Spatial–Spectral Squeeze-and-Excitation Residual Network for Hyperspectral Image Classification
Abstract
:1. Introduction
2. Spatial-Spectral Squeeze-and-Excitation Residual Network
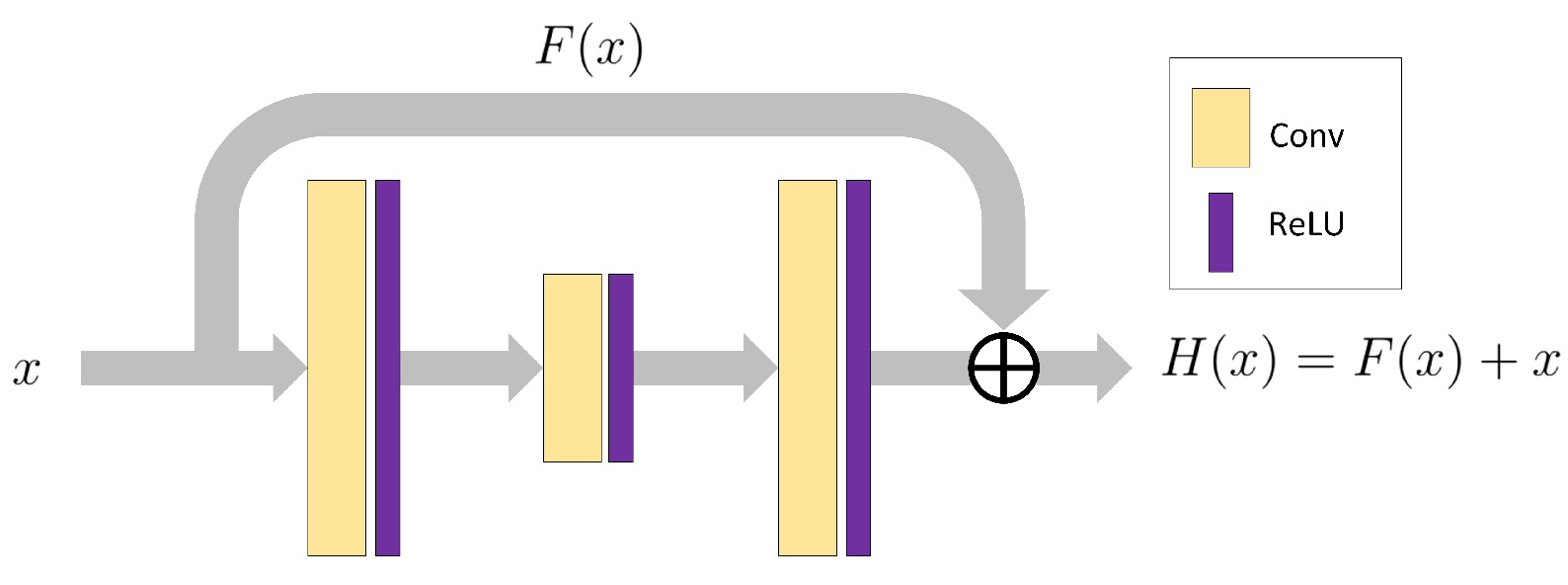
2.1. Residual Connections
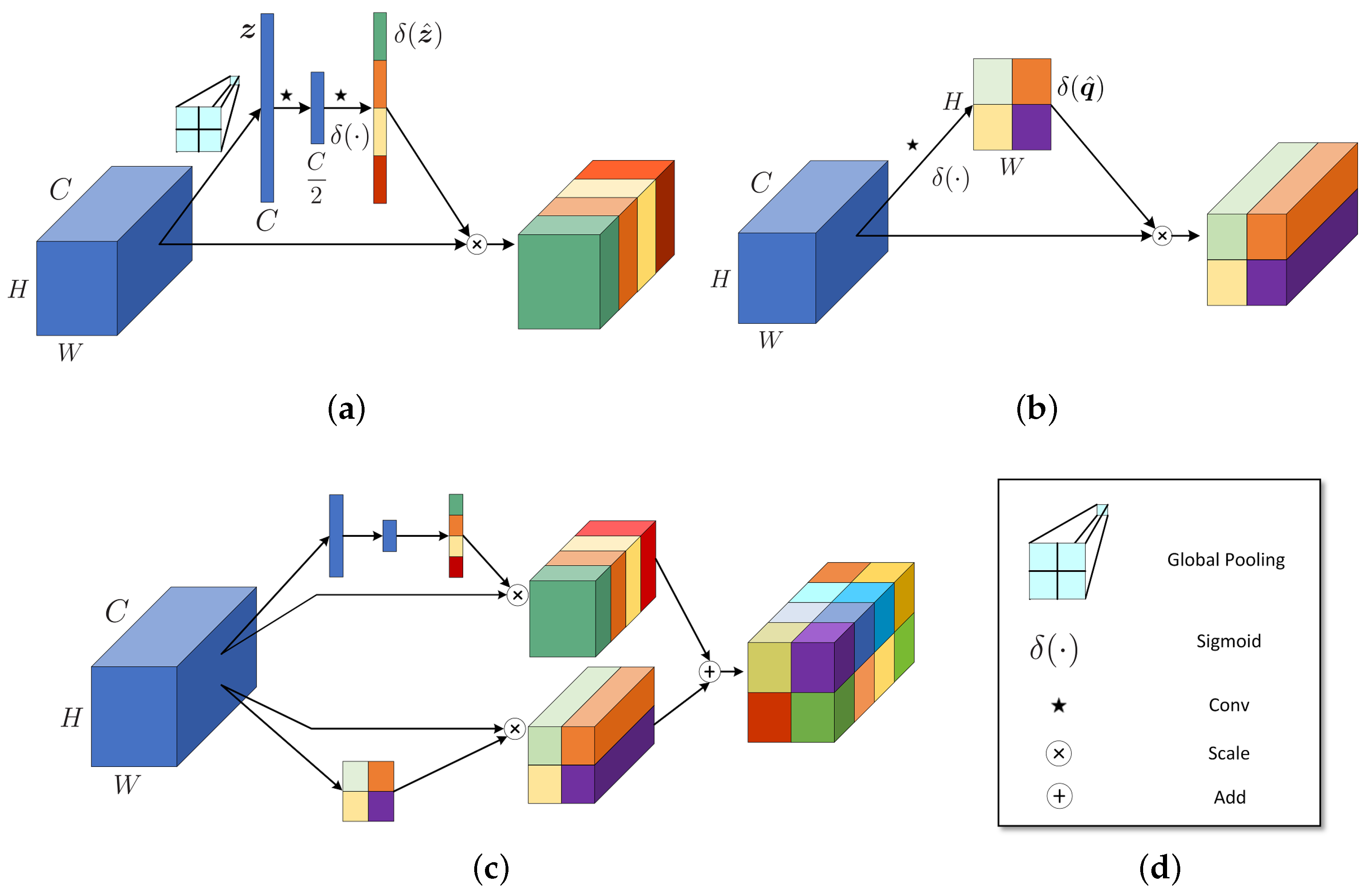
2.2. SpectralSE: Squeeze Spatial Information and Excite Spectral Features
2.3. SpatialSE: Squeeze Spectral Information and Excite Spatial Features
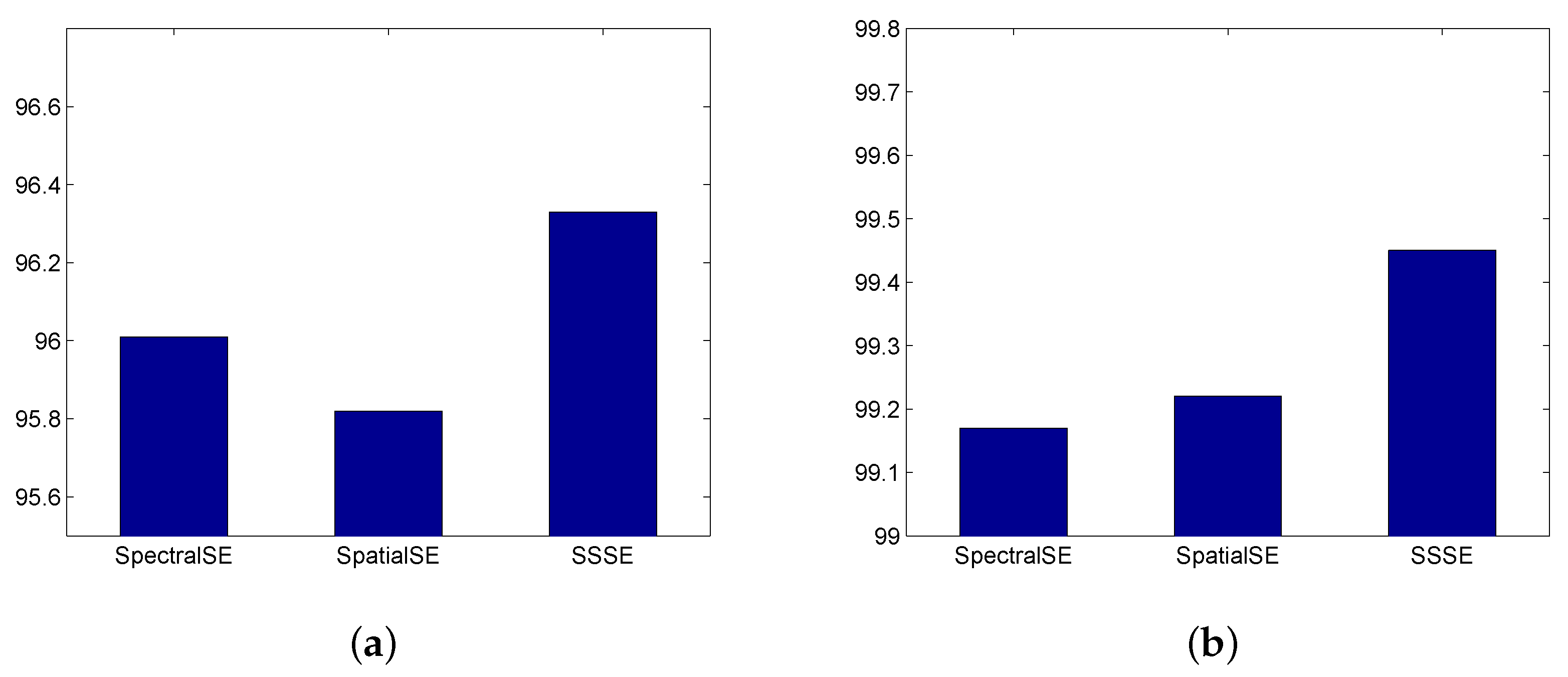
2.4. SSSE: Combination of SpectralSE and SpatialSE
2.5. SSSERN: Spatial-Spectral Squeeze-and-Excitation Residual Network
3. Experiments Results
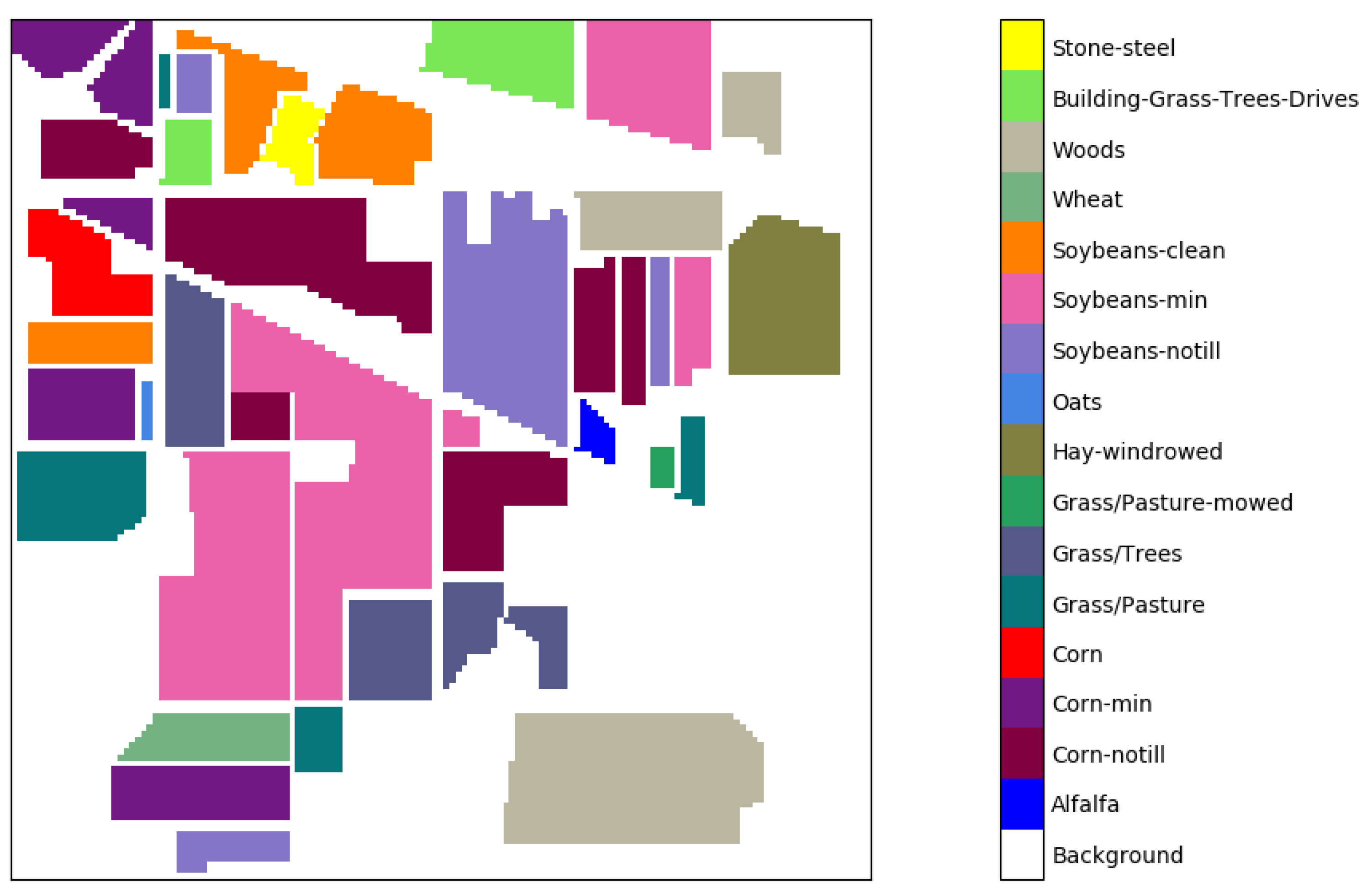
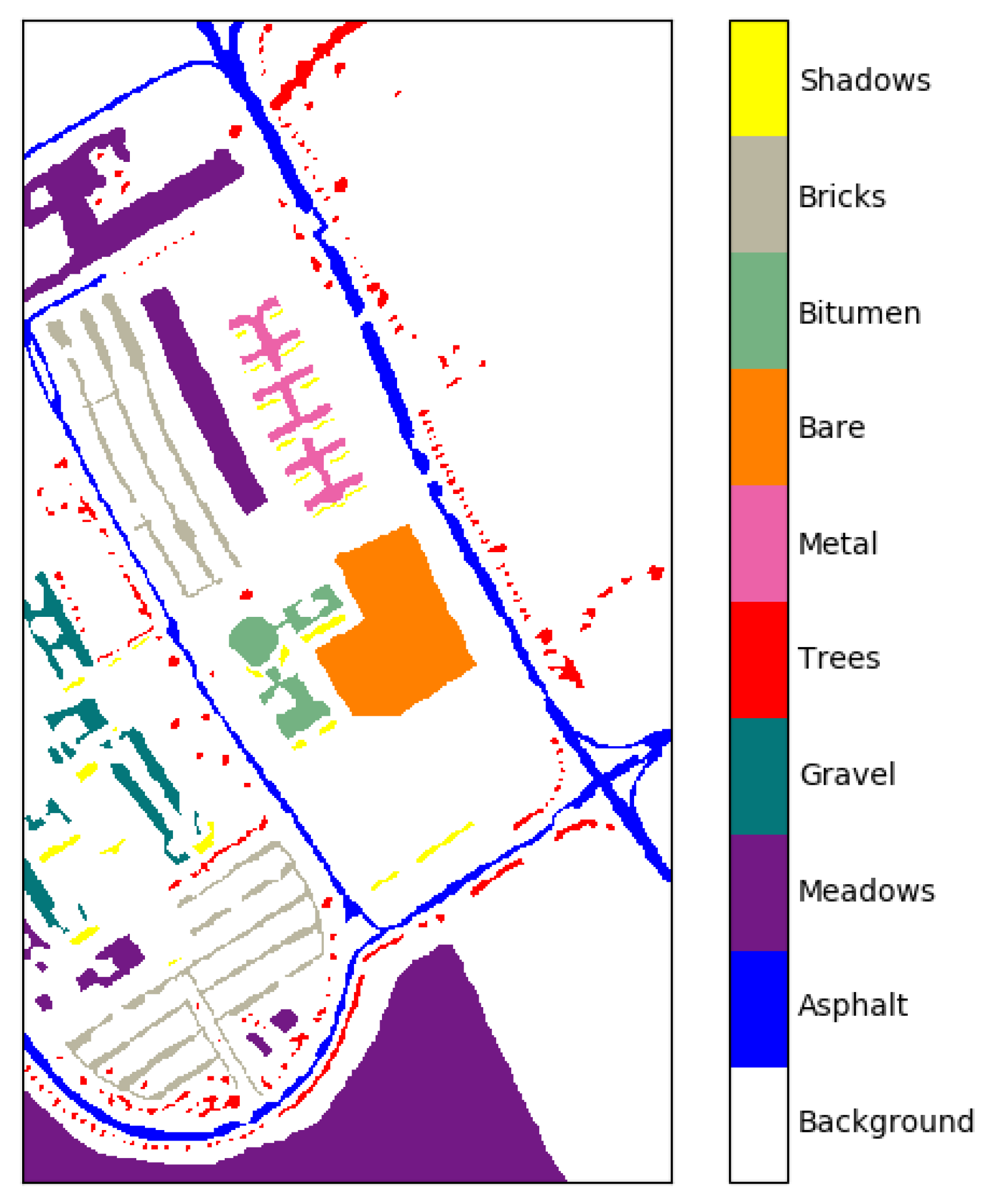
3.1. Datasets
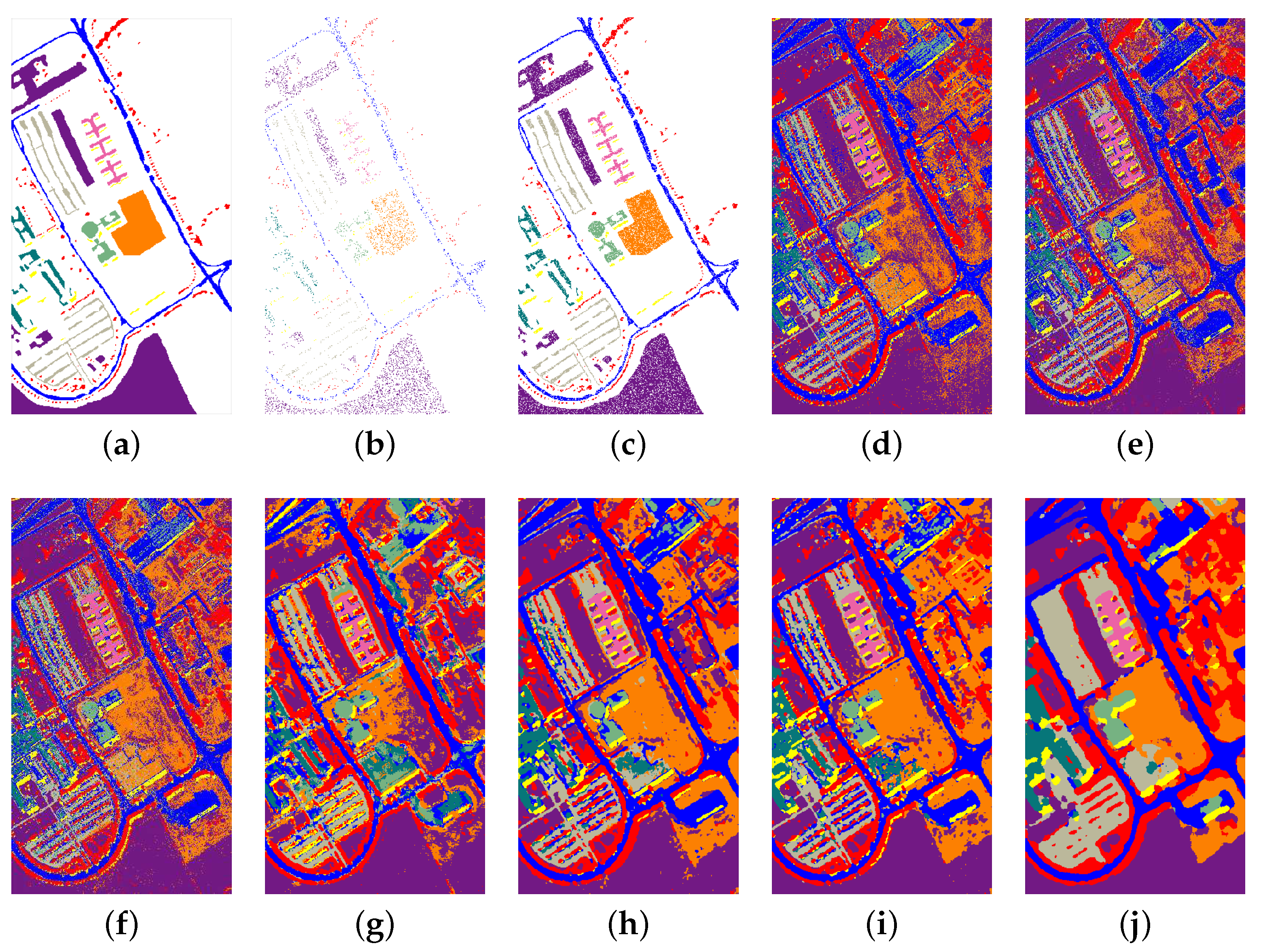
3.2. Classification Performance on Indian Pines and University of Pavia Data Sets
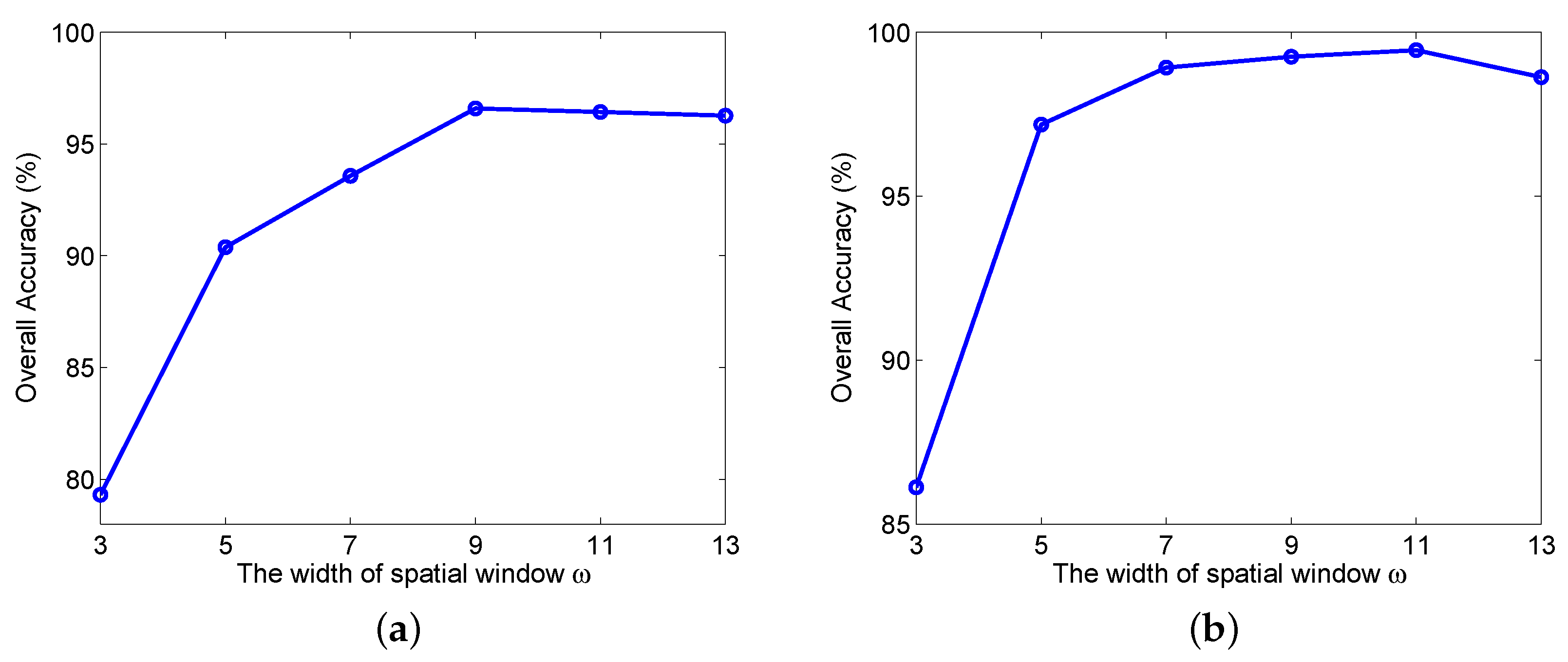
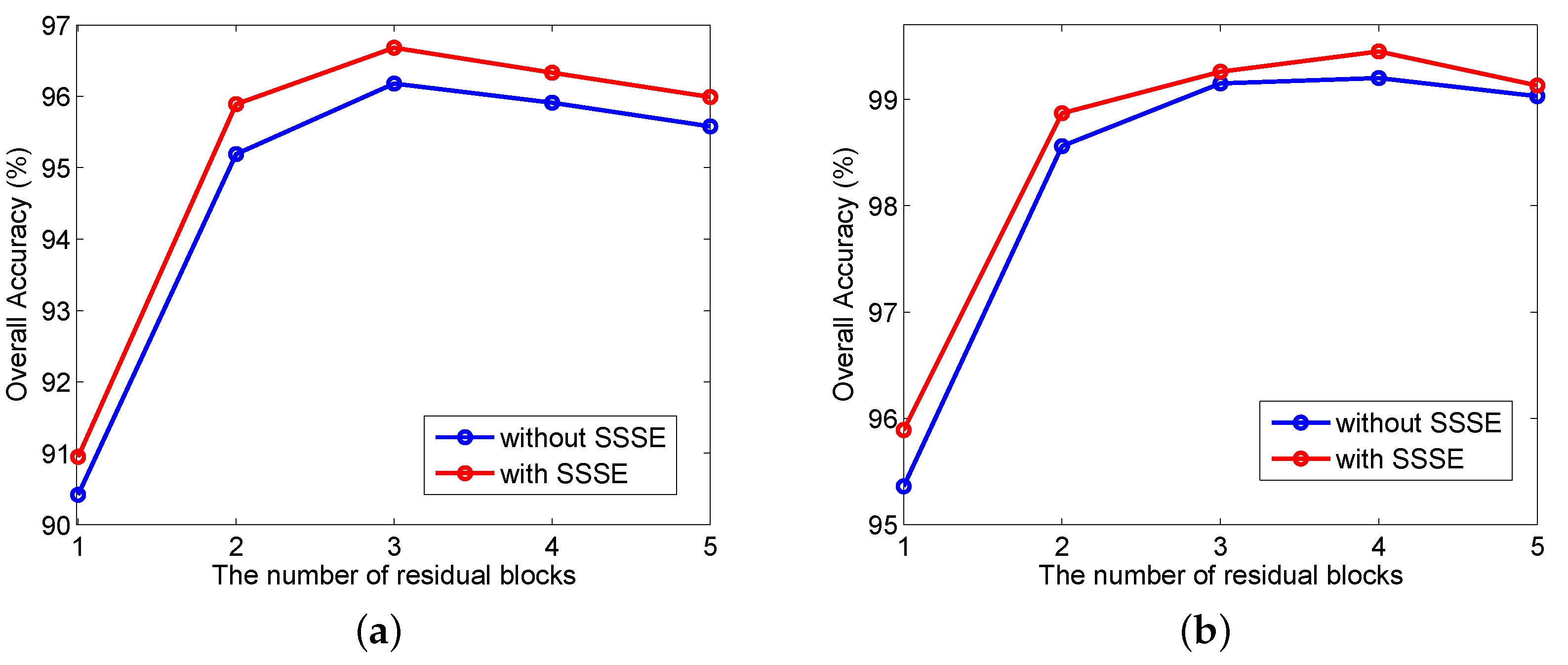
3.3. Investigation on the Effect of Network Parameters
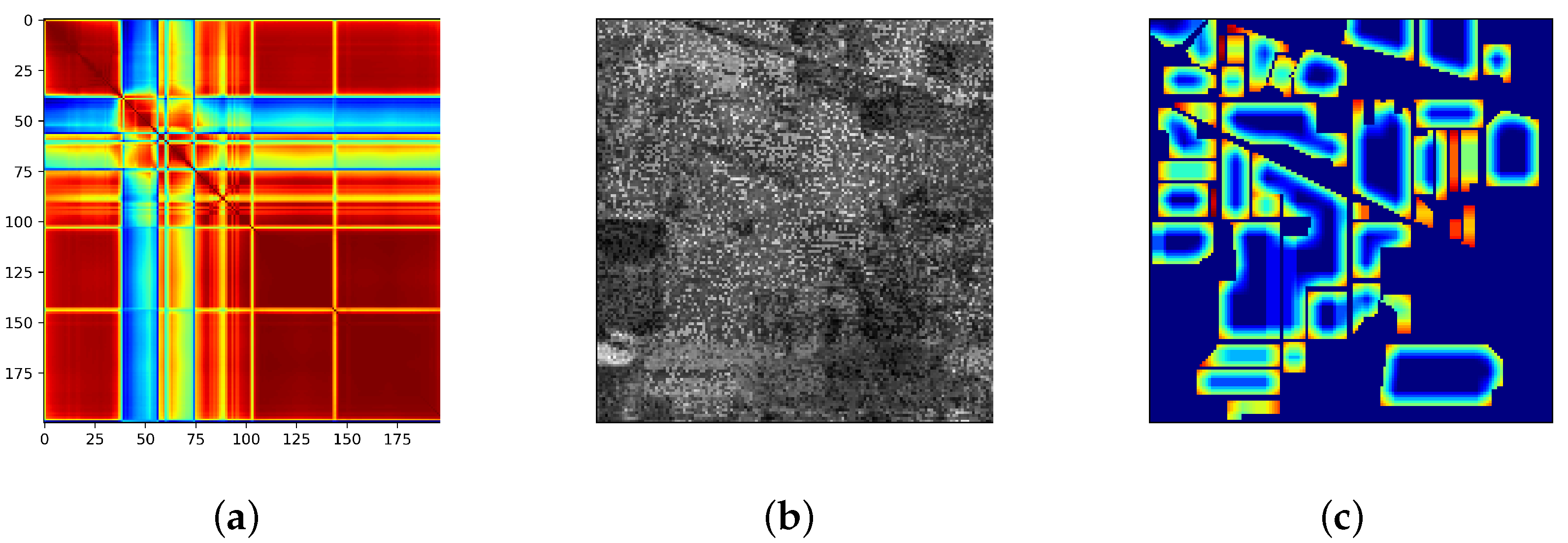
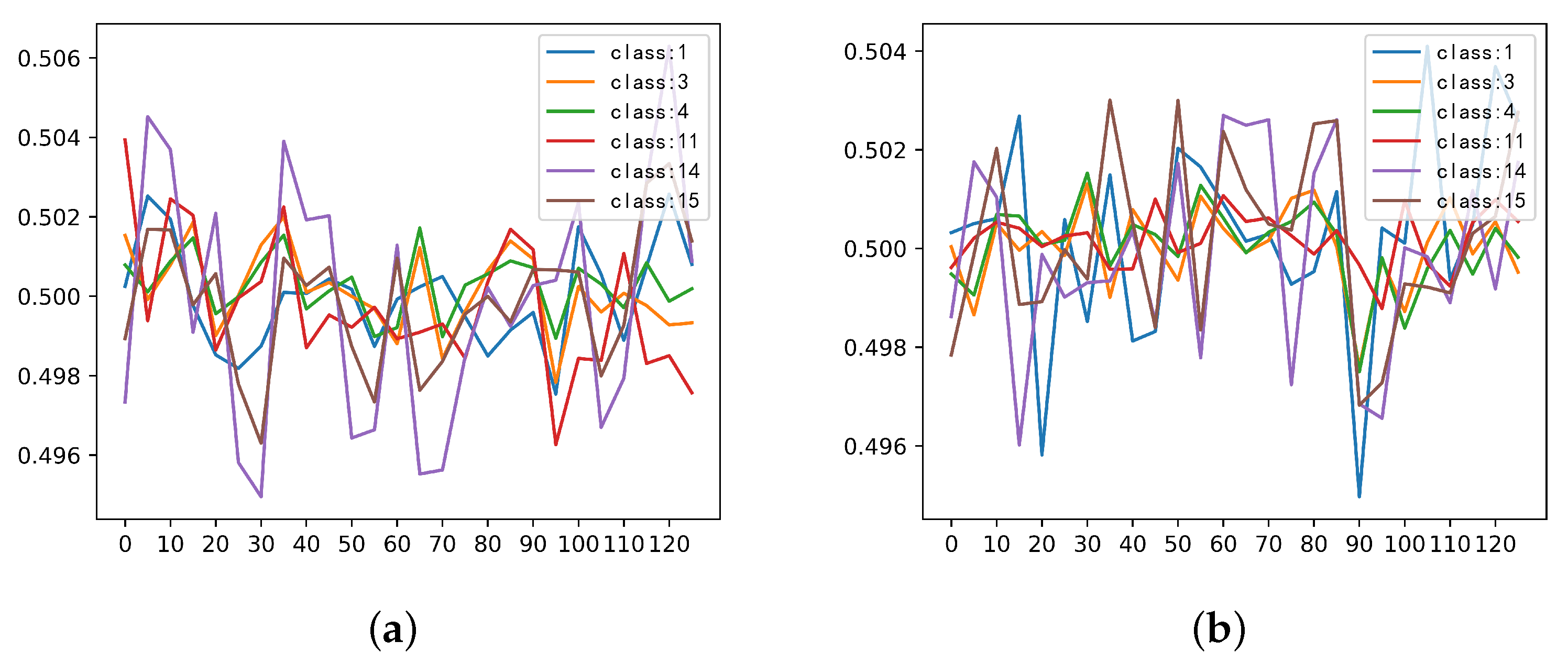
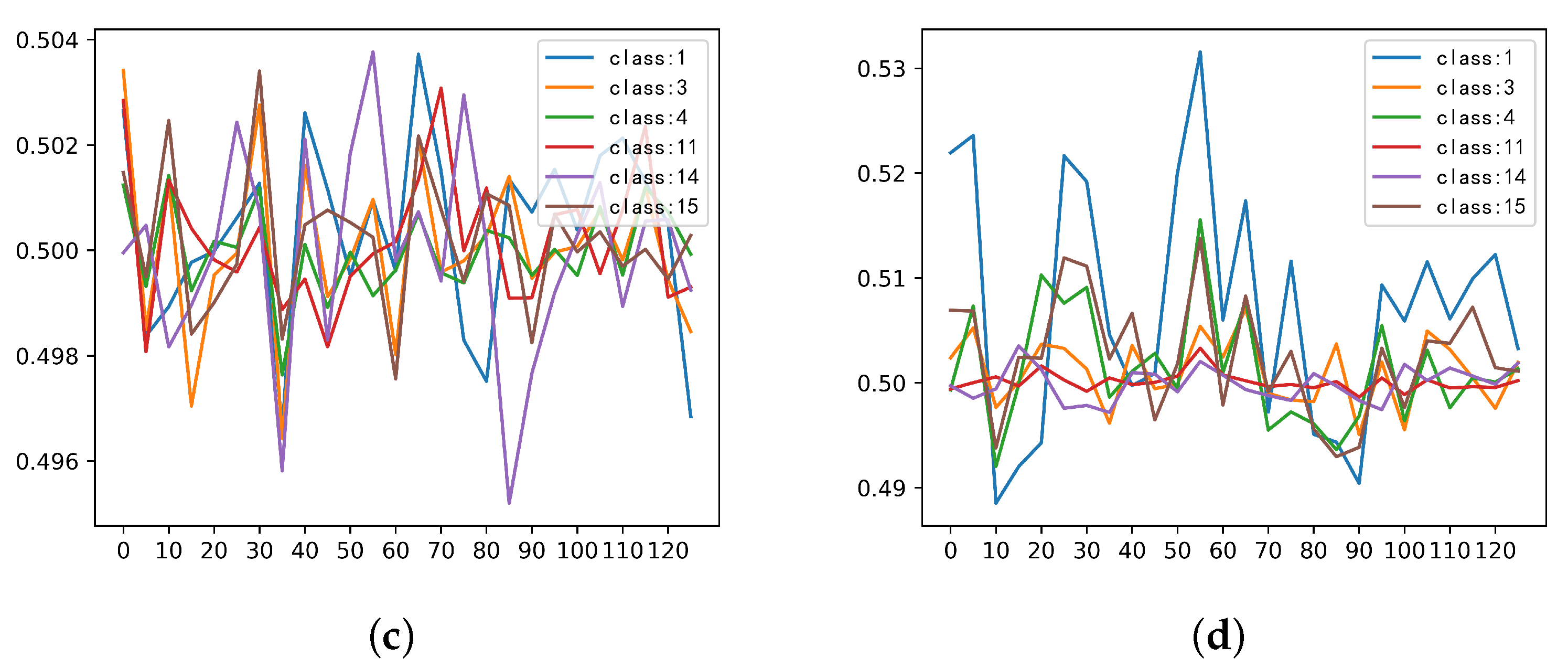
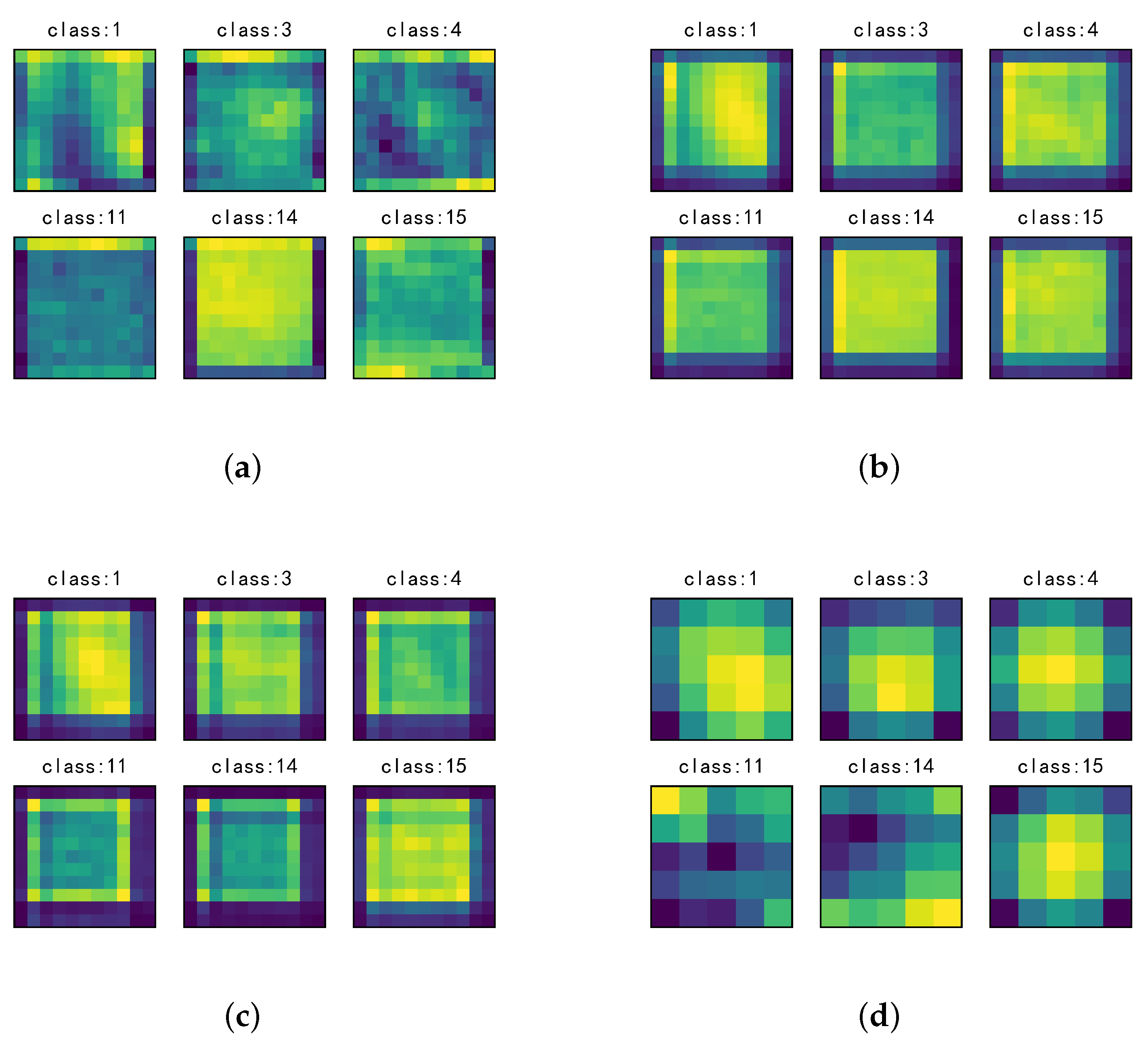
3.4. Investigation on the Stimulus Values by the SSSE Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HSI | Hyperspectral image |
| SE | Squeeze and excitation |
| SSSE | Spatial–spectral squeeze and excitation |
| SSSERN | Spatial–spectral squeeze and excitation residual network |
| CNN | Convolutional neural network |
| SAE | Stacked auto-encoder |
| DBN | Deep belief network |
References
