Learning. In Ensemble Machine Learning: Methods and Applications; Zhang, C., Ma, Y., Eds.; Springer: Boston, MA, USA, 2012; pp. 1–34. ISBN 978-1-4419-9326-7. [Google Scholar]
- Michalski, R.S. A theory and methodology of inductive learning. In Machine Learning; Springer: Berlin, Heidelberg, 1983; pp. 83–134. [Google Scholar]
- Hansen, L.K.; Salamon, P. Neural network ensembles. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 993–1001. [Google Scholar] [CrossRef]
- Mandler, E.; Schümann, J. Combining the classification results of independent classifiers based on the Dempster/Shafer theory of evidence. In Machine Intelligence and Pattern Recognition; Elsevier: Amsterdam, The Netherlands, 1988; Volume 7, pp. 381–393. [Google Scholar]
- Sharkey, A.J.C. On combining artificial neural nets. Conn. Sci. 1996, 8, 299–314. [Google Scholar] [CrossRef]
- Cunningham, P.; Carney, J. Diversity versus Quality in Classification Ensembles Based on Feature Selection BT—Machine Learning: ECML 2000; López de Mántaras, R., Plaza, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 109–116. [Google Scholar]
- Krogh, A.; Vedelsby, J. Neural network ensembles, cross validation, and active learning. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 1995; pp. 231–238. [Google Scholar]
- Battiti, R.; Colla, A.M. Democracy in neural nets: Voting schemes for classification. Neural Netw. 1994, 7, 691–707. [Google Scholar] [CrossRef]
- Deng, Z.; Sun, H.; Zhou, S.; Zhao, J.; Lei, L.; Zou, H. Multi-scale object detection in remote sensing imagery with convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2018, 145, 3–22. [Google Scholar] [CrossRef]
- Giacinto, G.; Roli, F.; Bruzzone, L. Combination of neural and statistical algorithms for supervised classification of remote-sensing images. Pattern Recognit. Lett. 2000, 21, 385–397. [Google Scholar] [CrossRef]
- Partridge, D.; Yates, W.B. Engineering Multiversion Neural-Net Systems. Neural Comput. 1996, 8, 869–893. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2016; Volume 9905, pp. 21–37. [Google Scholar]
- Thüringer Landesamt für Bodenmanagement und Geoinformation. Available online: https://www.geoportal-th.de/de-de (accessed on 12 December 2019).
- Corner, B.R.; Narayanan, R.M.; Reichenbach, S.E. Principal component analysis of remote sensing imagery: Effects of additive and multiplicative noise. In Proceedings of the SPIE on Applications of Digital Image Processing XXII, Denver, CO, USA, 18–23 July 1999; Volume 3808, pp. 183–191. [Google Scholar]
- Csillik, O.; Evans, I.S.; Drăguţ, L. Transformation (normalization) of slope gradient and surface curvatures, automated for statistical analyses from DEMs. Geomorphology 2015, 232, 65–77. [Google Scholar] [CrossRef]
- Howard, J.; Gugger, S. Fastai: A Layered API for Deep Learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Banada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Marcel, S.; Rodriguez, Y. Torchvision the machine-vision package of torch. In Proceedings of the MM’10—ACM Multimedia 2010 International Conference, Philadelphia, PA, USA, 12–16 October 2010; ACM Press: New York, NY, USA, 2010; pp. 1485–1488. [Google Scholar]
