1. Introduction
Hyperspectral images (HSIs) contain both spectral and spatial information and generally consist of hundreds of spectral bands for the same observed scene [
1]. Due to the vast amounts of information they contain, HSIs have found important applications in a variety of fields, such as the non-contact analysis of food materials [
2], the detection and identification of plant diseases [
3], multispectral change detection [
4], and medicine [
5]. HSI classification is the core technology in these applications. However, since HSIs include inherently high-dimensional structures, their classification remains a challenging task in the remote sensing community.
Traditional classification methods involve feature engineering using a classifier. This process aims to extract or select features from original HSI data, typically producing a classifier based on low-dimensional features. Support vector machines (SVMs) are the most commonly used method in the early stages of HSI classification, due to their low sensitivity to high dimensionality [
6]. Spectral–spatial classification methods have become predominant in recent years [
7]. Mathematical-morphology-based techniques [
8], Markov random fields (MRFs) [
9], and sparse representations [
10] are also commonly used branches. However, many of these techniques suffer from low classification accuracy due to shallow feature extraction.
Deep learning, a popular tool in multiple areas including remote sensing, has recently been applied to HSI classification [
11]. Traditional feature extraction methods have struggled to identify high-level features in HSIs. However, deep learning frameworks have been proposed, in which stacked auto-encoders (SAEs) were used to obtain useful deep features [
11]. Deep learning-based methods can extract deep spectral and spatial features from HSIs to obtain higher classification accuracies than those of most traditional methods [
12]. Consequently, in recent years, a variety of deep learning-based methods have been used for classification [
7]. For example, one study used a deep belief network (DBN) that combined PCA with logistic regression to perform HSI classification, achieving competitive classification accuracy [
13].
Among these methods, deep convolutional neural network (CNN) algorithms have achieved particularly high accuracy. Deep supervised methods using randomized PCA have also been proposed to reduce the dimensionality of raw HSIs. Additionally, two-dimensional (2D) CNNs have been used to encode spectral data, spatial information, and a multilayer perceptron (MLP) for classification tasks [
14]. Three-dimensional (3D) CNNs have also been used as feature extraction models to acquire spectral–spatial features from HSIs [
15]. Two-layer 3D CNNs have performed far better than 2D CNN-based methods [
16].
Recently, two deep convolutional spectral–spatial networks, the spectral–spatial residual network (SSRN) [
17] and the fast and dense spectral–spatial convolutional network (FDSSC) [
18], achieved unprecedented classification accuracy. This was due in part to the inclusion of deeper 3D CNN architectures. SSRN and FDSSC achieved an overall accuracy of above 99% across three widely used HSI data sets. As such, there appears to be little room for improvement in HSI classification. However, deep supervised methods require large quantities of data. For example, SAE logistic regression (SAE-LR) requires 60% of a data set to be labeled [
11] and DBNs [
13] and 3D CNNs [
16] require 50% to be labeled. In contrast, SSRN and FDSSC require only 20% and 10% of a data set to be labeled, respectively. However, even a minimal labeling requirement (e.g., 10%) typically includes more than a thousand samples. As a result, the cost of sample labeling remains high in remote sensing studies.
In contrast, semi-supervised methods require only limited labeled samples. Recently, a semi-supervised model was introduced that labels samples based on local, global, and self-decisions. As a result, test samples were labeled based on multiple decisions [
19]. Generative adversarial networks (GANs) can also be used for HSI classification. Real labeled HSIs and fake data generated by a generative network can be used as inputs to a discriminative network. Trained discriminative networks can then classify unlabeled samples [
20]. Although GANs require only 200 real labeled samples to train, their classification accuracy remains relatively low.
Attention mechanisms [
21], a popular research topic in network structures, have also proven to be effective for image classification [
22]. These mechanisms mimic the internal processes of biological systems by aligning internal experiences with objective sensations, thereby increasing the observational fineness of subregions. When humans view a digital image, they do not observe every pixel in the image simultaneously. Most viewers focus on specific regions according to their requirements. Additionally, while viewing, their attentional focus is influenced by previously observed images. Attention mechanisms implemented through feedback connections [
23] in a network structure can enable the network to re-weight target information and ignore background information and noise. Cross-entropy loss is the most commonly used loss function in multi-objective classification tasks and has achieved excellent performance. It increases the inter-class distance, yet neglects the intra-class distance. However, sometimes the intra-class distance is even greater than the inter-class distance, which reduces the discrimination of the extracted features. The objective function must ensure that these extracted features are distinguishable. Furthermore, the center loss function [
24], which is designed to reduce the intra-class distance, has been shown to help the network extract more discriminant features. However, to prevent the degradation of classification accuracy, center loss can only be used as an auxiliary loss function.
This study introduces an attention mechanism and a center loss function for HSI classification. Inspired by previous studies [
25], we propose a deep supervised method with an end-to-end alternately updated convolutional spectral–spatial network (AUSSC). Unlike 3D CNN, SSRN, and FDSSC, which include only forward connections in the convolutional layers, the AUSSC includes both forward and feedback connections. Additionally, the convolutional kernels of the AUSSC are smaller than those of 3D CNN, SSRN, or FDSSC, as the kernels are decomposed into smaller kernels. Deeper spectral and spatial features can be obtained in the AUSSC using a fixed number of parameters, due to the alternate updating of blocks.
Due to the inclusion of attention mechanisms and factorization into smaller convolutions, the AUSSC is more capable of spectral and spatial feature learning than other CNN-based methods. Both forward and feedback connections are densely connected within the alternately updated blocks. Consequently, spectral and spatial features are optimally learned and feature maps from different blocks are repeatedly refined by attention. The classification results obtained using the proposed method demonstrate that this AUSSC has been optimized for classification with a limited number of training samples. The four principal contributions of this study are as follows:
- (1)
The proposed method includes a recurrent feedback spectral–spatial structure with fixed parameters, in order to learn not only deep but also refined spectral and spatial features to improve HSI classification accuracy.
