Abstract
Hyperspectral sensing, measuring reflectance over visible to shortwave infrared wavelengths, has enabled the classification and mapping of vegetation at a range of taxonomic scales, often down to the species level. Classification with hyperspectral measurements, acquired by narrow band spectroradiometers or imaging sensors, has generally required some form of spectral feature selection to reduce the dimensionality of the data to a level suitable for the construction of a classification model. Despite the large number of hyperspectral plant classification studies, an in-depth review of feature selection methods and resultant waveband selections has not yet been performed. Here, we present a review of the last 22 years of hyperspectral vegetation classification literature that evaluates the overall waveband selection frequency, waveband selection frequency variation by taxonomic, structural, or functional group, and the influence of feature selection choice by comparing such methods as stepwise discriminant analysis (SDA), support vector machines (SVM), and random forests (RF). This review determined that all characteristics of hyperspectral plant studies influence the wavebands selected for classification. This includes the taxonomic, structural, and functional groups of the target samples, the methods, and scale at which hyperspectral measurements are recorded, as well as the feature selection method used. Furthermore, these influences do not appear to be consistent. Moreover, the considerable variability in waveband selection caused by the feature selectors effectively masks the analysis of any variability between studies related to plant groupings. Additionally, questions are raised about the suitability of SDA as a feature selection method, with it producing waveband selections at odds with the other feature selectors. Caution is recommended when choosing a feature selector for hyperspectral plant classification: We recommend multiple methods being performed. The resultant sets of selected spectral features can either be evaluated individually by multiple classification models or combined as an ensemble for evaluation by a single classifier. Additionally, we suggest caution when relying upon waveband recommendations from the literature to guide waveband selections or classifications for new plant discrimination applications, as such recommendations appear to be weakly generalizable between studies.
1. Introduction
The classification of reflectance spectra to determine broad plant type or species has been explored increasingly over the past two decades. This has been driven by the increased availability of hyperspectral sensing from imaging spectrometers and field spectroradiometers, and increasing need from environmental conservation, agriculture, and forestry groups [1]. High classification accuracies, particularly at fine taxonomic units such as species, or even clones for grapevine varieties [2], has in some cases been enabled by hyperspectral observation [3]. Hyperspectral measurements have been used to classify a variety of plant types including annual gramineous weeds [4], food crops [5], arid zone shrubs [6], and montane/sub-alpine trees [7], growing in equally varied environments, including tropical wetlands [8], urban streetscapes [9], savanna plains [10], and alpine forests [11]. Due to the scale required to map and monitor the world’s vegetation, fast, generalizable, and objective methods that provide results, that can be quickly and easily shared and analysed, are required. Hyperspectral imagery and data can fulfil these requirements, producing digital measurements that can be easily shared and quickly analysed with semi-automated procedures in a repeatable and objective manner. However, the potential generalisability of classification models has yet to be fully evaluated.
Hyperspectral measurements consist of numerous, finely spaced, contiguous measurements (wavebands) providing considerably more information about targets than broadband multispectral observations. These advantages come at the cost of high dimensionality and large data volumes. Hyperspectral instruments record radiance within the range of 350 to 2500 nm of the electromagnetic spectrum, with bandwidths often between 1 and 10 nm. The number of wavebands per observation varies from hundreds to thousands. Training a classification model with such large numbers of spectral features generally requires a large sample size. However, since the collection of samples for hyperspectral studies is onerous, with high costs for imagery and arduous fieldwork for gathering field measurements, sample sizes tend to be small. Data of this high dimensionality is prone to the Hughes phenomenon, also known as the curse of dimensionality, whereby an increasing number of features originally aids in improving classification, before the addition of more features decreases performance as noise and sparsity of the feature space increases [12]. This problem is exacerbated by small sample sizes [13].
In order to overcome this the ratio between sample size and data dimensionality must be improved. In this review, we focus on reducing dimensionality via feature selection, though methods of artificially increasing sample size through data augmentation, semi-supervised classification, and active learning can aid in countering the curse of dimensionality [14,15,16]. Hyperspectral measurements tend to include noisy or redundant features, with high levels of collinearity between wavebands. The elimination of collinearity can substantially improve classification efforts and is in fact a requirement of parametric statistical methods that assume the independence of all variables [17,18]. Additionally, feature selection inherently reveals the spectral regions that offer the greatest discriminatory power for a set of samples. Long held associations between specific spectral regions or individual wavebands and biophysical or biochemical foliar traits [19] have often guided researchers in the selection of features to differentiate species or plant types. The overall aim of this review is to assess these assumptions in light of the evidence from 22 years of hyperspectral plant studies.
Review Scope and Approach
Here, we address some important questions that motivate much hyperspectral plant research. Do the taxonomic, structural, or functional characteristics of plant types or species influence the spectral regions that are most important to classification, or are particular spectral regions consistently selected across a diversity of plant or ecological types? A review of selected features from the hyperspectral literature could identify best practices for feature selection methods, as well as detect wave-regions of high-utility, those that best generalize across taxonomic or ecological boundaries.
The search for literature spanned two decades, from January 1996 to December 2018, focusing on peer reviewed journals in the English language. Search was performed with Google Scholar using combinations of the keywords, namely Hyperspectral, Spectra, Vegetation, Plant, Tree, Species, Identi*, Discriminat*, Classif*, Map, Feature Select*, Waveband, Band, UAV, Drone. In order to be included, a study must have performed a feature selection technique on hyperspectral vegetation data with an aim to classify plant samples.
Many studies fulfilled the initial requirement, but did not report selected wavebands with sufficient specificity, and therefore could not be included. Here, we present waveband selections derived from 38 hyperspectral vegetation classification studies. When applicable, studies that included multiple feature selection techniques were broken into sub-studies, increasing the total number of reviewed studies to 61 (Table 1 and Table 2). These included studies are from a wide variety of scales (leaf, branch, and canopy), recording methods (lab, field, aerial, satellite), taxonomic units, and bandwidths.

Table 1.
Overview of Visible/Near Infra-red (VIS/NIR) studies included in this review.

Table 2.
Overview of Visible/Shortwave Infra-red (VIS/SWIR) studies included in this review.
Additionally, a dataset was synthesised from hyperspectral measurements of 22 species of New Zealand plants collected as field spectra from four locations on the North island [20,21]. This dataset was used to examine how study design (number of classes, number of samples, included species, and feature selection method) influenced waveband selection. This was performed with the aim of determining which elements of the study design most contributed to variation seen in selected wavebands.
The remainder of this paper is structured in the following way. Section 2 provides a meta-analysis of the selected wavebands, broken down by spectral region. Section 3 identifies and describes feature selection techniques from these studies, and where possible, highlights their effects on waveband selection. Section 4 examines study design influence on waveband selection, while Section 5 and Section 6 present a discussion of the results and conclusions.
2. Meta-Analysis
The delineation of spectral regions in this review follows that of [3], as adapted from [53]. Imaging and non-imaging hyperspectral instruments have different sampling intervals, so a direct comparison of selected wavebands between studies is not possible. This was resolved by aggregating selected wavebands into 50 nm bins based on their band centres (Figure 1). The design and presentation of the binned wavelengths is adapted from [1] with adjustments. Additionally, the bin size of the histogram has the benefit of grouping highly correlated and often redundant wavebands together, reducing noise from the selection of correlated features from the analysis. The percentage of studies that selected wavebands within each 50 nm region is presented in the histogram, giving the selection rate for each 50-nm spectral region. The binned table and selection rate histogram (Figure 1) only give an indication of the rate with which a spectral region was selected and do not include information on the number of bands selected in each 50 nm bin, nor the determined importance of a selected band for subject discrimination.