- Körez, A.; Barışçı, N.; Çetin, A.; Ergün, U. Weighted Ensemble Object Detection with Optimized Coefficients for Remote Sensing Images. ISPRS Int. J. Geo-Inf. 2020, 9, 370. [Google Scholar] [CrossRef]
- Qiu, S.; Wen, G.; Deng, Z.; Liu, J.; Fan, Y. Accurate non-maximum suppression for object detection in high-resolution remote sensing images. Remote Sens. Lett. 2018, 9, 237–246. [Google Scholar] [CrossRef]
- Tang, T.; Zhou, S.; Deng, Z.; Lei, L.; Zou, H. Arbitrary-Oriented Vehicle Detection in Aerial Imagery with Single Convolutional Neural Networks. Remote Sens. 2017, 9, 1170. [Google Scholar] [CrossRef]
- Wu, X.; Sahoo, D.; Hoi, S.C.H. Recent advances in deep learning for object detection. Neurocomputing 2020. [Google Scholar] [CrossRef]
- Powers, D. Evaluation: From Precision, Recall and F-Factor to ROC, Informedness, Markedness & Correlation. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in {P}ython. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Zhou, Z.-H. Ensemble Methods: Foundations and Algorithms; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Minetto, R.; Segundo, M.P.; Sarkar, S. Hydra: An Ensemble of Convolutional Neural Networks for Geospatial Land Classification. IEEE Trans. Geosci. Remote Sens. 2018, 57, 6530–6541. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Ran, Q.; Du, Q.; Gao, L.; Zhang, B. Multisource Remote Sensing Data Classification Based on Convolutional Neural Network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 937–949. [Google Scholar] [CrossRef]
- Stupariu, M.-S.; Pleșoianu, A.-I.; Pătru-Stupariu, I.; Fürst, C. A Method for Tree Detection Based on Similarity with Geometric Shapes of 3D Geospatial Data. ISPRS Int. J. Geo-Inf. 2020, 9, 298. [Google Scholar] [CrossRef]
- Peng, X.; Li, X.; Wang, C.; Zhu, J.; Liang, L.; Fu, H.; Du, Y.; Yang, Z.; Xie, Q. SPICE-based SAR tomography over forest areas using a small number of P-band airborne F-SAR images characterized by non-uniformly distributed baselines. Remote Sens. 2019, 11, 975. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.-K.; Yang, S.-I. An ensemble method of cnn models for object detection. In Proceedings of the 2018 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Korea, 17–19 October 2018; pp. 898–901. [Google Scholar]
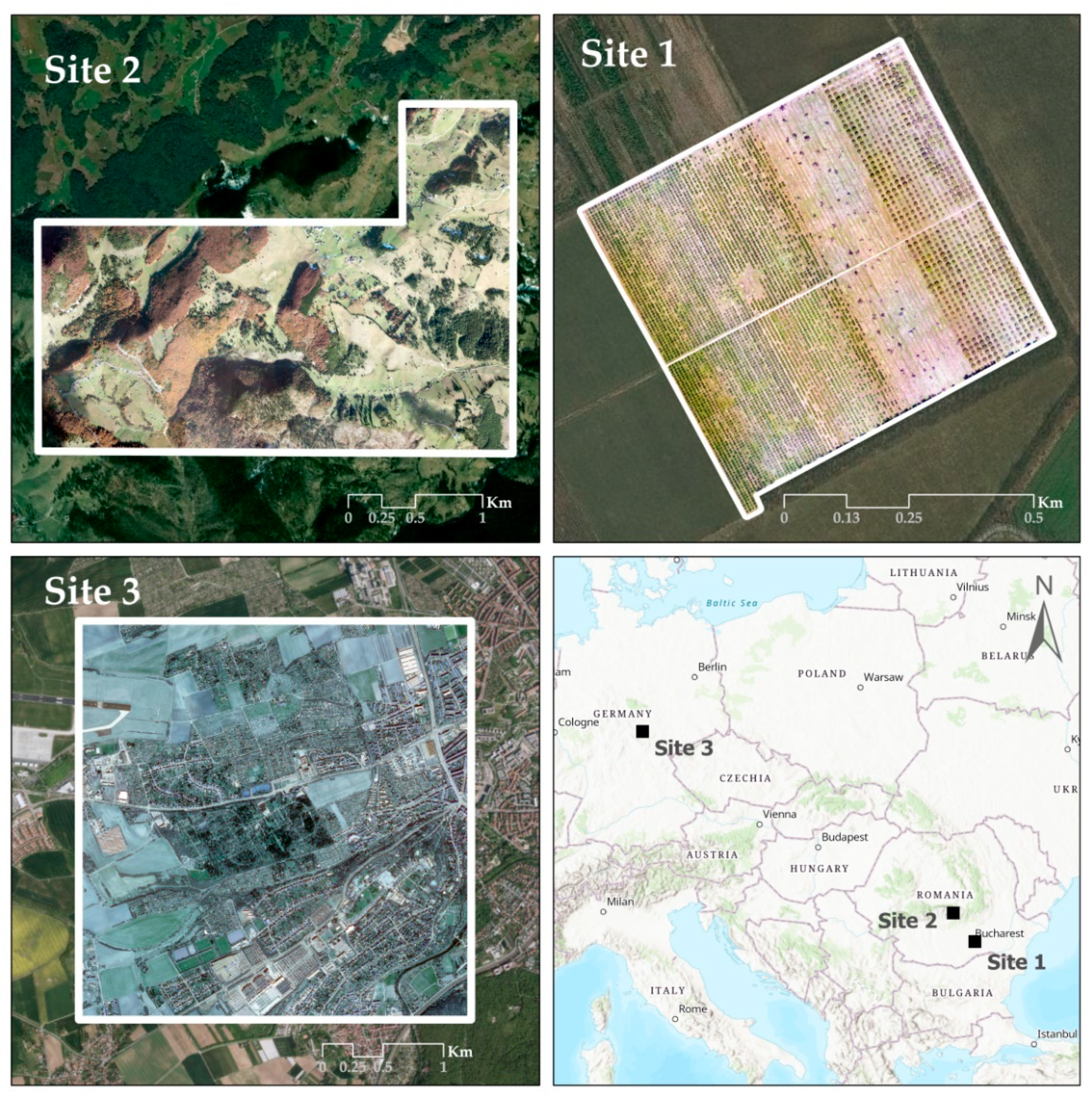




| Original Product | Input Product | Site 1 | Site 2 | Site 3 |
|---|---|---|---|---|
| RGB | RGB | 6 cm | 12 cm | 20 cm |
| Grayscale | 6 cm | 12 cm | 20 cm | |
| Principal Component Analysis (PCA) | 6 cm | 12 cm | 20 cm | |
| DSM | Digital Surface Model (DSM) | 10 cm | 100 cm | 100 cm |
| Canopy Height Model (CHM) | 10 cm | 100 cm | 100 cm | |
| Slope | 10 cm | 100 cm | 100 cm | |
| Hillshade | 10 cm | 100 cm | 100 cm | |
| Box Cox | 25 cm | 100 cm | 100 cm | |
| Combinations | DSM–Slope–Hillshade | 10 cm | 100 cm | 100 cm |
| Grayscale–DSM–Slope | 10 cm | 100 cm | 100 cm |
| Site | Site 1 | Site 2 | Site 3 | |||
|---|---|---|---|---|---|---|
| Species | Apricot | Plum | Walnut | Coniferous | Deciduous | No Species |
| Training Labels | 1420 | 2354 | 634 | 1500 | 1250 | 1200 |
| Validation Labels | 356 | 589 | 159 | 300 | 250 | 300 |