- (2)
The effectiveness of the center loss function is validated as an auxiliary loss function used to improve the results of hyperspectral image classification.
- (3)
The AUSSC decomposes a large 3D convolutional kernel into three smaller 1D convolutional kernels, thereby saving a large number of parameters and reducing overfitting.
- (4)
The AUSSC achieves state-of-the-art classification accuracy across four widely used HSI data sets, using limited training data with a fixed spatial size.
The remainder of this paper is organized as follows.
Section 2 presents the framework of the proposed AUSSC.
Section 3 describes the experimental data sets. The details of the experimental results and a discussion are given in
Section 4. Conclusions and suggestions for future work are presented in
Section 5.
2. Methods
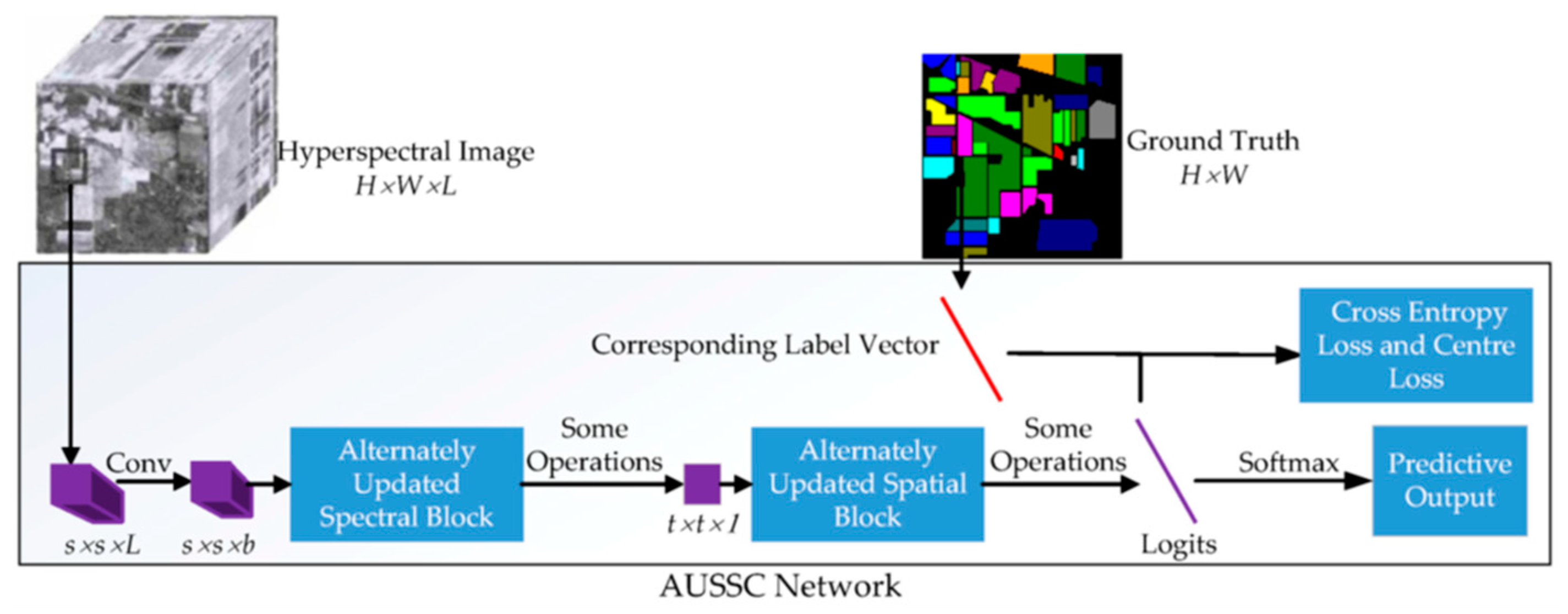
In this section, an alternately updated spectral–spatial convolutional network is proposed for HSI classification.
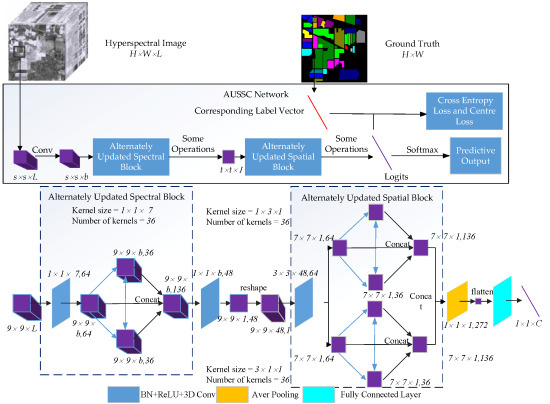
Figure 1 shows an overview of the proposed method. For HSI data with L channels and a size of
, a spatial size of
was selected from the raw HSI data and used as the input to the AUSSC network. First, the AUSSC uses three smaller convolutional kernels to learn spectral and spatial features from an original HSI patch. Second, the alternately updated spectral and spatial blocks refine the deep spectral and spatial features using recurrent feedback. Finally, the model parameters are optimized using the cross-entropy loss and center-loss loss functions. Details of each stage are elaborated in the following subsections.
2.1. Learning Spectral and Spatial Features with Smaller Convolutional Kernels
During HSI classification, deep CNN-based methods typically utilize preprocessing technology such as PCA. This is often followed by several convolutional layers with multiple activation functions and a classifier for obtaining classification maps. The convolution and activation can be formulated as
where
is the
th input feature map for the
th layer,
N is the number of feature maps in the
th layer, * is the convolution operation,
is an activation function, and
and
are learnable parameters that can be fine-tuned using the back-propagation (BP) algorithm.