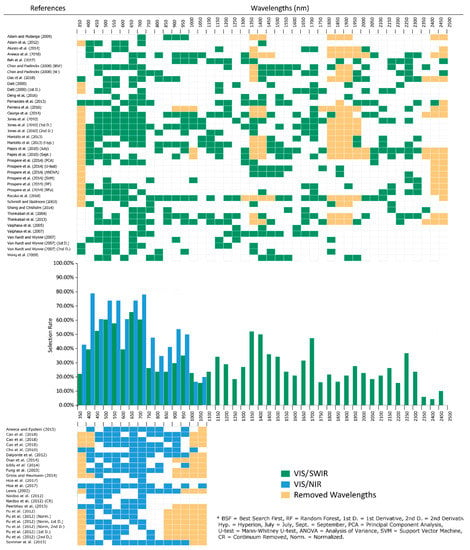
Figure 1.
Waveband selection binned at 50 nm intervals for the VIS/SWIR studies (3502500 nm) green, VIS/NIR studies (350–1100 nm) blue. Orange filled cells represent waveband regions removed from a study due to noise. Selection rate is the percentage of studies that selected a given 50 nm region for species classification. Each row of the table is an individual study, with each column being a 50 nm range bin. Green/blue shaded bins represent at least one waveband being selected from within that range, while orange shaded bins represent removed wavelength regions (e.g. major water absorption regions). Wavelength bins were only removed if the entire 50 nm region was removed due to noise/atmospheric effects in that particular study.
2.1. Spectral Range
Of the studies that met the rules for inclusion in this review, 38 used hyperspectral data spanning most of the range 350–2500 nm. However, a number of studies utilised devices that recorded a more restricted wavelength range between 350 and 1100 nm, generally from 400 to 800 nm or 1000 nm (Table 2). These studies are presented separately as the absence of Shortwave Infra-red(SWIR), and much of the Near Infra-red(NIR) has shown to have an influence on waveband selection for the Visible (VIS) and partial NIR [54]. Although selection rates in the VIS/NIR studies appear similar to those from broader wavelength hyperspectral studies there are some notable differences. The initial peak in selection rates present in both sets is shifted towards shorter blue wavelengths, and a greater importance of the red edge over the red minimum is evident for the VIS/NIR studies. However, the overall pattern is the same with two peaks in the rate of selection at both the blue/green and red reflectance minima, with yellow wavelength bands having the lowest selection rate, save for the sub-400 nm bands that appear in a very limited number of studies. Although the VIS/NIR studies do not cover the full NIR region, selection rates for the red edge and shorter wavelength NIR are closely matched between both groups (Figure 1). The overall higher rates present in the VIS/NIR table results from the smaller number of studies in that group, with selection rates tending to decrease as more studies are added. Additionally, the relatively small number of studies included in the VIS/NIR group prevents the analysis of specific subsets, such as canopy and leaf. The following discussion of selection rates refers to VIS/SWIR studies (Table 1) and is generally applicable to the VIS/NIR studies, although particular discussion of the VIS/NIR studies is included when required.
2.2. Visible (VIS; 400–700 nm)
Primarily a region of low reflectance in living foliage, typically as low as 5%–10% with the exception of the green peak at ~550 nm where reflectance can be more than twice that of surrounding wavelengths (Figure 2). Reflection in the visible wavelengths is dominated by absorptions from foliar pigments. Differences in leaf pigments between species have been identified by many studies as important factors for discrimination [39], despite variability in the VIS being generally low compared to longer wavelengths [53,55]. Of the pigments, chlorophyll a and b have the strongest influence over absorption in this region, followed by those of carotenoids and anthocyanins whose effects are predominantly masked by that of chlorophyll. The visible region is one of the most influential regions for classification, with the vast majority of studies in this review selecting bands from within it. The visible wavelengths can be divided into three regions of high discriminatory value, spanning almost the entire visible range: the blue/blue-green edge (400–499 nm), the green peak centred around 550 nm, and the red reflectance minimum (650–700 nm) (Figure 2). Of these, the red reflectance minimum, specifically bands near 680 nm has previously been identified as the most commonly selected and critical band centre for crop type discrimination [56]. The continued selection of 680 nm, along with neighbouring bands in later studies has validated the importance of this region amongst agricultural crop studies [5,25,47,48,57], as well as for other vegetation types [10,17,18,26,27,29,30,46,58,59]. In addition to the obvious relationship with chlorophyll, absorption in the red region has been related to anthocyanin content, a foliar pigment responsible for the red colouration in leaves [60], particularly evident in juvenile leaves of certain species [30].
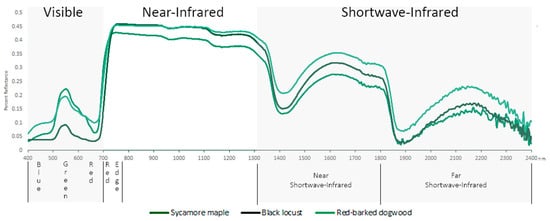
Figure 2.
Example hyperspectral reflectance of 3 species of tree and key broad regions of the electromagnetic spectrum (400–2400 nm).
The green region has the second highest selection rate amongst both the VIS and entire measured spectrum (Figure 1). Wavebands selected in this region tend to be focused around the green reflectance peak at approximately 550 nm, which is strongly correlated with chlorophyll content [61]. The green peak, either manually chosen as a spectral variable as a representation of chlorophyll content or selected via feature selection, has demonstrated importance in classifying species [9,30,42,62,63]. Additionally, absorption in wavebands within the green region adjacent to the reflectance peak is associated with xanthophylls and anthocyanins. Xanthophyll pigments protect against photo-oxidation of the photosynthesis reaction centres during high light conditions [64], resulting in short term changes in reflectance at 531 nm. This band, along with 570 nm, makes up the photochemical reflectance index [65]. Anthocyanins can be estimated by an index using anthocyanin’s absorption maximum near 550 nm, and a band from the red edge, usually 700 nm [66]. Although not necessarily associated with these additional pigments, studies have selected bands along the leading edge of the green reflectance peak between 500,550 nm [10].
Selection from the blue region (400–449 nm) has the third highest rate in the VIS region, though the blue-green edge (450–499 nm) has an almost equal rate of selection to the green region (55.8% and 58.8%, respectively). The importance of blue bands has been established for discriminating within groups of conifers, and between conifers and broadleaf species [67,68], though its inclusion in approximately half of the studies, many of which include non-coniferous species, indicates its importance in general for a wider range of vegetation types. Some of these non-coniferous studies focused on the savanna ecosystem, where blue bands along with the red reflectance minimum and red edge were informative [10,29]. Blue wavelengths are strongly influenced by chlorophyll absorption, along with carotenoid absorption features present in the 450–499 nm region. Carotenoids have proven important for the discrimination of senescent leaves, when the decay of chlorophyll and the diminishing of the strong chlorophyll-absorption feature reveal the carotenoid absorption feature [18].
However, studies have noted that strong similarities between the visible reflectance of different species can decrease the significance of VIS wavelengths for classification purposes. In one such study, the NIR region was more informative for distinguishing species than the VIS, with spectral differences in the VIS region being non-significant between species [69]. Additionally, in a study of tropical trees, Rivard et al. [54] performed feature selection and classification on various datasets derived from the same original spectra. One dataset included the wavelengths 350–2500 nm, another excluded the VIS, while another excluded the SWIR. Although it was found that the full spectrum produced greater overall classification accuracy, and both reduced datasets produced lower overall accuracies, individual accuracies for certain species remained high. The classification model excluding the VIS region maintained high accuracies for six out of 20 species, whereas the model excluding the SWIR maintained high accuracies for five out 20 species. Although the importance of the VIS region has been described by many authors and is clearly seen in the binned data, studies such as [54] demonstrate that wavelength importance is dependent on the species included in the study.
2.3. Red Edge (680–780 nm)
The red edge encompasses the region from the red reflectance minimum around 680 nm to the NIR shoulder at approximately 780 nm and indicates the sharp increase in reflectance from the VIS to NIR regions associated with strong chlorophyll absorptions and internal leaf structure (Figure 2). The inflection point of the slope in this region has been defined as the red edge position (REP) [70], and its strong correlation with chlorophyll concentration has seen it used as an indicator of stress and senescence in vegetation [71,72]. In the VIS-SWIR studies, the red edge region as represented by the 700–749 nm bin has the same rate of selection as the red minimum bin, whereas the VIS-NIR studies have a slightly higher red edge rate than red minimum. However, as previously stated, the delineation between the red minimum bin (650–699 nm) and the red edge bin (700–749 nm) means that bands selected from the lower point of the red edge would be included in the red minimum bin, potentially skewing red edge band selection rates.