- Landgrebe, D. Hyperspectral image data analysis. IEEE Signal Process. Mag. 2002, 19, 17–28. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral remote sensing data analysis and future challenges. IEEE Geosci. Remote. Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Wang, Q.; Meng, Z.; Li, X. Locality adaptive discriminant analysis for spectral-spatial classification of hyperspectral images. IEEE Geosci. Remote. Sens. Lett. 2017, 14, 2077–2081. [Google Scholar] [CrossRef]
- Donoho, D.L. High-dimensional data analysis: The curses and blessings of dimensionality. AMS Math Chall. Lect. 2000, 1, 32. [Google Scholar]
- Huang, Z.; Zhu, H.; Zhou, T.; Peng, X. Multiple marginal fisher analysis. IEEE Trans. Ind. Electron. 2018. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Dimension reduction using spatial and spectral regularized local discriminant embedding for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2015, 53, 1082–1095. [Google Scholar] [CrossRef]
- He, L.; Li, J.; Liu, C.; Li, S. Recent advances on spectral-spatial hyperspectral image classification: An overview and new guidelines. IEEE Trans. Geosci. Remote. Sens. 2018, 56, 1579–1597. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.L.P. Extreme learning machine with composite kernels for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2015, 8, 2351–2360. [Google Scholar] [CrossRef]
- Peng, J.; Zhou, Y.; Chen, C.L.P. Region-kernel-based support vector machines for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2015, 53, 4810–4824. [Google Scholar] [CrossRef]