The 3D CNN, SSRN, and FDSSC algorithms all demonstrate that an end-to-end 3D-CNN-based framework outperforms 2D-CNN-based methods that include preprocessing or post-processing, as well as other deep learning-based methods. One reason for this is that an end-to-end framework can reduce pre-processing and subsequent post-processing, allowing the connection between the original input and the final output to be as close as possible. The model then includes more space that can be adjusted automatically by the data, thereby increasing the degree of fitness. Additionally, when applied to HSIs with a 3D structure, 1D convolution operations focus on spectral features. 2D convolution operations focus on spatial features and 3D convolution operations can learn both spatial and spectral features. However, 3D kernel parameters are larger than 2D or 1D kernel parameters when the number of convolutional layers and kernels is the same. As such, a large number of model parameters can lead to overfitting.
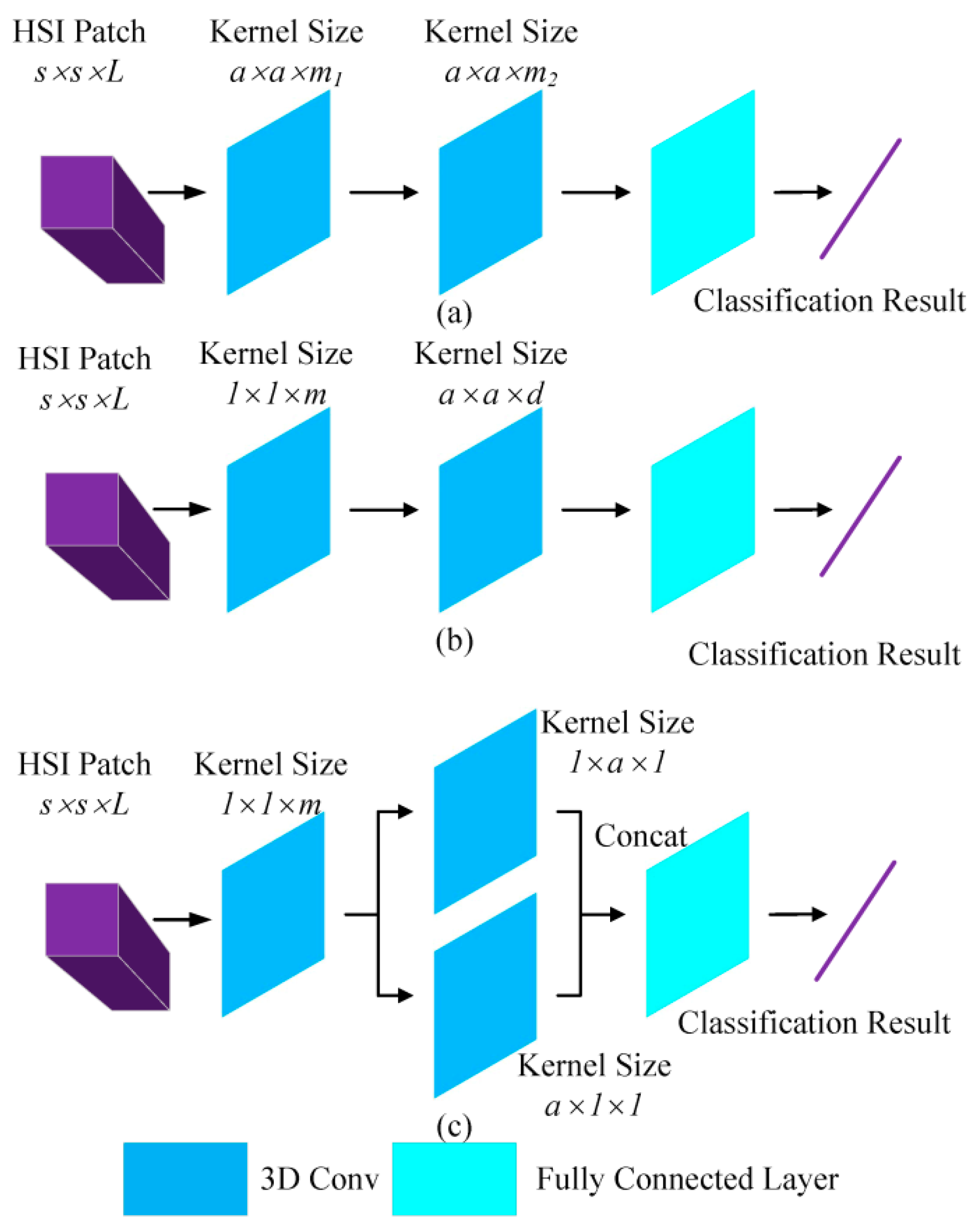
As such, we propose an end-to-end CNN-based framework that uses smaller convolutional kernels compared to other CNN-based methods. As shown in
Figure 2, the AUSSC utilizes kernels for HSI classification, ignoring other specific architectures. The 3D CNN method uses two similar convolutional kernels with sizes of
and
, with the two convolutional kernels differing only in spectral dimension. SSRN uses a spectral kernel with a size of
and a spatial kernel with a size of
to learn spectral and spatial representations, respectively. Convolutional kernels dictate model parameters and determine which features are learned by the CNN. In contrast, we introduce the idea of factorization into smaller convolutions from InceptionV3 [
26]. In this process, a larger 3D convolutional kernel with a size of
was divided into three smaller convolutional kernels with sizes of
,
, and
. This substantially reduced the number of parameters, accelerated the operation, and reduced the possibility of overfitting. As shown in
Table 1, in the absence of bias (with all other conditions remaining the same), the convolutional kernel with a size of
included
parameters. The smallest convolutional kernel only included parameters, which is more economical than the other two. This increased the nonlinear representation capabilities of the model due to the use of multiple nonlinear activation functions.
2.2. Refining Spectral and Spatial Features via Alternately Updated Blocks
Deep CNN architectures have been used for HSI classification and have produced competitive classification results [
17]. However, the connection structure in the convolutional layers is typically in the forward direction. Additionally, the convolutional kernels in SSRN and FDSSC increase with depth. Alternately updated cliques have a recurrent feedback structure and go deeper into the convolutional layers with a fixed number of parameters [
25]. Therefore, we propose combining small convolutional kernels with this loop structure and design two alternately updated blocks to learn refined spectral and spatial features separately from HSIs.