| Total Labels | 1776 | 2493 | 793 | 1800 | 1500 | 1500 |
| Invalid Product Pairs |
|---|
| DSM/DSM–Slope–Hillshade |
| DSM/Grayscale–DSM–Slope |
| Slope/DSM–Slope–Hillshade |
| Slope/Grayscale–DSM–Slope |
| Hillshade/DSM–Slope–Hillshade |
| Grayscale/Grayscale–DSM–Slope |
| DSM–Slope–Hillshade/Grayscale–DSM–Slope |
| RGB/Grayscale–DSM–Slope |
| RGB/Grayscale |
| RGB/PCA |
| PCA/Grayscale |
| PCA/Grayscale–DSM–Slope |
| Product | Detection Percentage—Site 1 | Detection Percentage—Site 2 | Detection Percentage—Site 3 |
|---|---|---|---|
| RGB | 21.61% | 64.73% | 73% |
| Grayscale | 27.56% | 38.55% | 60.33% |
| PCA | 27.87% | 37.45% | 38% |
| DSM | 20.03% | 0% | 15.67% |
| Hillshade | 28.17% | 17.27% | 14% |
| CHM | 27.32% | 24.36% | 12.33% |
| Slope | 25.63% | 22% | 10.67% |
| Box Cox | 56.73% | 21.82% | 12% |
| Grayscale–DSM–Slope | 1% | 4.73% | 1.33% |
| DSM–Slope–Hillshade | 24.71% | 14.36% | 0.67% |
| Site | Ensemble Model | Detection Percentage |
|---|---|---|
| Site 1 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 76.8% |
| DSM + Slope + Hillshade + Grayscale + Box Cox + CHM | 76.43% | |
| Slope + Hillshade + PCA + Box Cox + CHM | 76.25% | |
| Slope + Hillshade + Grayscale + Box Cox + CHM | 76% | |
| Site 2 | RGB + Slope + Hillshade + Box Cox + CHM | 71.82% |
| RGB + Slope + Hillshade + CHM | 71.64% | |
| RGB + Hillshade + Box Cox + CHM | 71.45% | |
| RGB + DSM-Slope-Hillshade + Box Cox + CHM | 71.27% | |
| Site 3 | RGB + DSM + Slope + CHM | 76.33% |
| RGB + DSM + Slope + Hillshade + CHM | 76.33% | |
| RGB + DSM + Slope + Hillshade + Box Cox + CHM | 76.33% | |
| RGB + DSM + Slope + Box Cox + CHM | 76.33% |
| Site | Species | Voting Strategy | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Majority | Unison | Weighted | Confidence | ||||||
| Ensemble Model | Det. per. (%) | Ensemble Model | Det. per. (%) | Ensemble Model | Det. per. (%) | Ensemble Model | Det. per. (%) | ||
| Site 1 | Plum | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 71.52 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 67.62 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 70.88 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 70.47 |
| Apricot | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 64.58 | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 59.68 | DSM + Slope + Hillshade + Grayscale + Box Cox + CHM | 63.12 | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 62.73 | |
| Walnut | DSM + Slope + Hillshade + Grayscale + CHM | 89.66 | DSM + Slope + Hillshade + Grayscale + CHM | 88.27 | DSM + Slope + Hillshade + Grayscale + Box Cox + CHM | 90.29 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 90.04 | |
| Site 2 | Coniferous | RGB + Hillshade + CHM | 67.00 | RGB + Hillshade | 66.00 | RGB + Hillshade + CHM | 67.33 | RGB + Hillshade + CHM | 67.67 |
| Deciduous | RGB + DSM–Slope–Hillshade + CHM | 71.2 | RGB + DSM–Slope–Hillshade | 69.60 | RGB + DSM–Slope–Hillshade + Box Cox + CHM | 72.00 | RGB + DSM–Slope–Hillshade + Box Cox + CHM | 71.60 | |
| Site | Species | Voting Strategy | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Majority | Unison | Weighted | Confidence | ||||||