- Peng, J.; Du, Q. Robust joint sparse representation based on maximum correntropy criterion for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2017, 55, 7152–7164. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.; Hu, J.; Zhang, L. Transferring deep convolutional neural networks for the scene classification of high-resolution remote sensing imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Du, B. Deep learning for remote sensing data: A technical tutorial on the state of the art. IEEE Geosci. Remote. Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Zhu, X.; Tuia, D.; Mou, L.; Xia, G.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote. Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef]
- Zhao, C.; Wan, X.; Zhao, G.; Cui, B.; Liu, W.; Qi, B. Spectral-spatial classification of hyperspectral imagery based on stacked sparse autoencoder and random forest. Eur. J. Remote. Sens. 2017, 50, 47–63. [Google Scholar] [CrossRef]
- Li, T.; Zhang, J.; Zhang, Y. Classification of hyperspectral image based on deep belief networks. In Proceedings of the 2014 IEEE International Conference on Image Processing, Paris, France, 27–30 October 2014; pp. 5132–5136. [Google Scholar]
- Zhong, P.; Gong, Z.; Li, S.; Schnlieb, C. Learning to diversify deep belief networks for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2017, 55, 3516–3530. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote. Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 1476–4687. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, J.; Yuan, Y. Embedding structured contour and location prior in siamesed fully convolutional networks for road detection. IEEE Trans. Intell. Transp. Syst. 2018, 19, 230–241. [Google Scholar] [CrossRef]