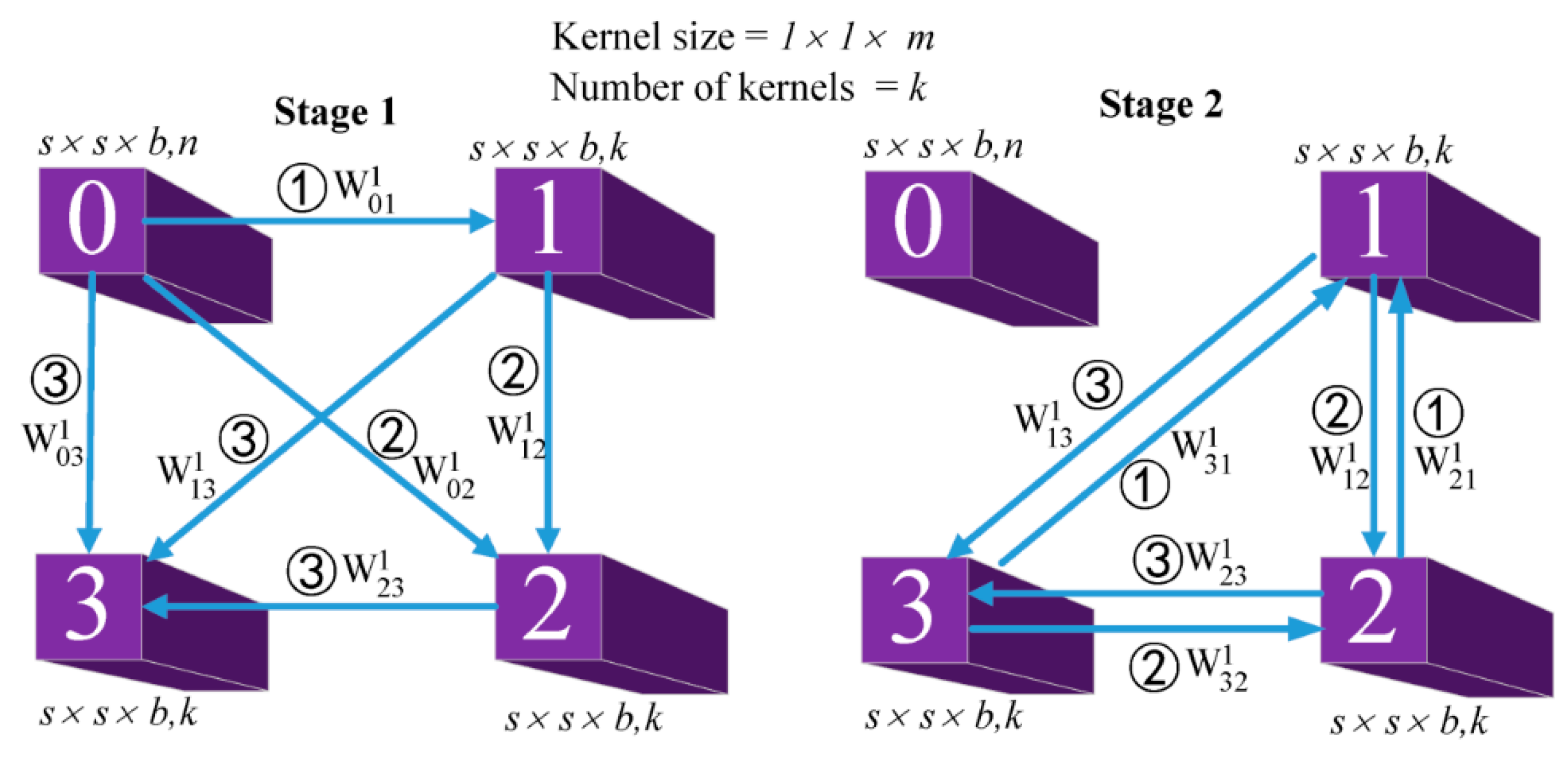
As shown in
Figure 3, there are two stages in the alternately updated spectral blocks. In the initialization stage (stage 1), the 3D convolutional layers use k kernels with sizes of
to learn deep spectral features. In stage 2, the 3D convolutional layers use k kernels with sizes of
to learn refined spectral features. A feature map with a size of
and a number,
, was input to the alternately updated spectral block. This input is denoted as
, where the subscript 0 represents the feature map in the initial position of the alternately updated spectral block. The superscript (1) indicates the feature map is in the first stage of the alternately updated process. In stage 1, the input of every convolutional layer is the output of all the previous convolutional layers. Stage 1 can be formulated as follows:
where
is the output of the lth
convolutional layer in stage 1 of an alternately updated spectral block,
is a nonlinear activation function,
is the convolutional operation using the padding method, and
is a parameter reused in stage 2.
In the looping stage (stage 2), each convolutional layer (except the input convolutional layer) is alternately updated to refine features. Stage 2 has a recurrent feedback structure, meaning that the feature map can be refined several times using the same weights. Therefore, any two convolutional layers in the alternately updated spectral block are connected bi-directionally. Stage 2 can then be formulated as follows:
where
since the feature map is in stage 2 and can be updated multiple times by the recurrent feedback structure. Similarly,
since the input feature map is not updated.
After learning refined deep spectral features, the input convolutional layer and the updated convolutional layer are concatenated in the alternately updated spectral block and transferred to the next block. Once spectral information from the HSI has been learned, the high dimensions of the feature map can be reduced by valid convolution and reshaping operations (see figure in
Section 2.4.). The resulting input to the alternately updated spatial block is a feature map with number,
and size
.