The red edge region has been described as one of the most informative and frequently selected regions in a number of studies, where the authors have attributed its importance to its correlation with chlorophyll abundance, nitrogen concentration, water content, and structural features such as leaf area index (LAI) [3,10,11,73]. Additionally, significant variation of the red edge region between species has been documented after a first derivative transformation has been applied to the spectra [74]. The red edge has proven especially important in studies discriminating species with high levels of chlorophyll and high LAI values such as the giant reed (Arundo donax), in which a distinctive “red shift” is seen where the Red Edge Position (REP) is located at higher wavelengths [32,39]. This “red shift” mirrors the “blue shift” of the REP where its position is shifted towards the shorter blue wavelengths associated with a decrease in chlorophyll and used to monitor senescence or stress [75].
2.4. Near Infrared (NIR) (700–1327 nm)
The NIR is often defined to include wavelengths within the red edge region (680–780 nm) [42]: As this region has been previously discussed, this section focusses on the NIR plateau (780–1327 nm). The high reflectance of the plateau results from the scattering of photons within the leaf structure due to a change in the refractive index from liquid water to air within the inter-cellular spaces [76]. Two minor water absorption features at ~980 nm and ~1200 nm are the only major features of the plateau. Along with water content, the depth and width of these absorptions can be influenced by the spectral recording method. Canopy scale spectra tend to produce deeper and wider absorption features compared to the leaf scale, at which absorption features can vary with leaf stack thickness [3]. High levels of intraspecific variability have been identified in the NIR and related to leaf age, water, and chlorophyll concentration, as well as herbivory, necrosis, and epiphyll cover [3,38]. Wavebands selected in studies reporting these high levels of intraspecific variation have generally been limited to the water absorption features [11,38], although it has been suggested to avoid band selection from within or near water absorption features due to this high level of within-class variability, specifically for Eucalypts [46,77,78]. Despite this, [3] reported greater interspecific variability in the NIR, particularly at the canopy scale, potentially related to species-specific photon scattering caused by differences in canopy architecture, a result also reported by other studies [68,69]. However, it has been suggested that the importance of the NIR and SWIR in [3] is linked to the time delay between leaf collection and spectral measurement, causing a decrease in water content and affecting waveband importance [58].
Even when the high selection rate of the red edge is included, the average selection rate of the NIR is close to half of that of the VIS, placing it third after the near SWIR. However, there are two small peaks in the rate of selection within the NIR, in bins 950–999 and 1150–1199, both of which are associated with water absorption features near 980 and 1200 nm. Despite having one of the lowest rates, some studies have reported that bands in the NIR plateau are the most strongly discriminating [45,52].
2.5. Shortwave Infrared (SWIR) (1328–2500 nm)
Based on the binned results (Figure 1) the SWIR can be divided into two distinct regions, the near SWIR (NSWIR) from 1350–1800 nm, including the strong water absorption feature at 1350–1450 nm, and the far SWIR (FSWIR) from 1800–2500 nm, including another strong water absorption feature from 1800–2000 nm. The wavebands associated with these water absorption features, that mark the start of the SWIR and separate the near and far SWIR, are often removed from spectra due to high levels of noise, as are the bands at the far end of the SWIR above 2400 nm. Selection rates within the NSWIR are on average the second highest, primarily caused by high rates of selection at 1350–1450 and 1700–1750 nm. This initial high selection rate, spanning two consecutive bins, is associated with the water absorption feature focused around 1400 nm. However, these bins are often removed in studies, primarily when hyperspectral imagery is used due to increased noise that is not as prevalent in lab or field spectra. Selection rates then drop in the mid-NSWIR bands before peaking again for the 1700–1750 nm bin, containing wavebands often associated with lignin, cellulose, tannins, and other biochemical constituents of foliar and non-foliar plant matter [19,79]. The FSWIR has the lowest average band selection rate, with its highest selection at bin 2250–2299 nm most likely associated with the weak absorption features of cellulose and lignin present at 2270 nm [19,79].
As the selection results suggest, wavebands selected from the SWIR are reported in the literature as being associated with water absorption [17,33,38,40,46,47,48,58] or the weak harmonic and overtone absorptions from biochemicals such as lignin, starch, and cellulose [9,40,42,46,47,48,52,58,80]. However, as described in regards to the NIR, the selection of bands in or near water absorption features may not be suitable for classification in field or lab spectra, due to high levels of intraspecific variance [46,77]. Additionally, bands selected from leaf scale spectra in the two major water absorption features would not be applicable to remotely sensed imagery as they coincide with low irradiance levels resulting from atmospheric water absorption. The observation of higher selection rates in the NSWIR compared to the FSWIR has previously been made with studies noting the importance of NSWIR bands and absence of selection from the FSWIR [9,42], even when visual differences between species were apparent [52]. Possible reasons for this reduced selection of the FSWIR could be high levels of LAI or leaf water content masking the biochemical features present in this region [81], or a high correlation between the FSWIR, NSWIR, and VIS bands [9].
2.6. Canopy and Leaf Scale Spectral Selection Rates
The red edge has been demonstrated as one of the most frequently selected regions (Figure 1), though the remainder of the NIR (consisting of 12 bins from 750–1349 nm) has the second lowest mean selection rate, only slightly higher than the FSWIR. As the literature has identified an increase in importance of the NIR for canopy spectra, a comparison of band selection rates for each bin was made between canopy and leaf scale spectral studies (Figure 3). Leaf spectra were defined as only containing pure leaf reflectance, with canopy being primarily leaf spectra, though also containing non-photosynthetic vegetation and potentially background reflectance. This comparison shows a clear increase in selection rates for the NIR bins associated with water absorption features for the canopy studies, and a related decrease amongst the leaf scale spectra. Differences are also apparent in the visible regions, with a substantial increase in the selection of the leading edge of the green peak, and a decrease in selection of the trailing edge of the green peak for leaf scale studies compared to canopy level (Figure 3). This would indicate a blue-shift for green bands selected in leaf scale spectra, and a red-shift of selected bands for canopy spectra. Differences in spectral reflectance for the VIS region have been identified at different scales, with branch/canopy spectra including reflectance characteristics from non-foliar sources, shadows and uneven lighting, as well as generally displaying an increase in pigment absorption features [3,53]. Variation in selection rates is also evident in the SWIR, most notably a broad region of increased selection for canopy spectra across four bins from 1950 to 2149 nm, and a sudden peak at 1800–1850 nm. The selection peaks of the canopy spectra correspond to regions of water absorption which have demonstrated an increase in depth and width in canopy studies. However, the disparity between canopy and leaf scale spectra is potentially exaggerated by the fact that a majority of canopy studies eliminate these wavebands due to noise concerns, with the remaining few studies selecting these wavebands as being discriminatory. Increased selection of the broader region could also be related to water absorption, as well as structural components such as lignin and cellulose, particularly from non-photosynthetic material in the canopy [3]. The NSWIR however demonstrates the highest degree of conformity for a large region, covering nine bins from 1300–1750 nm.
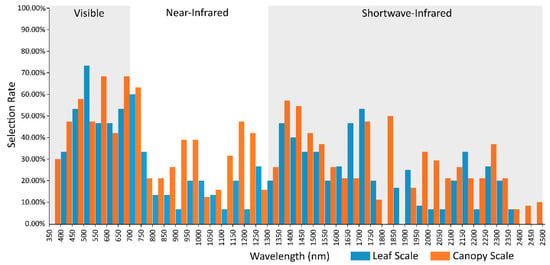
Figure 3.
Feature selection rates for 350–2500 nm studies (Table 2) per 50-nm bins of both canopy and leaf scale spectra.