| Ensemble Model | f1-S. | Ensemble Model | f1-S. | Ensemble Model | f1-S. | Ensemble Model | f1-S. | ||
| Site 1 | Plum | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 0.814 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 0.793 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 0.809 | DSM + Slope + Hillshade + PCA + Box Cox + CHM | 0.806 |
| Apricot | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 0.766 | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 0.736 | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 0.750 | RGB + DSM + Slope + Hillshade + Box Cox + CHM | 0.747 | |
| Walnut | DSM + Slope + Hillshade + Grayscale + CHM | 0.925 | Slope + Hillshade + Grayscale + CHM | 0.921 | DSM + Slope + Hillshade + Grayscale + CHM | 0.922 | DSM + Hillshade + Grayscale + CHM | 0.920 | |
| Site 2 | Coniferous | RGB + CHM | 0.786 | RGB + CHM | 0.782 | RGB + CHM | 0.792 | RGB + Slope + CHM | 0.791 |
| Deciduous | RGB + DSM–Slope–Hillshade + CHM | 0.809 | RGB + DSM–Slope–Hillshade | 0.813 | RGB + DSM–Slope–Hillshade + Box Cox + CHM | 0.828 | RGB + DSM–Slope–Hillshade + Box Cox + CHM | 0.821 | |
| Input Data Product | Site 1 | Site 2 | Site 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Majority | Unison | Weighted | Confidence | Majority | Unison | Weighted | Confidence | No Voting | |
| RGB | 6 | 8 | 6 | 7 | 22 | 22 | 22 | 22 | 22 |
| Grayscale | 9 | 7 | 8 | 7 | 0 | 0 | 0 | 0 | 0 |
| PCA | 7 | 6 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| DSM | 13 | 13 | 13 | 13 | 9 | 10 | 9 | 9 | 16 |
| Hillshade | 17 | 19 | 18 | 19 | 2 | 4 | 8 | 6 | 12 |
| CHM | 19 | 18 | 18 | 17 | 12 | 8 | 18 | 16 | 12 |
| Slope | 16 | 17 | 17 | 17 | 10 | 10 | 10 | 12 | 14 |
| Box Cox | 23 | 23 | 23 | 23 | 10 | 9 | 12 | 12 | 11 |
| Grayscale–DSM–Slope | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DSM–Slope–Hillshade | 1 | 0 | 0 | 0 | 4 | 2 | 4 | 4 | 0 |
| Voting Strategy | Site 1 | Site 2 | ||
|---|---|---|---|---|
| Precision | Recall | Precision | Recall | |
| Majority | 0.93 | 0.7 | 0.96 | 0.66 |
| Unison | 0.95 | 0.66 | 0.98 | 0.63 |
| Weighted | 0.92 | 0.69 | 0.97 | 0.68 |
| Confidence | 0.92 | 0.69 | 0.97 | 0.68 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pleșoianu, A.-I.; Stupariu, M.-S.; Șandric, I.; Pătru-Stupariu, I.; Drăguț, L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sens. 2020, 12, 2426. https://doi.org/10.3390/rs12152426
Pleșoianu A-I, Stupariu M-S, Șandric I, Pătru-Stupariu I, Drăguț L. Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sensing. 2020; 12(15):2426. https://doi.org/10.3390/rs12152426
Chicago/Turabian StylePleșoianu, Alin-Ionuț, Mihai-Sorin Stupariu, Ionuț Șandric, Ileana Pătru-Stupariu, and Lucian Drăguț. 2020. "Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model" Remote Sensing 12, no. 15: 2426. https://doi.org/10.3390/rs12152426
APA StylePleșoianu, A.-I., Stupariu, M.-S., Șandric, I., Pătru-Stupariu, I., & Drăguț, L. (2020). Individual Tree-Crown Detection and Species Classification in Very High-Resolution Remote Sensing Imagery Using a Deep Learning Ensemble Model. Remote Sensing, 12(15), 2426. https://doi.org/10.3390/rs12152426