- Peng, X.; Feng, J.; Xiao, S.; Yau, W.; Zhou, T.; Yang, S. Structured autoEncoders for aubspace clustering. IEEE Trans. Image Process. 2018, 27, 5076–5086. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Ji, J.; Bi, Q.; Hou, J.; Du, Q.; Li, W. Integrating spectral and spatial information into deep convolutional neural networks for hyperspectral classification. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium, Beijing, China, 10–15 July 2016; pp. 5067–5070. [Google Scholar]
- Yang, J.; Zhao, Y.; Chan, J.C.; Yi, C. Hyperspectral image classification using two-channel deep convolutional neural network. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium, Beijing, China, 10–15 July 2016; pp. 5079–5082. [Google Scholar]
- Zhang, H.; Li, Y.; Zhang, Y.; Shen, Q. Spectral-spatial classification of hyperspectral imagery using a dual-channel convolutional neural network. Remote Sens. Lett. 2017, 8, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, H.; Shen, Q. Spectral-spatial classification of hyperspectral imagery with 3D convolutional neural network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Plaza, J.; Plaza, A. A new deep convolutional neural network for fast hyperspectral image classification. ISPRS J. Photogramm. Remote. Sens. 2018, 145, 120–147. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral-spatial residual network for hyperspectral image classification: A 3-D deep learning framework. IEEE Trans. Geosci. Remote. Sens. 2018, 56, 847–858. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 630–645. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv, 2015; arXiv:1502.03167. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the 14th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; pp. 315–323. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv, 2014; arXiv:1412.6980. [Google Scholar]
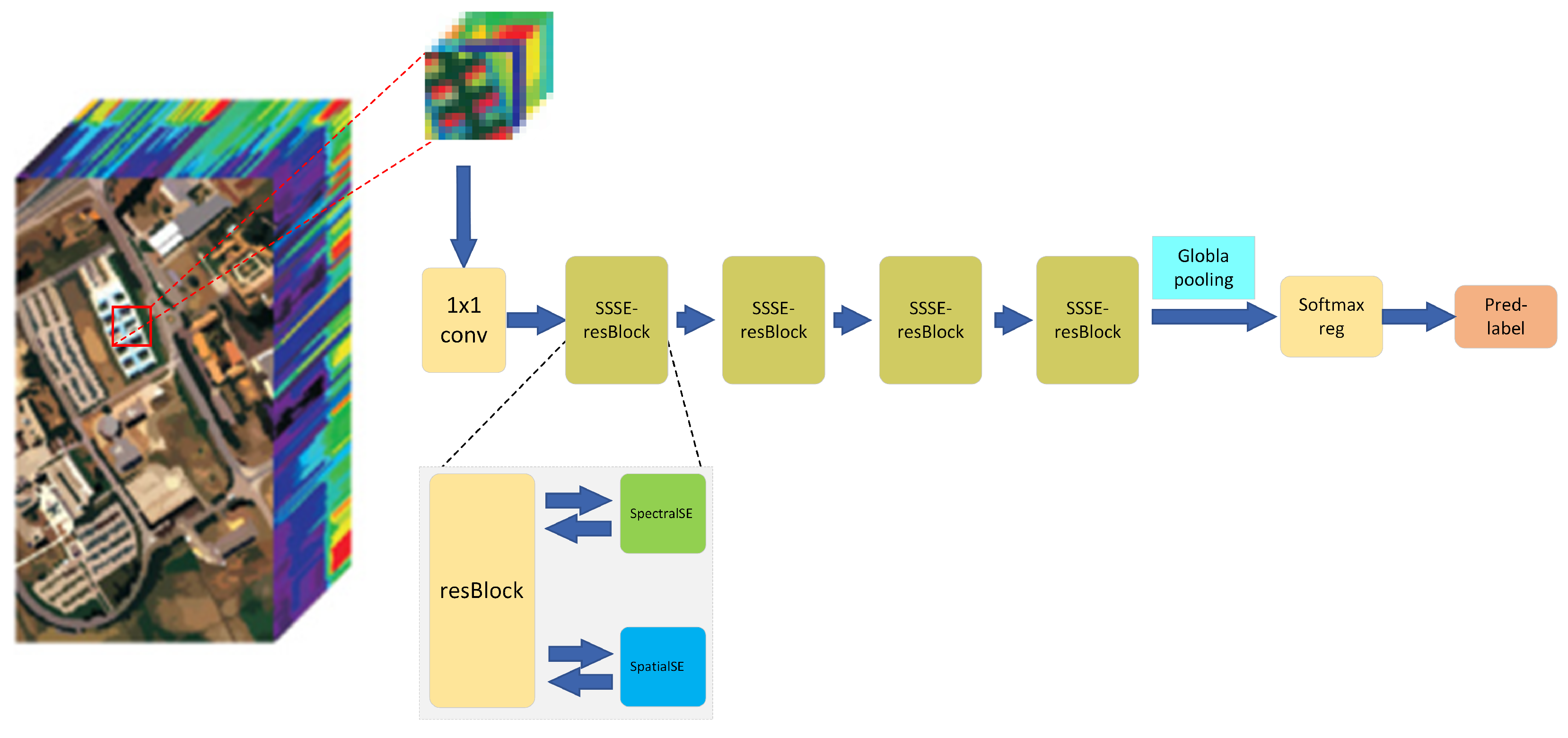

| Name | Details | Kernel Size |
|---|---|---|
| Input | - | - |
| Conv1 | - | 1, 1, 200, 128 |
| SSSE-resBlock | resBlock | 1, 1, 128, 32 |
| 3, 3, 32, 32 | ||
| 1, 1, 32, 128 | ||
| SpectralSE | 128, 32 | |
| 32, 128 | ||
| SpatialSE | 128, 1 | |
| ⋯Repeat the Block 4 Times | ||
| Global pooling | - | - |
| Softmax Reg | - | 128, 16 |
| Class | Samples | |
|---|---|---|
| Number | Name | Number of Samples |
| 1 | Alfalfa | 46 |
| 2 | Corn-notill | 1428 |
| 3 | Corn-min | 830 |
| 4 | Corn | 237 |
| 5 | Grass/Pasture | 483 |
| 6 | Grass/Trees | 730 |
| 7 | Grass/Pasture-mowed | 28 |
| 8 | Hay-windrowed | 478 |
| 9 | Oats | 20 |
| 10 | Soybeans-notill | 972 |
| 11 | Soybeans-min | 2455 |
| 12 | Soybeans-clean | 593 |
| 13 | Wheat | 205 |
| 14 | Woods | 1265 |
| 15 | Building-Grass-Trees-Drives | 386 |
| 16 | Stone-steel Towers | 93 |
| Total | 10,249 | |
| Class | Samples | |
|---|---|---|
| Number | Name | Number of Samples |
| 1 | Asphalt | 6631 |
| 2 | Meadows | 18,649 |
| 3 | Gravel | 2099 |
| 4 | Trees | 3064 |