3. Feature Selection
Feature selection is implemented to select a subset of features to improve generalization and computation requirements while preserving or improving classification accuracy. In this review, feature and waveband selection are used interchangeably. Feature selection techniques are generally divided into three categories: filter, wrapper, and embedded methods. Filter methods are named as such as they act as a pre-processing step that filters out irrelevant features. Filter methods are known to be computationally fast and efficient, though they are generally outperformed by the other methods, as well as not able to handle nonlinear relationships [82].
3.1. Filter Methods
Analysis of variance (ANOVA) is a parametric statistical filter method to determine significant differences between group means. Related to ANOVA is the non-parametric Mann-Whitney U-test, and the Kruskal–Wallis test which extends the Mann–Whitney U-test for more than two groups [45]. Following initial dimensionality reduction by one of these methods a secondary feature selection step to further reduce the number of selected features is used, such as Linear Discriminant Analysis (LDA) [35,52], classification and regression trees (CART) [32,37,39], or the manual selection of known influential bands [8,45,46]. This secondary selection step found important bands in the VIS and SWIR, with a reduced selection of NIR bands [39,52]. However, the reverse was found by [32] where CART secondary selection was restricted to NIR wavelengths. The remainder of the studies manually selected bands that differentiated the greatest number of species pairs [8], or selected known influential bands from the wavelengths that demonstrated high levels of pairwise group variance [45,46].
3.2. Wrapper Methods
Wrapper methods search for a subset of features that gives the best classification performance, with the best performing subset being selected. Although generally considered to outperform filter methods, wrappers are known to be computationally demanding and can suffer from overfitting [82].
Two of the studies reviewed implemented genetic algorithms (GA), in which wavebands are encoded as genes that are subsequently grouped into chromosomes. These chromosomes are allowed to evolve over many generations where their fitness, as determined by a classifier, controls their likelihood to reproduce and pass their genes onto the next generation. Fitness of chromosomes is determined each generation by a chosen classifier, and with the classification accuracy of each chromosome being its fitness score, chromosomes with increased fitness are more likely to reproduce. Both studies used the same dataset of lab measured tropical mangrove leaves [49,50]. The selection of bands differed between the two studies, despite the use of the same dataset and feature selector, though methodologies did differ. The variability of selected bands with similar classification performance seen between these studies demonstrates that multiple band selections can perform classification equally well. The ensemble of chromosomes used in [50] helped to identify key regions for discriminating target species related to biophysical and biochemical aspects of the vegetation that may have been missed if a study was reliant upon the first single chromosome to reach the stopping criterion. This is apparent when comparing the bands selected in both studies, with [49] selecting no VIS bands, resulting in the authors concluding that pigments were not significant for the discrimination of the target species. However, the importance of the VIS, particularly the green region became apparent in [50] where 21 out of 120 total bands were selected from 513 ±19 nm.
Forward feature selection (FFS) is a wrapper method of feature selection that begins with a model containing a single feature that best discriminates the classes, with new features iteratively added to the model based on their ability to improve class discrimination [83]. FFS was implemented by [27] in their comparison between floral and leaf spectra, however, only the results for leaf spectra are discussed here. The leaf spectra within this study were constrained to 475–900 nm at 1 nm increments, with only eight wavebands being selected. These bands came from narrow regions of the spectra, occurring at 450–499 nm in the blue, and the red minimum and red edge from 650–749 nm. In a similar spectral range of 402.9 to 989.1 nm of airborne collected spectra, a very different feature selection trend was observed by [11] following the use of the FFS variant sequential floating feature selection (SFFS). Wavebands were selected from across the entire reduced spectrum, with a notable gap in selection occurring in the NIR between 800 and 849 nm. Selection differences exhibited between these studies could be related to the differences in target species, leaf or canopy scale spectra, or version of FFS used. The only VIS-SWIR study in this review to use FFS applied it to AVIRIS imagery of urban street trees [9]. However, feature selection was only performed to identify spectral regions responsible for species separability, with all bands used for classification. These informative spectral regions matched a number of known informative regions from the literature, such as water absorption in the NIR, cellulose and lignin features in the SWIR, and bands associated with photosynthetic pigments in the VIS. Interestingly however, the highly selected red minimum and red edge were not selected in this study, along with the majority of the NIR.
3.3. Embedded Methods
Despite being described as a wrapper method in [8], recursive feature elimination with a support vector machine (SVM-RFE) is considered to be an embedded method [84]. Embedded methods differ from wrappers, as they do not treat the classifier as a black box, rather, features are selected using information gained whilst training the classifier [85]. A claimed strength of SVM as a classifier is its reported independence of the Hughes effect, or curse of dimensionality [86,87]. However, it has been shown that SVM classifications can be affected by the Hughes effect and can benefit the from dimensionality reduction of its inputs, especially when sample sizes are small [88].
In order to be used as a feature selection method, [8] implemented recursive feature elimination (RFE) with a SVM, determining that from the original 401 bands the optimal number of features to include for classification is 20, after 1–5, 10, 15, 20, and 30 were all evaluated. The 20 bands selected demonstrated a number of trends that were not apparent in the other feature selection methods implemented in the same study. Firstly, the bands formed four distinct contiguous clusters at 520–530 nm, 745–775 nm, 1005–1030 nm, 2295–2305 nm, and then a final single band at 2345 nm. Secondly, the wavelengths of certain selected bands were also unique amongst the methods used, with SVM-RFE being the only method to select bands from the NIR plateau out of all feature selection methods implemented in [8]. Additionally, being the only method to not select bands from the NSWIR. Although not reported in a manner suitable for inclusion in Table 1, [17] also performed feature ranking with a SVM. As with [8], [17] identified the optimal number of features to be between 15 and 20, depending on the dataset, pre-processing, and feature selection methods used. Unlike [8], where the SVM selected bands from distinct contiguous regions, [17] report the SVM selecting bands evenly spread over the entire spectrum.
Random forest (RF) is an ensemble classification method, in which a number of decision tree classifiers are trained from a sub-sample of the dataset, with their results combined via a voting system. One third of samples are retained for validation purposes known as the out-of-bag (OOB) samples, with the remaining in-the-bag samples being used to construct the decision tree [89].
Of the original 72 bands in [29] between 384.8 nm and 1054.3 nm, eight were selected for classification via RF. Although no other feature selection method was implemented in this study, a previous study by [10] performed feature selection with the spectral angle mapper (SAM) add-on Selector using the same data. This resulted in the selection of a far greater 31 bands. Upon binning of the bands at 50 nm, a clear difference in the selection methods are evident (Figure 1). The RF selected bands of [29] are focused in the 400–550 nm region with a single band from the red edge at 706 nm, whereas the SAM bands are focused along the red edge and NIR plateau between 650 and 950 nm, with additional bands in the 350–450 and 1000–1050 nm regions.
As with the bands selected in [29], the RF selected bands in [36] fell within four bins in the VIS and VNIR regions. However, in [29], band selection was focused on the green region with limited selection apparent in the red and NIR plateau with the exception of a single band near the red edge inflection point. This focus was seemingly switched in [36] with bands falling into the bins along the red edge up to the NIR plateau shoulder, with the remaining bin occurring at the blue/green edge. The Chan and Paelinckx study [36] also offers a comparison to an alternative feature selection method using the best-first search (BFS) algorithm as a wrapper. The band selection techniques differ greatly in the VIS and VNIR regions with only the bins at 450–499 and 700–749 in common. However, band selection is more similar at longer wavelengths where the majority of bands were selected by both methods.
The wavebands selected via RF in [8] are in direct opposition to those selected by RF in [36]. Selected bands in [36] mainly occurred along the red edge and NIR plateau shoulder, no band was selected in this region by [8]. Instead, focus was placed on the green, yellow, and red regions of the VIS wavelengths, an area completely ignored by [36] RF selector, though significant for their BFS selection. Additionally, [8] provided the top 20 informative bands determined by a RF classifier using the full 201 waveband dataset. Although these two implementations of RF differed in selecting bands, the overall trend was very similar, with high selection rates in the VIS, low in the NIR, and similar selection throughout the SWIR.