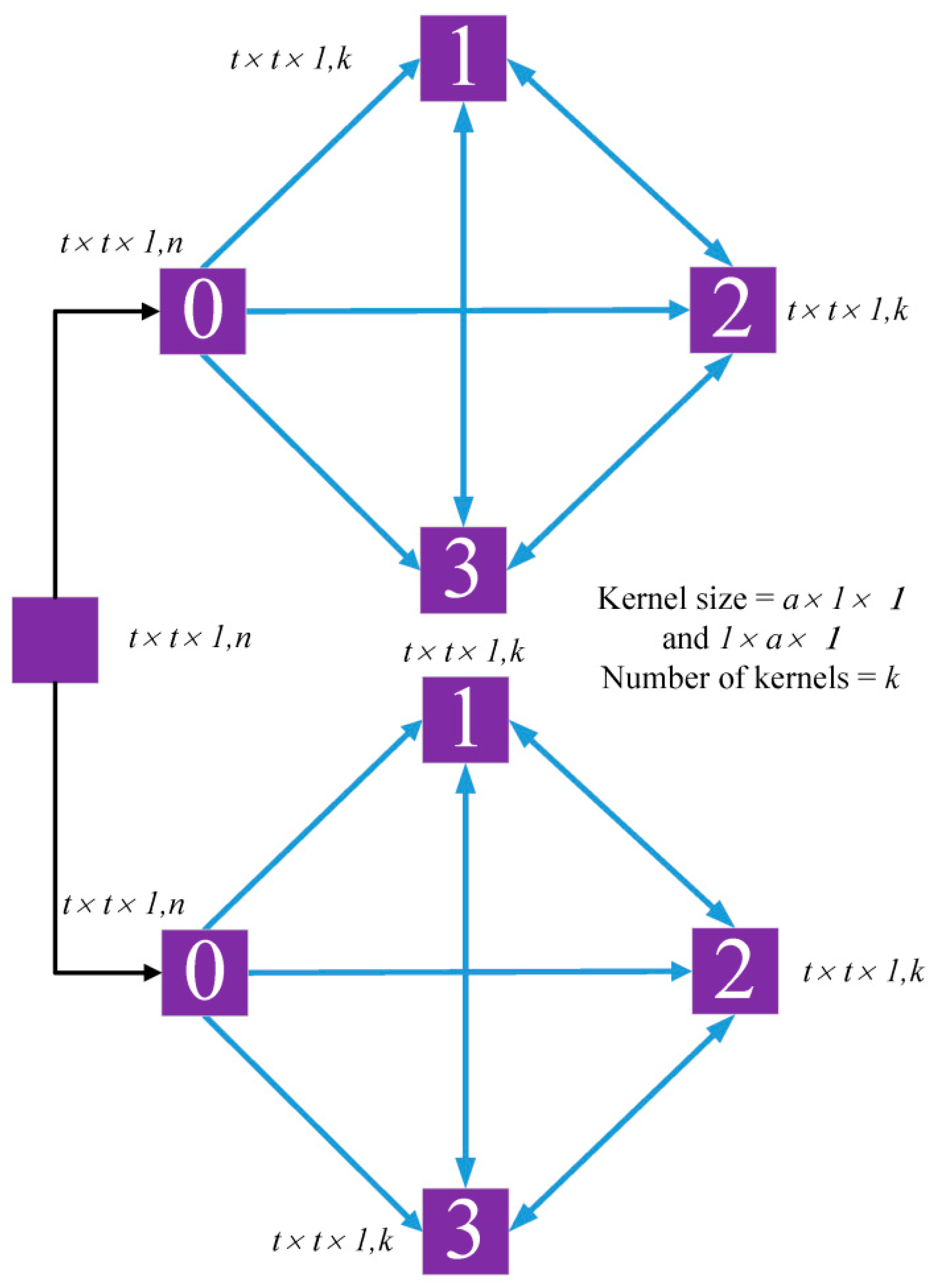
As shown in
Figure 4, there are two different convolutional kernels in the alternately updated spatial block. The 3D convolutional layers use
and
convolutional kernels to learn deep refined spatial features with an alternately updated structure that is also used for the alternately updated spectral block. In the spatial block, two different convolutional kernels learn spatial features in parallel rather than in series. The convolutional relationship between the spatial block is the same as for the previous block.
These alternately updated blocks achieve spectral and spatial attention due to the presence of refined features obtained in the looping stages. Densely connected forward and feedback structures allow the spectral and spatial information to flow in convolutional layers within the blocks. These alternately updated blocks also include weight sharing. In stage 1, the weights increase linearly as the number of convolutional layers increases. However, in stage 2, the weights are fixed since they are shared. The partial weights from stage 1, such as
,
, and
(see
Figure 2), are reused in stage 2. As features are cycled repeatedly in stage 2, the number of parameters remains unchanged.
2.3. Optimization by the Cross-Entropy Loss and Center Loss Functions
HSI classification is inherently a multi-classification task and cross-entropy loss with a softmax layer is a well-known objective function that is used for such problems. The softmax cross-entropy loss can be written in the following form:
where
m is the size of the mini-batch,
n is the number of classes,
is the
th deep feature belonging to the
th class,
is the
th column of the weights
W in the last fully connected layer, and
b is the bias. The last layer of the CNN-based model is typically fully connected, as it is difficult to make the dimensions of the last layer equal to the number of categories without a fully connected layer. Intuitively, one would expect that learning more discriminatory features would improve the generalization performance. As such, we introduce an auxiliary loss function [
24] to improve the discrimination of features acquired by the model. This function can be formulated as follows:
where
is the central feature in the
th class. The function decreases the quadratic sum of the distance from the center of the feature to the features of each sample in one batch, which decreases the intra-class distance. The center of feature
can then be updated through iterative training.