| 5 | Metal sheets | 1345 |
| 6 | Bare soil | 5029 |
| 7 | Bitumen | 1330 |
| 8 | Bricks | 3682 |
| 9 | Shadows | 947 |
| Total | 42,776 | |
| Class | SVM | RF | MLP | 2D-CNN | 3D-CNN | SSRN | SSSERN |
|---|---|---|---|---|---|---|---|
| 1 | 85.19 ± 3.02 | 73.15 ± 9.26 | 83.76 ± 9.00 | 70.94 ± 10.68 | 95.14 ± 7.98 | 97.53 ± 1.39 | 98.12 ± 0.97 |
| 2 | 82.68 ± 0.78 | 73.22 ± 1.74 | 71.78 ± 5.63 | 73.40 ± 3.19 | 96.96 ± 1.58 | 98.45 ± 0.26 | 99.63 ± 0.56 |
| 3 | 71.53 ± 2.21 | 72.13 ± 2.21 | 69.93 ± 1.13 | 74.85 ± 0.94 | 97.05 ± 1.90 | 97.70 ± 0.33 | 99.57 ± 0.54 |
| 4 | 65.67 ± 5.28 | 69.01 ± 5.98 | 74.96 ± 2.74 | 88.56 ± 5.24 | 89.68 ± 2.46 | 89.46 ± 2.78 | 99.41 ± 0.72 |
| 5 | 94.03 ± 1.53 | 90.92 ± 1.28 | 88.94 ± 2.03 | 69.35 ± 1.49 | 96.95 ± 1.65 | 99.16 ± 0.54 | 100.00 ± 0.00 |
| 6 | 97.54 ± 0.88 | 97.43 ± 0.51 | 94.89 ± 2.28 | 92.10 ± 3.52 | 98.71 ± 1.02 | 99.80 ± 0.29 | 99.74 ± 0.28 |
| 7 | 82.81 ± 9.38 | 73.44 ± 16.44 | 94.20 ± 2.51 | 65.22 ± 15.06 | 97.73 ± 4.55 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| 8 | 98.08 ± 1.29 | 99.13 ± 0.45 | 97.29 ± 2.26 | 97.29 ± 1.37 | 99.21 ± 1.25 | 99.80 ± 0.25 | 100.00 ± 0.00 |
| 9 | 70.45 ± 13.64 | 72.73 ± 7.42 | 75.00 ± 6.25 | 81.25 ± 12.50 | 78.57 ± 24.74 | 94.64 ± 6.84 | 100.00 ± 0.00 |
| 10 | 73.20 ± 2.58 | 79.89 ± 3.44 | 84.42 ± 1.10 | 77.12 ± 4.97 | 95.52 ± 1.41 | 96.75 ± 0.37 | 99.52 ± 0.77 |
| 11 | 80.79 ± 1.16 | 90.23 ± 1.13 | 86.31 ± 2.78 | 86.19 ± 1.05 | 97.33 ± 1.02 | 98.13 ± 0.23 | 99.85 ± 0.69 |
| 12 | 78.17 ± 1.53 | 76.34 ± 2.10 | 74.21 ± 6.15 | 74.27 ± 1.27 | 97.46 ± 4.10 | 99.00 ± 0.61 | 96.54 ± 0.68 |