Additionally, a study by [33] produced waveband selections similar to those in [8] with similar results in the VIS with the exception of no selection in the early green (500–549 nm), and selection of the red edge bin rather than the red minimum. The biggest difference between [33] and all other RF studies is the reduced selection at longer wavelengths, although all studies essentially ignored the NIR, [33] only selected two bands from the SWIR, both within the same NSWIR bin at the water absorption feature near 1400–1449 nm.
3.4. Comparison of Stepwise Discriminant Analysis (SDA) with non-SDA Feature Selectors
Stepwise discriminant analysis is a filter method that selects a subset of features by attempting to minimise within-class variation while simultaneously maximising between-class variation [90]. Although a number of metrics are available to determine class separability, Wilk’s lambda is by far the most frequently used to enter and remove variables from the selection in a stepwise manner. Some studies reported Wilk’s lambda approaching zero and becoming asymptotic, indicating near perfect separation of classes [48]. Features selected after this point can be safely removed from the model as they will not substantially increase classification accuracy. This normally resulted in the selection of 10–20 wavebands [5,38,47,48,51].
SDA in general selects wavebands more uniformly across the spectrum than other methods, though the greatest number of selected bands is still found in the VIS (Figure 4). The most significant difference for selection rates is the increased importance of the NIR beyond the red edge. The NIR demonstrates significant selection with the use of SDA in all bar a first derivative dataset from [51], and [38], with the author of the latter suggesting high levels of intraspecific variance due to differences in leaf maturity as the reason no bands were selected in this region.
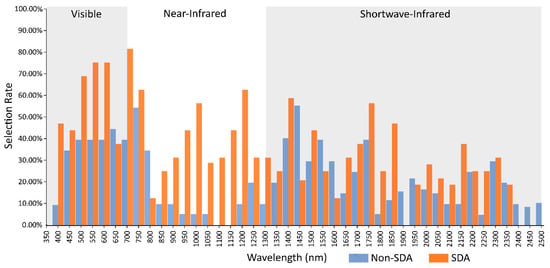
Figure 4.
Feature selection rates for 350–2500 nm studies that used SDA feature selection, and the selection rate of all other feature selection methods combined.
Upon comparing the selection rates of SDA studies compared to non-SDA, a clear difference in selection of NIR bands is apparent. As with the difference between canopy and leaf scale spectra, the increased selection is focused around the NIR water absorption features (Figure 4). Additionally, in the VIS, there is significantly higher selection for the blue, green, and red regions in SDA studies. In order to determine if the spectral acquisition scale or feature selection technique had a greater influence on band selection, the selection rates were further subset into canopy studies using SDA and non-SDA feature selection, and leaf scale studies using SDA and non-SDA selection (Figure 5). It is apparent that the feature selection method has a greater impact on band selection rates, with SDA selecting from the NIR with far greater rates than the non-SDA methods in both canopy and leaf scale studies. The non-SDA methods demonstrated minimal selection in the NIR beyond the red edge for leaf scale spectra, with only a slight increase in selection for canopy spectra focused around the water absorption wavelengths from 1150–1250 nm. The studies that did select from the NIR with leaf scale samples via non-SDA methods stated that the selected bands represented differences in internal reflectance for leaf scale spectra [50]. The blue and red shifts around the green peak for canopy and leaf scale spectra are still evident once the data has been subset into SDA/non-SDA, although it becomes apparent that the high rates of selection in many parts of the VIS is driven by the SDA studies. However, the use of SDA does not explain the selection rates of the VIS for the reduced spectral domain VIS/NIR studies, as only a single study used SDA for feature selection, perhaps indicating an alternate driving force. The red edge demonstrates its robustness to variations in measurement scale and band selection technique as it was frequently selected for all study subsets, although slightly less frequently for leaf scale spectra with non-SDA feature selection.
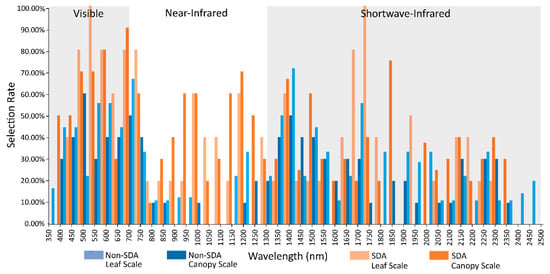
Figure 5.
Feature selection rates for 350–2500 nm studies that used SDA feature selection subset by canopy and leaf scale spectra, and the selection rate of all other feature selection methods combined.
According to [91], “Stepwise analytic methods may be among the most popular research practices employed in both substantive and validity research”. Despite this statement being made in the late 1980s, the use of SDA in approximately a third of the studies included in this review demonstrates its continued popularity, being by far the most used method encountered. However, the widespread use of stepwise methods has prompted strong arguments against its usage [90,92,93,94], particularly when utilised in a predictive discriminant analysis application such as feature selection for classification [95]. The studies that utilised SDA in this review made no mention of these criticisms and therefore no direct attempt to mitigate them. Despite this, [25] did validate their model with 20 repetitions of 1000 random samples, with the final feature subset being based on the selection rates of features across the repetitions, the consideration of important features identified in the literature from [6] and [47], as well as the results from principal component analysis (PCA). PCA is a mathematical transformation used to produce uncorrelated features from the spectral features, reducing dimensionality whilst retaining the most informative spectral data. Additionally, [47], and [5] included SDA as part of an ensemble of feature selection methods, again determining the final feature subset based on the selection rates of features across all methods within the ensemble. Although one of these ensemble methods (Lambda– Lambda plots) allows for the identification and removal of correlated features, in both cases, it was run in parallel to SDA with the removal of correlated features occurring after features had been selected. The remaining studies reported no efforts to mitigate the concerns of using SDA for feature selection [38,41,42,43,48,51].
It must be acknowledged that the sub-setting of reviewed studies into canopy and leaf scale, and then into SDA and non-SDA, meant each class was only represented by a small number of samples (~8 per class), though leaf-SDA was only represented by five studies extracted from two papers. As a result of this, a few outliers are evident, such as the 100% selection in bin 1700–1749 nm, and the 100% selection of the 500–549 nm bin, both associated with the low leaf-SDA sample size. Additionally, the comparison of SDA to non-SDA may disguise selection biases of the non-SDA methods as they are often only represented by one or two studies, with any bias they may exhibit being masked by the selection rates of the other methods.
4. Study Design Influence
All aspects of a study design influence waveband selection. However, many of these aspects may be outside the control or be heavily constrained for the researcher, such as target classes, number of samples and collection method, though the researcher often has control over data pre-processing, feature selection, and classification methods. Due to this, and the apparent influence of feature selectors previously described, we focus on how the choice of feature selection method effects waveband selection.
In order to ascertain any influence feature selection may have over waveband selection, some of the most common feature selection methods were applied to a synthesised dataset. A key requirement for these experiments is the need for a dataset with many species with a large number of samples, something generally lacking in vegetation hyperspectral data. To accomplish this, a hyperspectral synthesis method was created [20] to allow for the creation of any number of samples from 22 species of New Zealand plants. The synthesised dataset consisted of 500 samples per class with 540 wavebands from 350–2450 nm at 3-nm bandwidths, excluding regions of high noise.
Hyperparameters for the feature selectors were tuned via a holdout dataset, with the parameters that selected features resulting in the highest classification accuracy being used for all experiments (Table 3). This is crucial to ensure that the only variables that could affect waveband selection were constrained to either the feature selector (svm_*, sda_*,sffs_*,rf_*) or the dataset (*_0 … *_9).

Table 3.
Software packages and hyperparameters for each feature selection method.
Three experiments were devised. First, each feature selection method was performed on the same dataset, cross-validated 10 times (eg. rf_0, rf_1 … rf_9), selecting the top 30 discriminative wavebands, thus revealing any possible biases in waveband selection resulting from the choice of feature selection method (Figure 6 and Figure 7). Secondly, feature selection was performed on datasets consisting of different classes and samples to simulate many different studies, giving an idea if attributes of the samples affect the wavebands being selected, which will impact generalizability and transferability. Variants of this experiment were performed wherein the classes used remained the same as did the number of samples, though actual samples were randomly selected. Additionally an experiment with the same classes though with differing numbers of samples. Results for these variants did not significantly differ and therefore aren’t shown here.