When two loss functions are used together for HSI classification, the softmax cross-entropy loss is considered to be responsible for increasing the inter-class distance. The center loss is then responsible for reducing the intra-class distance, thus increasing the discriminant degree and generalization abilities of learned features. Consequently, the objective function for the AUSSC can be written in the following form:
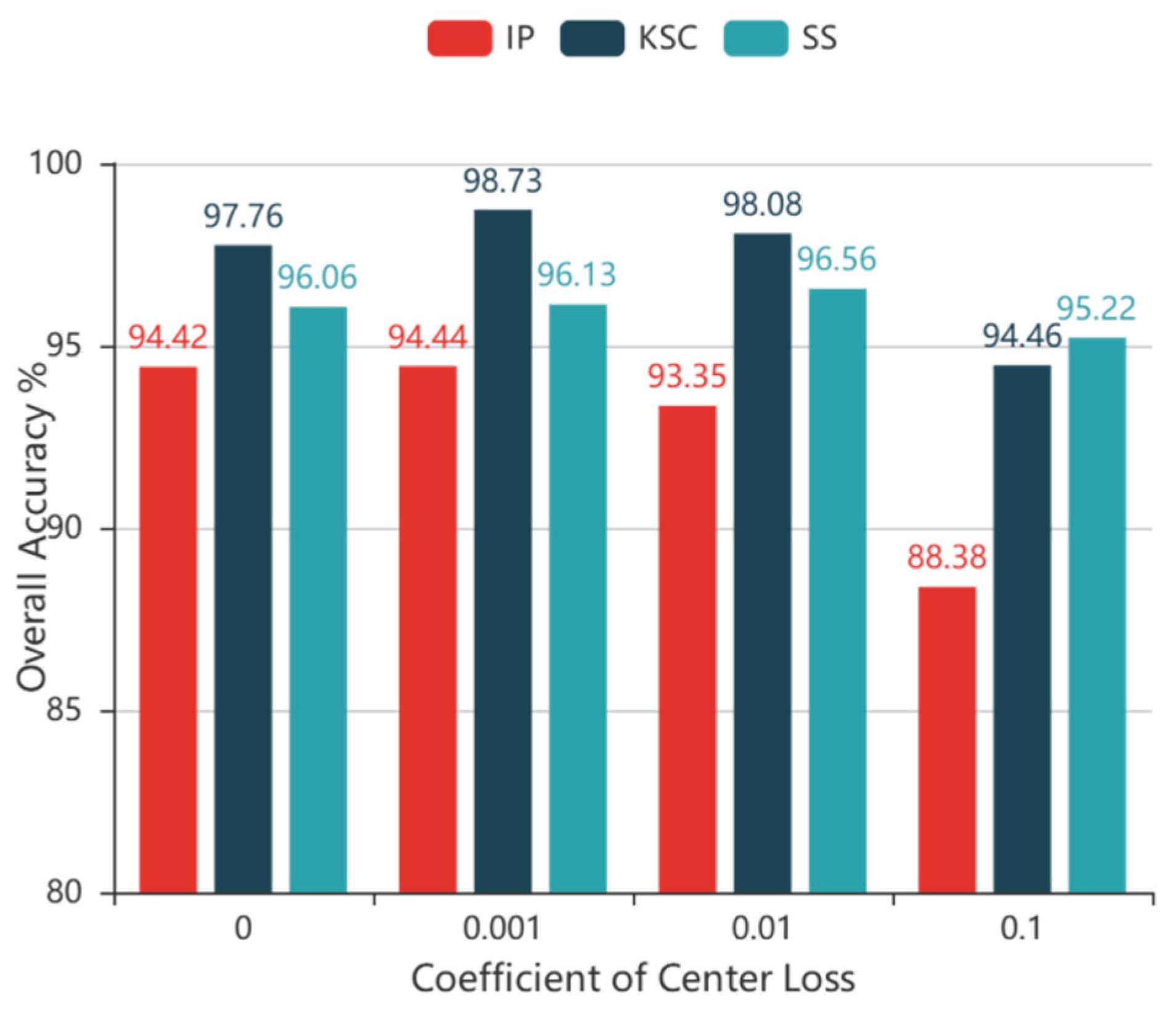
where λ ∈ [0, 1) controls the proportion of center loss and the value of λ is determined experimentally, as discussed in the following section. In summary, the cross-entropy loss is the principal objective function and the inter-class distance is the principal component. The center loss is the auxiliary used to reduce the intra-class distance.
2.4. Alternatively Updated Spectral–Spatial Convolutional Network
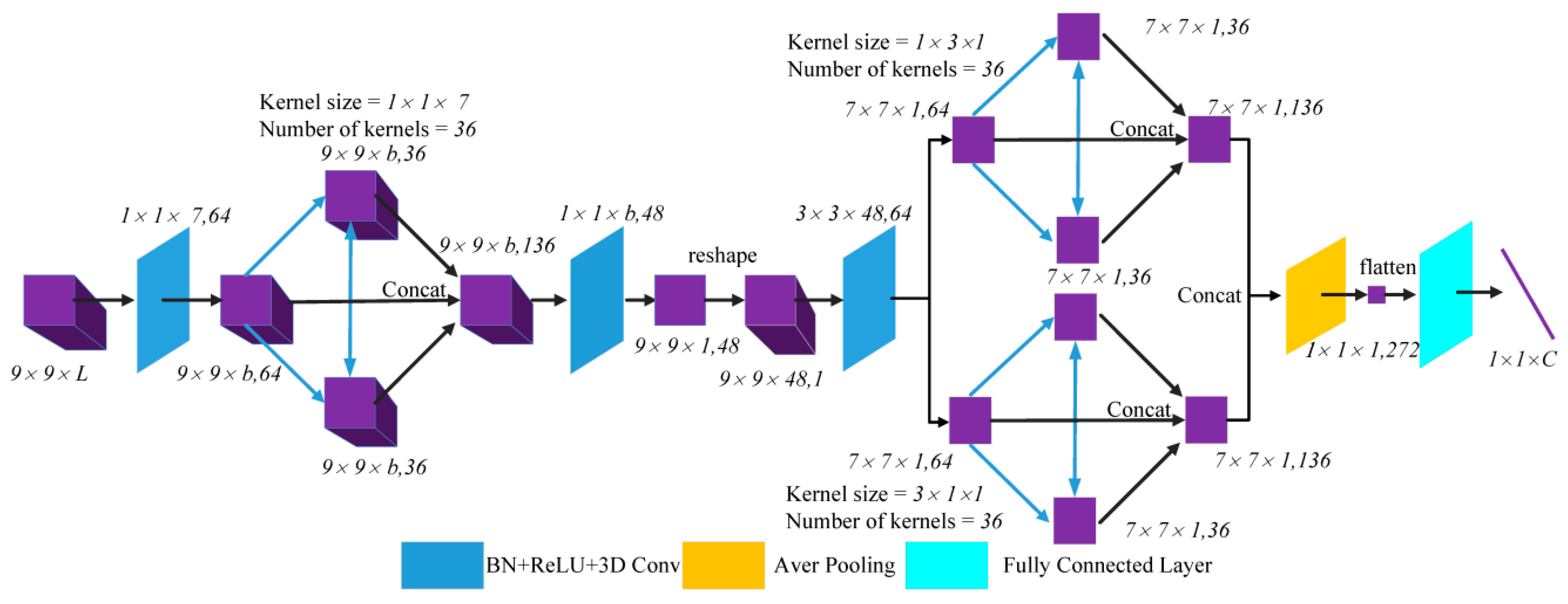
A flowchart is included below to explain the steps in the AUSSC end-to-end network. Considering the cost and time requirements of the collection of HSI labeled samples, we propose a 3D CNN-based framework that maximizes the flow and circulation of spectral and spatial information.