| 13 | 97.54 ± 1.50 | 96.72 ± 1.50 | 97.32 ± 0.33 | 98.85 ± 0.57 | 100.00 ± 0.00 | 100.00 ± 0.00 | 97.45 ± 0.82 |
| 14 | 94.82 ± 1.34 | 96.17 ± 0.81 | 96.16 ± 1.11 | 94.82 ± 2.07 | 99.38 ± 0.09 | 99.23 ± 0.28 | 99.91 ± 0.13 |
| 15 | 73.38 ± 2.93 | 58.87 ± 2.94 | 58.43 ± 2.83 | 80.89 ± 13.29 | 90.18 ± 3.76 | 94.07 ± 2.26 | 100.00 ± 0.00 |
| 16 | 93.64 ± 3.48 | 88.18 ± 5.65 | 90.72 ± 1.46 | 76.62 ± 4.10 | 89.73 ± 7.46 | 88.36 ± 4.26 | 95.94 ± 0.63 |
| OA | 83.61 ± 0.69 | 84.59 ± 0.55 | 83.48 ± 0.33 | 82.98 ± 0.78 | 97.01 ± 1.29 | 98.07 ± 0.17 | 99.44 ± 0.14 |
| AA | 83.72 ± 0.31 | 81.72 ± 1.24 | 83.64 ± 0.61 | 80.95 ± 1.54 | 96.98 ± 1.95 | 97.07 ± 0.68 | 98.89 ± 0.11 |
| 81.29 ± 0.79 | 82.31 ± 0.63 | 81.09 ± 0.41 | 80.54 ± 0.90 | 96.59 ± 1.47 | 97.79 ± 0.19 | 99.03 ± 0.21 |
| Class | SVM | RF | MLP | 2D-CNN | 3D-CNN | SSRN | SSSERN |
|---|---|---|---|---|---|---|---|
| 1 | 90.72 ± 0.69 | 89.45 ± 0.01 | 89.91 ± 1.09 | 91.83 ± 0.33 | 99.10 ± 0.49 | 99.74 ± 0.11 | 100.00 ± 0.00 |
| 2 | 94.42 ± 0.63 | 97.83 ± 0.27 | 96.67 ± 0.75 | 97.11 ± 0.99 | 98.29 ± 0.68 | 99.35 ± 0.37 | 100.00 ± 0.00 |
| 3 | 70.34 ± 0.93 | 64.65 ± 0.83 | 79.32 ± 1.05 | 89.46 ± 0.68 | 90.01 ± 0.35 | 97.50 ± 0.50 | 98.39 ± 0.31 |
| 4 | 92.20 ± 0.56 | 90.52 ± 0.90 | 91.54 ± 0.58 | 91.89 ± 1.08 | 94.58 ± 0.16 | 98.68 ± 0.09 | 98.38 ± 0.11 |
| 5 | 98.87 ± 0.97 | 98.94 ± 0.89 | 98.87 ± 0.72 | 97.45 ± 0.70 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 |
| 6 | 57.71 ± 0.78 | 63.39 ± 2.77 | 77.85 ± 1.09 | 68.09 ± 0.76 | 97.06 ± 0.26 | 98.50 ± 0.26 | 100.00 ± 0.00 |