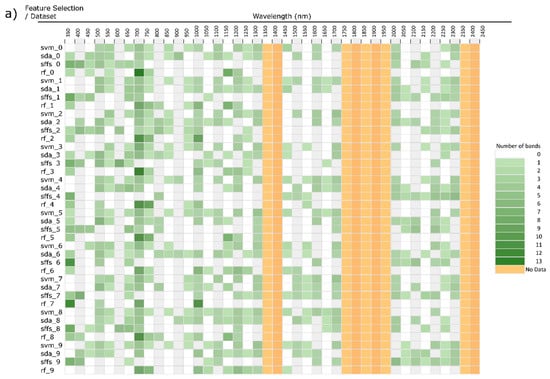
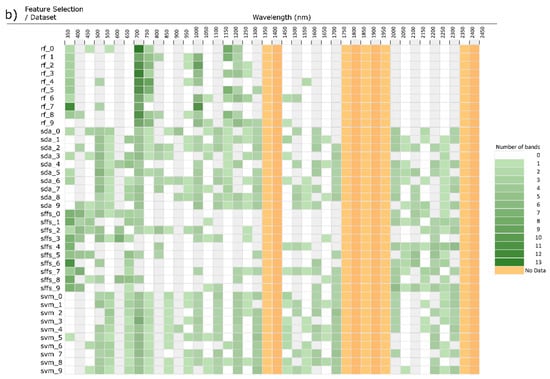
Figure 6.
(a): Histogram of band feature selection binned at 50nm, ordered by dataset. Four feature selectors run on the same dataset 10 cross-validation (new dataset consisting of 10 classes and 200 samples for each cross-val.). (b): Results of Figure 5a ordered by feature selection method. (RF = random forest, SDA = stepwise discriminant analysis, SFFS = sequential floating feature selection, SVM = support vector machine).
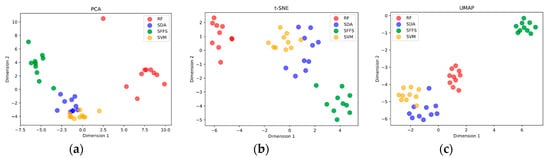
Figure 7.
(a) PCA dimensional reduction of histogram waveband feature selection. (b) t-Distributed Stochastic Neighbor Embedding (T-SNE) dimensional reduction of histogram waveband feature selection. (c) Uniform Manifold Approximation and Projection (UMAP) dimensional reduction of histogram waveband feature selection.
Each dataset produced significantly different waveband selections. This is especially evident in Figure 6b were the histogram is ordered by feature selector, placing each repetition with a new dataset next to each other. Here, it is clear the RF favours the red edge and NIR bands, essentially ignoring the SWIR. SFFS demonstrated higher selection in the VIS, especially at shorter wavelengths, minimal selection in the NIR, and medium selection in late SWIR. SDA and SVM are the most similar due to both selecting broadly and relatively evenly along the entire spectrum. Dimensionality reduction techniques offer a way to visualize the relationship between selection histograms (Figure 7). Due to their broad general selection, SDA and SVM are grouped close to each other with SFFS and RF adjacent though separate. Further, the histograms are clearly grouped by feature selection method rather than dataset, indicating that feature selection method is a dominant factor affecting the selection of wavebands.
5. Discussion
This review of hyperspectral vegetation classification literature has determined that every aspect of a study can greatly influence selected wavebands and classification performance. However, despite this, we have identified some important and consistent patterns that appear throughout the literature. Visible wavelengths and their associations with photosynthetic pigments have played important discriminatory roles in a wide range of studies, their high levels of selection clearly evident in this review (Figure 1). Selection rates in the VIS showed only minor variations between VIS/SWIR studies and the VIS/NIR (Figure 1), although the comparisons between canopy and leaf scale spectra demonstrated significant differences (Figure 3). The discriminatory value of the red edge has been well documented with its close relationship to chlorophyll concentration and structural features. This is reflected in the consistently high rates of selection of the red edge as well as the robustness of its selection with only minor variation in magnitude between the comparisons. The inclusion of structural features in canopy spectra can provide high levels of interspecific variation in the NIR, primarily in the form of differences in albedo, rather than spectral shape [68]. However, selection rates from the non-red edge NIR are low, with the selected bands generally being related to water absorption features and potentially high levels of within-class variability. Additionally, the NIR has demonstrated the greatest degree of variability between the canopy and leaf scale spectra studies. Wavebands selected in the SWIR are associated with water absorption and non-photosynthetic biochemicals, with selection rates heavily skewed towards the NSWIR over the FSWIR.
The reported importance of NIR bands [45,52] seems to be contentious, primarily being driven by the use of a single feature selection technique. Comparisons between selection rates for the NIR with and without the use of SDA as the feature selector are starkly contrasted, with the importance of NIR being significantly higher with the use of SDA. The criticisms of SDA and stepwise methods in general perhaps offer an answer to the selection biases presented in this review.
It is apparent that there is no single best feature selection method, with the same method performing very differently within and between studies. This suggests that either multiple methods should be applied to the data, or an ensemble of multiple methods may be the best practice, a conclusion recognized by this review, and previously suggested by some studies [36]. Additionally, multiple subsets of selected features have proven to discriminate species equally well [8], or alternatively, no feature selection, with the original data outperforming feature selected subsets [7,9,36]. Additionally, as computation power, dataset sizes, and machine learning techniques all increase, the need for feature selection as a data reduction technique becomes less necessary.
6. Conclusions
This review has established that the variability in waveband selection seen between studies, driven by study parameters beyond the characteristics of the target samples, prevents the determination of generalizable, high utility spectral regions for specific taxonomic discrimination. Broad trends such as the importance of VIS and red edge wavelengths are apparent, independent of plant groupings, though in and of themselves they are not sufficiently specific for taxonomic discrimination. The possibility of discriminatory spectral regions being associated with specific taxonomic, structural, or functional groupings of plants is inconclusive due to the large degree of variability between studies. This is further highlighted by the apparent dominance of feature selector choice over other parameters for waveband selection (Figure 6 and Figure 7). Building on this review, future works could investigate variance in waveband selection caused by the hyperparameter choice of feature selectors, data preprocessing, as well as the inclusion of vegetation indices.