Figure 5 shows a
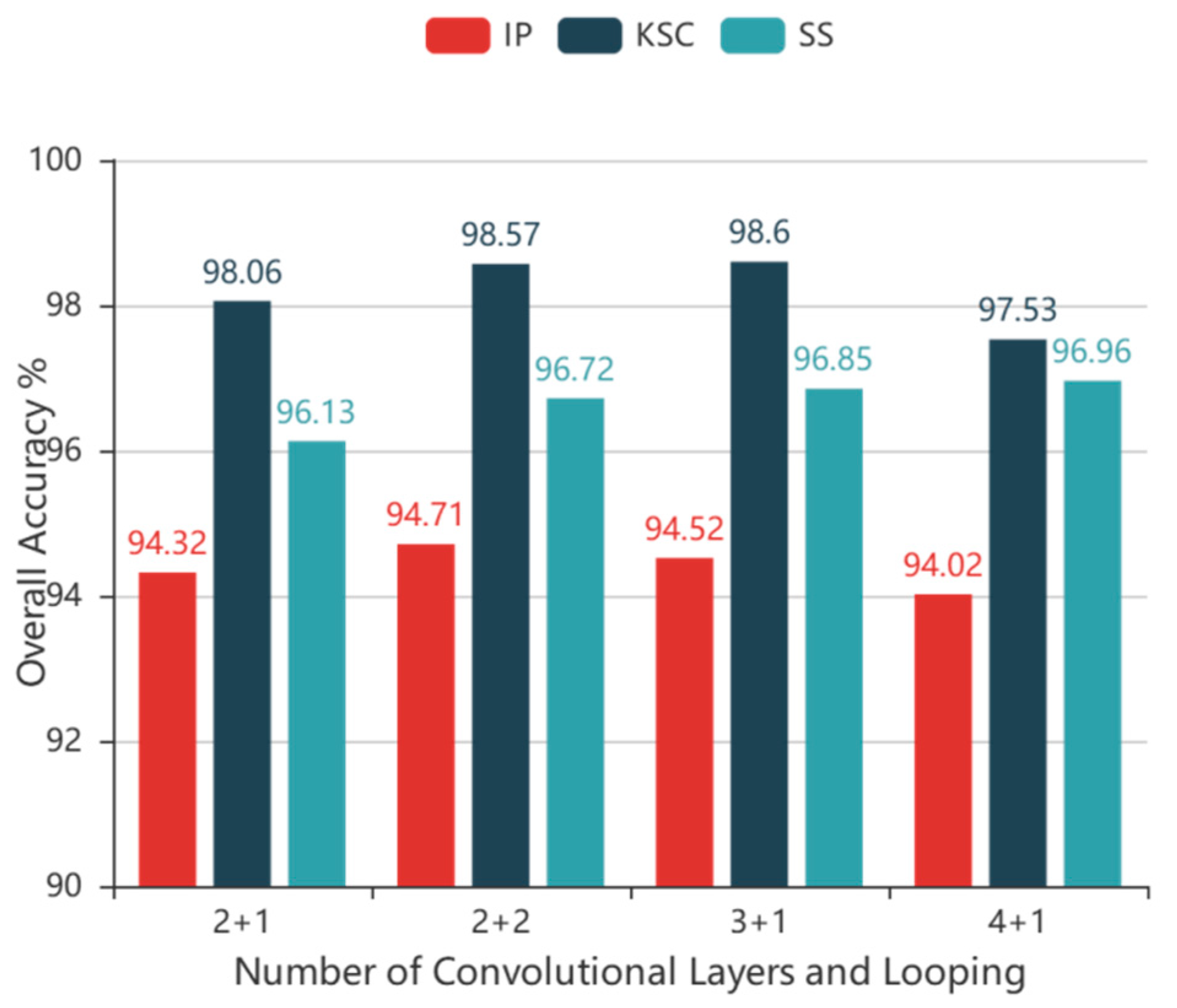
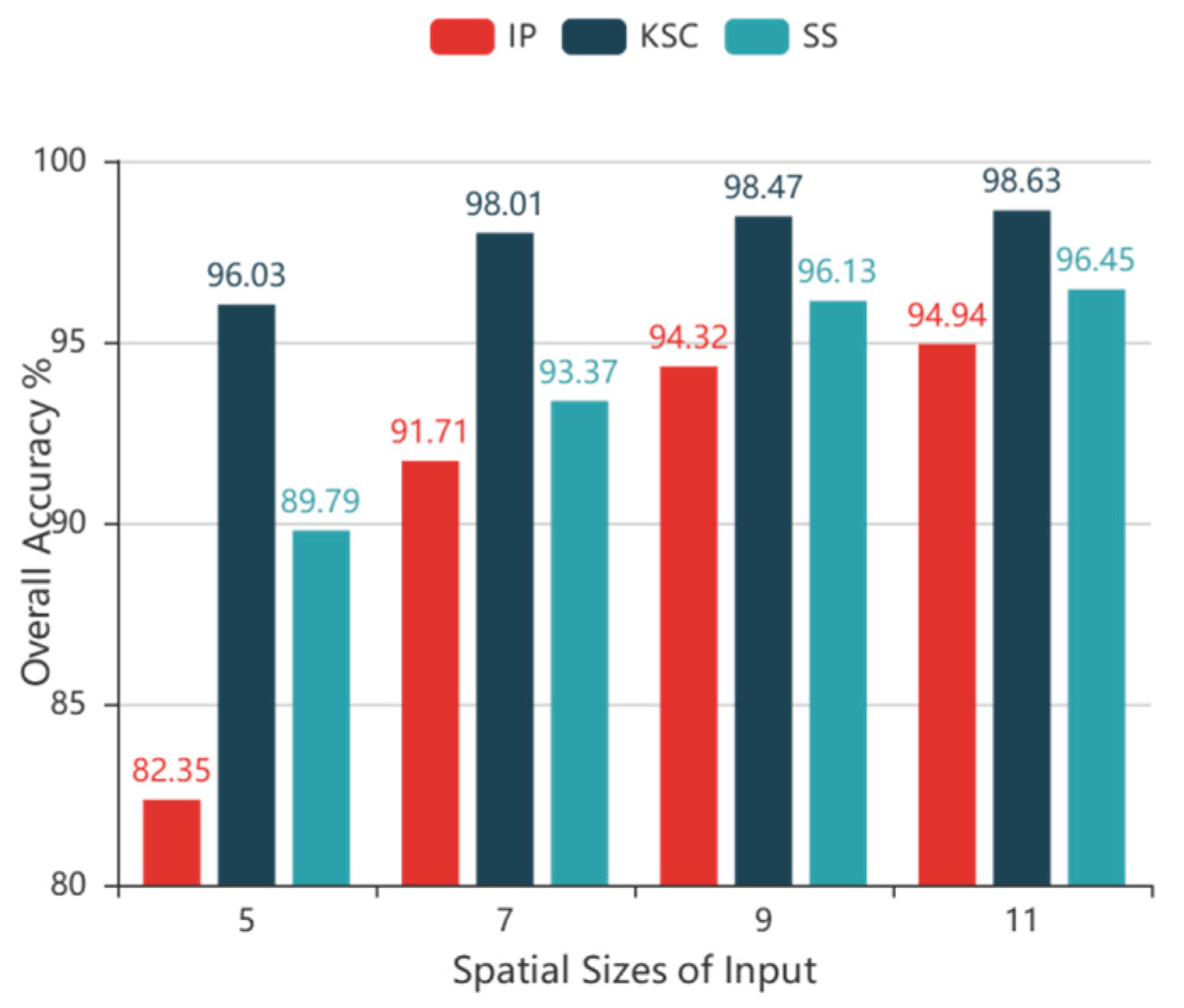
cube, which is used as input in our technique, where L is the number of HSI bands. Due to high computational costs, two convolutional layers were used in the alternately updated blocks and a single loop was used in stage 2.
L2 loss and batch normalization (BN) [
27] were used to improve the normalization of our model. In a broad sense, L2 and other regularization parameter terms added to the loss function in machine learning are essentially weighted norms. The goal of normalization with L2 loss is to effectively reduce the size of the original parameter values in the model, with BN performing normalization operations on input neuron values. The normalization target regularizes its input value to a normal distribution with a mean value of zero and a variance of one. The blue layers and blue lines both refer to the BN, rectified linear units (ReLU), and the convolution operation. The first convolutional layer lacks both a BN and a ReLU.
The original HSI input, which has a size of , flows to the first convolutional layer with a kernel size of and a stride of to generate feature maps with a size of . The number of kernels in the convolutional layers of alternately updated spectral block was 36, the kernel size was , and the convolutional padding method was the same. As a result, the output size for each layer remained , which was unchanged in stage 1 and stage 2. After concatenating the input and updated feature maps, the output of the alternately updated spectral blocks had a size of .
A valid convolutional layer with 48 channels and a kernel size of was included between alternately updated spectral and spatial blocks. This reduced the dimensions of the output of alternately updated spectral blocks, resulting in 48 feature maps with a size of 9 × 9 × 1. After reshaping the third dimension and the channel dimension, 48 channels with a size of 9 × 9 × 1 were merged into a single channel. A valid convolutional layer with a kernel size of and 64 kernels transformed the feature map into 64 channels with a size of .