| 7 | 77.73 ± 0.85 | 70.33 ± 0.97 | 81.77 ± 0.88 | 96.14 ± 0.74 | 89.54 ± 0.46 | 98.61 ± 0.18 | 99.74 ± 0.26 |
| 8 | 80.44 ± 0.69 | 86.36 ± 0.45 | 78.70 ± 0.98 | 95.27 ± 0.29 | 90.25 ± 0.28 | 95.76 ± 0.44 | 99.43 ± 0.35 |
| 9 | 92.39 ± 0.80 | 92.05 ± 0.51 | 93.87 ± 0.82 | 86.16 ± 0.14 | 99.51 ± 0.46 | 99.81 ± 0.54 | 96.19 ± 0.89 |
| OA | 86.17 ± 0.93 | 87.59 ± 0.35 | 90.64 ± 0.11 | 92.20 ± 0.16 | 96.59 ± 0.52 | 98.79 ± 0.26 | 99.62 ± 0.31 |
| AA | 83.78 ± 0.73 | 83.48 ± 0.21 | 87.61 ± 0.17 | 90.96 ± 0.70 | 95.12 ± 0.09 | 98.58 ± 0.26 | 99.13 ± 0.19 |
| 81.63 ± 0.60 | 83.91 ± 0.33 | 87.36 ± 0.07 | 89.79 ± 1.02 | 95.37 ± 0.39 | 98.76 ± 0.54 | 99.35 ± 0.32 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Peng, J.; Sun, W. Spatial–Spectral Squeeze-and-Excitation Residual Network for Hyperspectral Image Classification. Remote Sens. 2019, 11, 884. https://doi.org/10.3390/rs11070884
Wang L, Peng J, Sun W. Spatial–Spectral Squeeze-and-Excitation Residual Network for Hyperspectral Image Classification. Remote Sensing. 2019; 11(7):884. https://doi.org/10.3390/rs11070884
Chicago/Turabian StyleWang, Li, Jiangtao Peng, and Weiwei Sun. 2019. "Spatial–Spectral Squeeze-and-Excitation Residual Network for Hyperspectral Image Classification" Remote Sensing 11, no. 7: 884. https://doi.org/10.3390/rs11070884
APA StyleWang, L., Peng, J., & Sun, W. (2019). Spatial–Spectral Squeeze-and-Excitation Residual Network for Hyperspectral Image Classification. Remote Sensing, 11(7), 884. https://doi.org/10.3390/rs11070884