Author Contributions
Conceptualization, A.H.; methodology, A.H.; data curation, A.H.; formal analysis, A.H.; writing—original draft preparation, A.H., K.C. and M.L.; writing—review and editing, A.H., K.C., and M.L.; supervision, K.C., M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
Financial support for this research was provided by the Australian Government Research Training Program Scholarship and the University of Adelaide School of Biological Sciences.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Fernandes, A.; Melo-Pinto, P.; Millan, B.; Tardaguila, J.; Diago, M. Automatic discrimination of grapevine (Vitis vinifera L.) clones using leaf hyperspectral imaging and partial least squares. J. Agric. Sci. 2015, 153, 455–465. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Deng, W.; Huang, Y.; Zhao, C.; Chen, L.; Wang, X. Bayesian discriminant analysis of plant leaf hyperspectral reflectance for identification of weeds from cabbages. Afr. J. Agric. Res. 2016, 11, 551–562. [Google Scholar]
- Mariotto, I.; Thenkabail, P.S.; Huete, A.; Slonecker, E.T.; Platonov, A. Hyperspectral versus multispectral crop-productivity modeling and type discrimination for the HyspIRI mission. Remote Sens. Environ. 2013, 139, 291–305. [Google Scholar] [CrossRef]
- Lewis, M. Spectral characterization of Australian arid zone plants. Can. J. Remote Sens. 2002, 28, 219–230. [Google Scholar] [CrossRef]
- Sommer, C.; Holzwarth, S.; Heiden, U.; Heurich, M.; Müller, J.; Mauser, W. Feature based tree species classification using hyperspectral and lidar data in the Bavarian Forest National Park. In Proceedings of the 9th EARSeL Imaging Spectroscopy Workshop, Luxembourg, France, 14–16 April 2015; Volume 14, pp. 49–70. [Google Scholar]
- Prospere, K.; McLaren, K.; Wilson, B. Plant species discrimination in a tropical wetland using in situ hyperspectral data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Cho, M.A.; Debba, P.; Mathieu, R.; Naidoo, L.; Van Aardt, J.; Asner, G.P. Improving discrimination of savanna tree species through a multiple-endmember spectral angle mapper approach: Canopy-level analysis. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4133–4142. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Alonso, M.C.; Malpica, J.A.; de Agirre, A.M. Consequences of the Hughes phenomenon on some classification techniques. In Proceedings of the ASPRS 2011 Annual Conference, Milwaukee, WI, USA, 1–5 May 2011; pp. 1–5. [Google Scholar]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Settles, B. Active Learning Literature Survey; University of Wisconsin-Madison Department of Computer Sciences: Madison, WI, USA, 2009. [Google Scholar]
- Zhu, X.J. Semi-Supervised Learning Literature Survey; University of Wisconsin-Madison Department of Computer Sciences: Madison, WI, USA, 2005. [Google Scholar]
- Van Dyk, D.A.; Meng, X.-L. The art of data augmentation. J. Comput. Graph. Stat. 2001, 10, 1–50. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Förster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of feature reduction algorithms for classifying tree species with hyperspectral data on three central European test sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Richter, R.; Reu, B.; Wirth, C.; Doktor, D.; Vohland, M. The use of airborne hyperspectral data for tree species classification in a species-rich central European forest area. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 464–474. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Hennessy, A.; Lewis, M.; Clarke, K. Generative adversarial network synthesis of hyperspectral vegetation data. 2019; Unpublished manuscript, last modified 20 October 2019. [Google Scholar]
- Hueni, A. Field Spectroradiometer Data: Acquisition, Organisation, Processing and Analysis on the Example of New Zealand Native Plants. Master’s Thesis, Massey University, Palmerston North, New Zealand, 2006. [Google Scholar]
- Aneece, I.; Epstein, H. Distinguishing early successional plant communities using ground-level hyperspectral data. Remote Sens. 2015, 7, 16588–16606. [Google Scholar] [CrossRef]
- Cao, J.; Liu, K.; Liu, L.; Zhu, Y.; Li, J.; He, Z. Identifying mangrove species using field close-range snapshot hyperspectral imaging and machine-learning techniques. Remote Sens. 2018, 10, 2047. [Google Scholar] [CrossRef]
- Dian, Y.; Fang, S.; Le, Y.; Xu, Y.; Yao, C. Comparison of the different classifiers in vegetation species discrimination using hyperspectral reflectance data. J. Indian Soc. Remote Sens. 2014, 42, 61–72. [Google Scholar] [CrossRef]
- Eddy, P.; Smith, A.; Hill, B.; Peddle, D.; Coburn, C.; Blackshaw, R. Weed and crop discrimination using hyperspectral image data and reduced bandsets. Can. J. Remote Sens. 2014, 39, 481–490. [Google Scholar] [CrossRef]
- Fung, T.; Yan Ma, H.F.; Siu, W.L. Band selection using hyperspectral data of subtropical tree species. Geocarto Int. 2003, 18, 3–11. [Google Scholar] [CrossRef]
- Gross, J.W.; Heumann, B.W. Can flowers provide better spectral discrimination between herbaceous wetland species than leaves? Remote Sens. Lett. 2014, 5, 892–901. [Google Scholar] [CrossRef]
- Hoa, P.; Giang, N.; Binh, N.; Hieu, N.; Trang, N.; Toan, L.; Long, V.; Ai, T.; Hong, P.; Hai, L. Mangrove species discrimination in southern Vietnam based on in-situ measured hyperspectral reflectance. Int. J. Geoinform. 2017, 13, 25–35. [Google Scholar]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the greater Kruger national park region, by integrating hyperspectral and LiDAR data in a random forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Peerbhay, K.Y.; Mutanga, O.; Ismail, R. Commercial tree species discrimination using airborne AISA Eagle hyperspectral imagery and partial least squares discriminant analysis (PLS-DA) in KwaZulu–Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2013, 79, 19–28. [Google Scholar] [CrossRef]
- Pu, R.; Bell, S.; Baggett, L.; Meyer, C.; Zhao, Y. Discrimination of seagrass species and cover classes with in situ hyperspectral data. J. Coast. Res. 2012, 28, 1330–1344. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O. Spectral discrimination of papyrus vegetation (Cyperus papyrus L.) in swamp wetlands using field spectrometry. ISPRS J. Photogramm. Remote Sens. 2009, 64, 612–620. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D.; Ismail, R. Discriminating the papyrus vegetation (Cyperus papyrus L.) and its co-existent species using random forest and hyperspectral data resampled to HYMAP. Int. J. Remote Sens. 2012, 33, 552–569. [Google Scholar] [CrossRef]
- Aneece, I.; Thenkabail, P. Accuracies achieved in classifying five leading world crop types and their growth stages using optimal earth observing-1 hyperion hyperspectral narrowbands on google earth engine. Remote Sens. 2018, 10, 2027. [Google Scholar] [CrossRef]
- Beh, B.C.; Tan, K.C.; Jafri, M.Z.M.; San Lim, H. Comparison of different discriminant functions for mangrove species analysis in Matang Mangrove Forest Reserve (MMFR), Perak based on statistical approach. In Proceedings of the Remote Sensing for Agriculture, Ecosystems, and Hydrology XIX, Maspalomas, Spain, 14–16 September 2004; p. 104211U. [Google Scholar]
- Chan, J.C.-W.; Paelinckx, D. Evaluation of random forest and adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Das, B.; Sahoo, R.N.; Biswas, A.; Pargal, S.; Krishna, G.; Verma, R.; Chinnusamy, V.; Sehgal, V.K.; Gupta, V.K. Discrimination of rice genotypes using field spectroradiometry. Geocarto Int. 2018, 35, 64–77. [Google Scholar] [CrossRef]
- Datt, B. Recognition of eucalyptus forest species using hyperspectral reflectance data. In Proceedings of the IGARSS 2000. IEEE 2000 International Geoscience and Remote Sensing Symposium, Honolulu, HI, USA, 24–28 July 2000; pp. 1405–1407. [Google Scholar]
- Fernandes, M.R.; Aguiar, F.C.; Silva, J.M.; Ferreira, M.T.; Pereira, J.M. Spectral discrimination of giant reed (Arundo donax L.): A seasonal study in riparian areas. ISPRS J. Photogramm. Remote Sens. 2013, 80, 80–90. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- George, R.; Padalia, H.; Kushwaha, S. Forest tree species discrimination in western Himalaya using EO-1 Hyperion. Int. J. Appl. Earth Obs. Geoinf. 2014, 28, 140–149. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Employing ground-based spectroscopy for tree-species differentiation in the Gulf Islands National Park Reserve. Int. J. Remote Sens. 2010, 31, 1121–1127. [Google Scholar] [CrossRef]
- Papeş, M.; Tupayachi, R.; Martinez, P.; Peterson, A.; Powell, G. Using hyperspectral satellite imagery for regional inventories: A test with tropical emergent trees in the Amazon basin. J. Veg. Sci. 2010, 21, 342–354. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Tree species classification of the UNESCO man and the biosphere karkonoski national park (poland) using artificial neural networks and APEX hyperspectral images. Remote Sens. 2018, 10, 1111. [Google Scholar] [CrossRef]