Similar to the alternately updated spectral blocks, the alternately updated spatial block featured two convolutional kernels with sizes of and . In stage 1 and stage 2, the output of each layer was 36 kernels with a size of 7 × 7 × 1. The results of two convolutional kernels were concatenated into 272 kernels with a size of 7 × 7 × 1. Finally, the output passed through a 3D average pooling layer with a pooling size of , which was converted into 272 feature maps with a size of 1 × 1 × 1. After the flattening operation, a vector with a size of was produced by the fully connected layer, where C is the number of classes. Trainable AUSSC parameters were optimized by iterative training using Equation (6) and used to compute the loss between the predicted and real values.
The following sections provide a summary of the advantages of this proposed AUSSC architecture. First, the use of three different small convolutional kernels reduced both the number of parameters and overfitting, thereby increasing the nonlinear representation ability of the model and the diversity of features. Compared with symmetric splitting into several identical small convolutional kernels, this asymmetric splitting can handle more and richer features. Second, refined deep features learned by both forward and feedback connections between convolutional layers are more robust and have more high-level spectral and spatial information. Additionally, SSRN and FDSSC learn deeper features by increasing the number of convolutional layers in the blocks. However, unlike these conventional models, AUSSC can go deeper with fixed parameters due to its loop structure and shared weights. Finally, an auxiliary loss function was used to reduce the intra-class distance and increase the distinction between features of different categories.