- Schmidt, K.; Skidmore, A. Spectral discrimination of vegetation types in a coastal wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Shang, X.; Chisholm, L.A. Classification of Australian native forest species using hyperspectral remote sensing and machine-learning classification algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2481–2489. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Van Der Meer, B. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of hyperspectral narrowbands (HNBs) and composition of hyperspectral twoband vegetation indices (HVIs) for biophysical characterization and discrimination of crop types using field reflectance and Hyperion/EO-1 data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Ongsomwang, S.; Vaiphasa, T.; Skidmore, A.K. Tropical mangrove species discrimination using hyperspectral data: A laboratory study. Estuar. Coast. Shelf Sci. 2005, 65, 371–379. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Skidmore, A.K.; de Boer, W.F.; Vaiphasa, T. A hyperspectral band selector for plant species discrimination. ISPRS J. Photogramm. Remote Sens. 2007, 62, 225–235. [Google Scholar] [CrossRef]
- Van Aardt, J.; Wynne, R. Examining pine spectral separability using hyperspectral data from an airborne sensor: An extension of field-based results. Int. J. Remote Sens. 2007, 28, 431–436. [Google Scholar] [CrossRef]
- Wang, J.; Xu, R.; Yang, S. Estimation of plant water content by spectral absorption features centered at 1450 nm and 1940 nm regions. Environ. Monit. Assess. 2009, 157, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P. Biophysical and biochemical sources of variability in canopy reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Rivard, B.; Sanchez-Azofeifa, G.; Foley, S.; Calvo-Alvarado, J. Species classification of tropical tree leaf reflectance and dependence on selection of spectral bands. Hyperspect. Remote Sens. Trop. Sub-Trop. For. 2008, 6, 141–159. [Google Scholar]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Thenkabail, P.; Smith, R.; De Pauw, E. Hyperspectral Vegetation Indices for Determining Agricultural Crop Characteristics, CEO Research Publication Series No. 1; Center for Earth Observation, Yale University Press: New Haven, CT, USA, 1999. [Google Scholar]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Evaluation of narrowband and broadband vegetation indices for determining optimal hyperspectral wavebands for agricultural crop characterization. Photogramm. Eng. Remote Sens. 2002, 68, 607–622. [Google Scholar]
- Ferreira, M.P.; Grondona, A.E.B.; Rolim, S.B.A.; Shimabukuro, Y.E. Analyzing the spectral variability of tropical tree species using hyperspectral feature selection and leaf optical modeling. J. Appl. Remote Sens. 2013, 7, 073502. [Google Scholar] [CrossRef]
- Galvão, L.S.; Roberts, D.A.; Formaggio, A.R.; Numata, I.; Breunig, F.M. View angle effects on the discrimination of soybean varieties and on the relationships between vegetation indices and yield using off-nadir Hyperion data. Remote Sens. Environ. 2009, 113, 846–856. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2006, 58, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Gausman, H. Leaf reflectance vs. leaf chlorophyll and carotenoid concentrations for eight crops. Agron. J. 1977, 69, 799–802. [Google Scholar] [CrossRef]
- Castro-Esau, K.L.; Sánchez-Azofeifa, G.A.; Rivard, B.; Wright, S.J.; Quesada, M. Variability in leaf optical properties of Mesoamerican trees and the potential for species classification. Am. J. Bot. 2006, 93, 517–530. [Google Scholar] [CrossRef]
- Pu, R. Broadleaf species recognition with in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 2759–2779. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W. The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996, 1, 21–26. [Google Scholar] [CrossRef]
- Gamon, J.; Penuelas, J.; Field, C. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Gong, P.; Pu, R.; Yu, B. Conifer species recognition: An exploratory analysis of in situ hyperspectral data. Remote Sens. Environ. 1997, 62, 189–200. [Google Scholar] [CrossRef]
- Van Aardt, J.A. Spectral separability among six southern tree species. Photogramm. Eng. Remote Sens. 2001, 67, 1367–1375. [Google Scholar]
- Karlovska, A.; Grīnfelde, I.; Alsiņa, I.; Priedītis, G.; Roze, D. Plant reflected spectra depending on biological characteristics and growth conditions. In Proceedings of the International Scientific Conference Rural Development, Akademija, Lithuania, 23–24 November 2017. [Google Scholar]
- Clevers, J.; De Jong, S.; Epema, G.; Van Der Meer, F.; Bakker, W.; Skidmore, A.; Scholte, K. Derivation of the red edge index using the MERIS standard band setting. Int. J. Remote Sens. 2002, 23, 3169–3184. [Google Scholar] [CrossRef]
- Dawson, T.; Curran, P. Technical note A new technique for interpolating the reflectance red edge position. Int. J. Remote Sens. 1998, 19, 2133–2139. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Mišurec, J.; Kopačková, V.; Mielke, C.; Rogass, C. Assessment of red-edge position extraction techniques: A Case study for norway spruce forests using hymap and simulated sentinel-2 data. Forests 2016, 7, 226. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Vescovo, L.; Gianelle, D. The role of spectral resolution and classifier complexity in the analysis of hyperspectral images of forest areas. Remote Sens. Environ. 2009, 113, 2345–2355. [Google Scholar] [CrossRef]
- Cochrane, M. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2000, 21, 2075–2087. [Google Scholar] [CrossRef]
- Rock, B.; Hoshizaki, T.; Miller, J. Comparison of in situ and airborne spectral measurements of the blue shift associated with forest decline. Remote Sens. Environ. 1988, 24, 109–127. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Kumar, L. A comparison of reflectance characteristics of some Australian eucalyptus species based on high spectral resolution data—Discriminating using the visible and NIR regions. J. Spat. Sci. 2007, 52, 51–64. [Google Scholar] [CrossRef]
- Kumar, L.; Skidmore, A.K.; Mutanga, O. Leaf level experiments to discriminate between eucalyptus species using high spectral resolution reflectance data: Use of derivatives, ratios and vegetation indices. Geocarto Int. 2010, 25, 327–344. [Google Scholar] [CrossRef]
- Elvidge, C.D. Visible and near infrared reflectance characteristics of dry plant materials. Remote Sens. 1990, 11, 1775–1795. [Google Scholar] [CrossRef]
- Lehmann, J.R.K.; Große-Stoltenberg, A.; Römer, M.; Oldeland, J. Field spectroscopy in the VNIR-SWIR region to discriminate between Mediterranean native plants and exotic-invasive shrubs based on leaf tannin content. Remote Sens. 2015, 7, 1225–1241. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Alonso-Atienza, F.; Rojo-Álvarez, J.L.; Rosado-Muñoz, A.; Vinagre, J.J.; García-Alberola, A.; Camps-Valls, G. Feature selection using support vector machines and bootstrap methods for ventricular fibrillation detection. Expert Syst. Appl. 2012, 39, 1956–1967. [Google Scholar] [CrossRef]
- Pudil, P.; Novovičová, J.; Kittler, J. Floating search methods in feature selection. Pattern Recognit. Lett. 1994, 15, 1119–1125. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Deng, H.; Runger, G. Feature selection via regularized trees. In Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–8. [Google Scholar]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Assessment of the effectiveness of support vector machines for hyperspectral data. Future Gener. Comput. Syst. 2004, 20, 1215–1225. [Google Scholar] [CrossRef]
- Pal, M.; Foody, G.M. Feature selection for classification of hyperspectral data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Huberty, C.J. Applied Discriminant Analysis; Wiley-Interscience: New York, NY, USA, 1994. [Google Scholar]
- Thompson, B. Why Wont Stepwise Methods Die; American Counseling Association: Alexandria, VA, USA, 1989. [Google Scholar]
- Thompson, B. Stepwise Regression and Stepwise Discriminant Analysis Need Not Apply Here: A Guidelines Editorial; SAGE Publications: Thousand Oaks, CA, USA, 1995. [Google Scholar]
- Flom, P.L.; Cassell, D.L. Stopping stepwise: Why stepwise and similar selection methods are bad, and what you should use. In Proceedings of the NorthEast SAS Users Group Inc 20th Annual Conference, Baltimore, MD, USA, 11–14 November 2007. [Google Scholar]
- Whitaker, J.S. Use of Stepwise Methodology in Discriminant Analysis; The Annual Meeting of the Southwest Educational Research Association; Texas A & M University: College Station, TX, USA, 1997; p. 18. [Google Scholar]
- Huberty, C.J.; Barton, R.M. An Introduction to Discriminant Analysis. Meas. Eval. Couns. Dev. 1989, 22, 158–168. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).